Abstract
A total of 77 giant Pacific octopus, Enteroctopus dofleini, tissue samples were collected from the Oregon Coast (OR), Neah Bay Washington (NB), Puget Sound Washington (PS) and the southeast coast of Vancouver Island, British Columbia, Canada (BC) for genetic analyses. A suite of eight variable microsatellite markers developed from giant Pacific octopuses were amplified in these samples to determine population diversity, structure, relatedness and paternity. The majority of loci met Hardy-Weinberg equilibrium expectations within each population. We found moderate genetic diversity (average observed heterozygosity = 0.445, range = 0.307–0.515 and average expected heterozygosity = 0.567, range = 0.506–0.696) and moderate population structuring with distinct separation of groups (FST values ranged from 0.101 between BC and PS to 0.237 between BC and NB). Several egg strings from the BC population were collected from three female octopus dens for relatedness and paternity analyses. Results suggest strong support for multiple paternity within one egg clutch with progeny sired by between two to four males.
1. Introduction
Octopuses are found within every ocean throughout the world []. They belong to the phylum Mollusca, the class Cephalopoda, the order Octopoda, the family Octopodidae and are closely related to squid and cuttlefish and distantly related to clams, mussels and snails []. As a group they are thought to be highly intelligent and have been described as one of the most highly evolved invertebrates having lobed and folded brains similar to intelligent vertebrates [,]. The giant Pacific octopus, Enteroctopus doeflini, is found in the east Pacific Ocean along the Pacific Rim from Baja, California to Korea []. This is the largest octopus species, growing up to a maximal length of 9 m and a verified weight of 71 kg and an unverified weight of 272 kg [,]. The giant Pacific octopus is thought to have one of the longest lifespans of any octopus living between 3–5 years []. It is a terminal phase spawner with the female laying between 120,000 and 400,000 eggs at the end of her life cycle []. The female stops eating during this final phase to care for her eggs until they hatch in approximately six months []. At hatching the larvae are about the size of a grain of rice and it is thought that out of the hundreds of thousands that hatch only one to two survive to adulthood []. Their growth rate is one of the fastest in the animal kingdom with hatchlings starting out weighing less than a gram and growing approximately 0.9% per day to end up an average of 20–40 kg at adulthood []. Because of its large size and interesting behaviors, it is also one of the most popular species for display in public aquariums [,]. Although observations of both captive and wild giant Pacific octopuses have illuminated many aspects of its biology, behavior and husbandry [,,], the population genetics of this higher invertebrate in many areas of it’s range remain relatively unknown.
Population genetic studies have, however, been conducted on other species of octopuses. Several microsatellite markers have been isolated and characterized for the common octopus, Octopus vulgaris, [] and used to evaluate the genetic structure of its populations along the northwest coast of Africa [], the Mediterranean Sea [], the Iberian Peninsula and the Canary Islands [], and off the coast of Brazil []. All of these studies have reported relatively high genetic diversity and most report significant population structuring among sampled groups. However, Cabranes et al. (2008) [] found no significant differences between populations separated by approximately 200 km around the Iberian Peninsula and the Canary Islands. Other octopus species that have been investigated using microsatellite markers include the western Pacific common octopus, Octopus minor, in Korea and China [] and the Mexican four eyed octopus, Octopus maya, along the Yucatan Peninsula []. Both studies demonstrated significant stock structuring among sampled groups.
In addition to population diversity and structure, a small number of studies have investigated multiple paternity within cephalopods. Quinteiro et al. (2011) [] sampled egg clutches of the common octopus, O. vulgaris, and found evidence of between two to four males contributing to each clutch. Voight and Feldheim (2009) [] sampled young of the deep-sea octopus, Graneledone boreopacifica, and found at least two genetically distinct sires contributed to the hatchlings analyzed. Finally Squires et al. (2011) [] also found evidence of multiple paternity in dumpling squid, Euprymna tasmanica, egg clutches.
In 2012, eighteen microsatellite molecular markers were developed for use in giant Pacific Octopuses with many found to be sufficiently diverse and thus favorable for use in population genetics, individual identification, and parentage studies []. In this study we used a subset of the microsatellite markers developed by Toussaint et al., 2012 to measure genetic variability within and between giant Pacific octopuses captured and sampled from Oregon (OR), the south side of the Strait of Juan de Fuca near Neah Bay (NB), throughout central Puget Sound (PS) and the southeast coast of Vancouver Island, British Columbia, Canada (BC). Specific questions were: how much genetic variability do giant Pacific octopuses within these regions have, what is their population structure, and are eggs from a female’s clutch fertilized by multiple males?
2. Experimental Section
A total of 77 giant Pacific octopus samples were collected from the Oregon Coast (OR, N = 15); Neah Bay, Washington (NB, N = 5); Puget Sound, Washington (PS, N = 11); and the southeast coast of Vancouver Island, British Columbia, Canada (BC, N = 46). For the paternity analysis, eggs (N = 43) from multiple strings, and three adults (two resident females and one resident male) were sampled at Wain Rock in Saanich Inlet, within a fjord just north of Victoria on the east side of Vancouver Island, British Columbia. A total of three dens were sampled (den 1, den 7 and den 9) with only one female and one male sitting in or near dens sampled. Catch locations for octopuses sampled from OR, NB and PS were not recorded as specific sites but rather general sampling regions such as off the Oregon Coast, near Neah Bay or within Central Puget Sound.
Octopuses are invertebrates and thus not subject to the standard regulations that apply to sampling of vertebrates. Thus this work was not subject to review by an Institutional Animal Care and Use Committee (IACUC) or equivalent animal ethics committee and the work was not required to be reviewed by governmental agencies responsible for the management of octopuses in the wild. Samples for population genetics analysis were taken from captive animals held at the Seattle Aquarium for display (OR, NB and PS regions) and from wild octopuses that were sampled after natural death (BC). Collections of octopuses were permitted under the Washington Department of Fish and Wildlife (WDFW), the Oregon Department of Fish and Wildlife (ODFW), and the Canadian Department of Fish and Oceans (DFO) fishery regulations which allow taking of octopuses for recreational and scientific purposes. Specifically wild octopuses from OR were collected as a result of by-catch from crab pots after which they were placed in captivity at the Seaside Aquarium in Oregon or the Seattle Aquarium in Washington. Wild octopuses from Washington waters (NB and PS) were collected for public display by hand via SCUBA divers per WDFW fishery regulations and the Seattle Aquarium scientific collecting permit issued annually from WDFW. Wild octopuses and egg strings from Canadian waters (BC) were sampled after death or before hatching per DFO fishery regulations. All tissue sampling was either non-invasive in nature (such as the collection of naturally sloughed sucker molts while held in captivity), occurred after the animals died of natural causes or, as in the case of the sampled eggs, before hatching and after the adult stopped caring for them.
Tissue samples were preserved in 100% ethanol or frozen at −20 °C or −55 °C until analysis. DNA was extracted from tissue using the standard methods described by the manufacturer in the DNeay Blood and Tissue Kit (QIAGEN, Valenica, CA, USA). A suite of eight variable microsatellite markers developed previously for this species were amplified in octopus tissue to determine population diversity, structure, and paternity []. Loci included in this study were Edou6, Edou11, Edou110, Edou118, Edou122, Edou124, Edou129 and Edou216.
Microsatellites were amplified using a GeneAmp PCR 9700 thermocycler (ABI Life Technologies, Grand Island, NY, USA). Polymerase chain reaction (PCR) was performed on 10 µL samples that contained 1 µL of 100–250 ng/µL purified genomic DNA, 1 µL each of 0.5 µM/µL forward and reverse primers, and 7 µL Taq polymerase (Promega) mastermix containing manufacturer supplied buffer including dNTPs and MgCl2. There was no multiplexing of loci. The amplification profile for all loci was as follows: DNA was denatured at 94 °C for 4 min followed by thirty-five cycles of 94 °C (30 s), 53° (1 min), 72 °C (30 s), followed by a final polymerization step of 5 min at 72 °C. PCR products were analyzed on an ABI 3100 16-capillary system in GeneScan mode. Forward primers of these sets were labeled using fluorescent dyes compatible with the ABI GeneScan system and the filter wheel installed in the ABI 3100. Allele scoring for each locus was performed using Peakscanner software, version 2.0 (ABI). Tests for departures from Hardy-Weinberg equilibrium (p values) was performed using GenePop 3.1 software []. The FIS was calculated according to Weir and Cockerman (1984) [] using GenePop software []. Observed and expected heterozygosities, number of alleles and population structure such as FST values and Principal Coordinates Analysis (PCoA) was determined using GenAlEx []. Significance of FST vales was calculated using FSTAT []. STRUCTURE [] software was also used to determine population assignment and structure. Ten trials for each possible number of populations (K, ranged from 1–4) were run with a burn in period of 10,000 and 100,000 MCMC repetitions. Finally Colony [], Gerud [] and MLRelate [] was used to measure relatedness and multiple paternity within and among octopus egg strings.
3. Results and Discussion
The authors recognize that sample sizes for this analysis were small thus population genetics estimates for these four populations should be considered preliminary. The majority of the eight loci sampled within the populations were found to be within Hardy Weinberg Equilibrium (HWE) using Genepop 3.1 (when related individuals were excluded from analyses such as in BC) except for Edou11 and Edou216 within both OR and PS and Edou216 within PS. Since these loci were in HWE in the other populations, and since analysis run without them did not significantly change the results, these two loci were included in all analyses. Genetic diversity results from GenAlEx were moderate: Number of alleles (A) over all eight loci was 4.219 (range = 2.875–6.000), average observed heterozygosity (Ho) was 0.445 (range = 0.307–0.515) and average expected heterozygosity (He) was 0.567 (range = 0.506–0.696, Table 1).

Table 1.
Diversity measured within giant Pacific octopuses.
| POP | BC | NB | OR | PS |
|---|---|---|---|---|
| A | 6.000 | 2.875 | 3.000 | 5.000 |
| N | 46.000 | 5.000 | 15.000 | 11.000 |
| Ne | 2.817 | 2.446 | 2.286 | 3.837 |
| Ho | 0.514 | 0.515 | 0.307 | 0.444 |
| He | 0.559 | 0.508 | 0.506 | 0.696 |
| uHe | 0.568 | 0.542 | 0.540 | 0.749 |
| Fis | 0.142 | 0.448 | 0.788 | 0.488 |
| p value | 0.069 | 0.337 | 0.105 | 0.086 |
A = number of alleles; N = sample size; Ne = number of effective alleles; Ho = observed heterozygosity; He = expected heterozygosity; uHe = unbiased expected heterozygosity; Fis = average fixation index; p value = average p value for fixation index.
The moderate genetic diversity within giant Pacific octopuses measured here fall within the range measured in microsatellites within other octopuses. Genetic diversity within other octopus species has been found to range between 0.647 and 0.987 [,]. The genetic diversity measured within giant Pacific octopuses in this study is lower but within the range measured by Touissant et al. (2012) [] (Ho range of 0.069 to 0.930 and He range of 0.131–0.968) and is similar to the low end of the range reported for the common octopus. This is encouraging for giant Pacific octopuses in the Northeast Pacific as it is legal to harvest them throughout this region yet they do not appear to have suffered a significant loss of genetic diversity from mortalities associated with the targeted or incidental catch. However the differences between the diversity found here and that found within Prince William Sound by Touissant et al. (2012) requires further analysis potentially with larger sample sizes.
Population structure analyses results from GenAlEx suggests moderate population structuring among the four sampled groups. FST values were high to moderate ranging from a high of 0.237 between BC and NB to a low of 0.101 between BC and PS (Table 2). All FST values were found to be significant using FSTAT.

Table 2.
Population structure: FST matrix for sampled populations.
| BC | NB | OR | PS | |
|---|---|---|---|---|
| BC | * | |||
| NB | 0.237 | * | ||
| OR | 0.218 | 0.168 | * | |
| PS | 0.101 | 0.182 | 0.112 | * |
Population structuring is visually represented with distinct groupings in the Principal Coordinate Analysis (PCoA) graph from GeneAlEx describing 57.11% of the genetic variation and suggesting four distinct clusters. Even though there were four distinct groupings there was also overlap of individuals between all groups most notably with the NB cluster overlapping all other groups although distinct in its own segment of the graph (Figure 1).
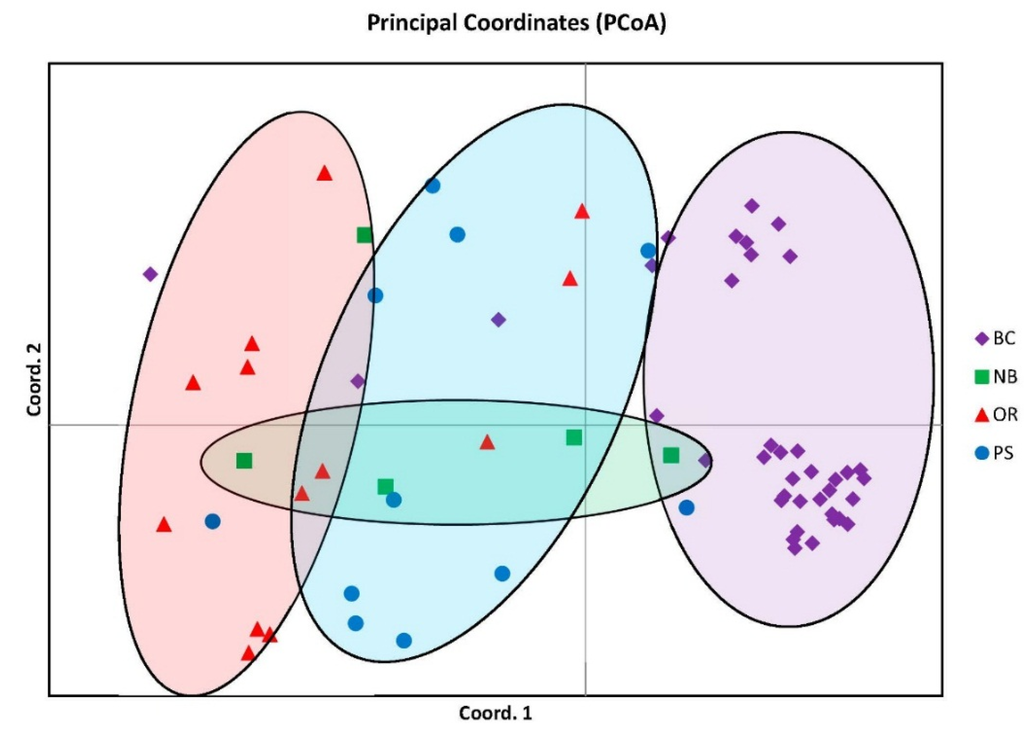
Figure 1.
Principal coordinate analysis similarity matrix (PCoA) accounting for 57.11% of the genetic variation. Note four distinct clusters but with NB overlapping over the other three.
In addition to the PCoA analysis STRUCTURE also suggested four distinct populations (lowest LnP(d) of all possible K values = 4) with some similarity between groups, primarily within PS, as indicated by the shared color schemes representing unique genetic signatures (Figure 2).
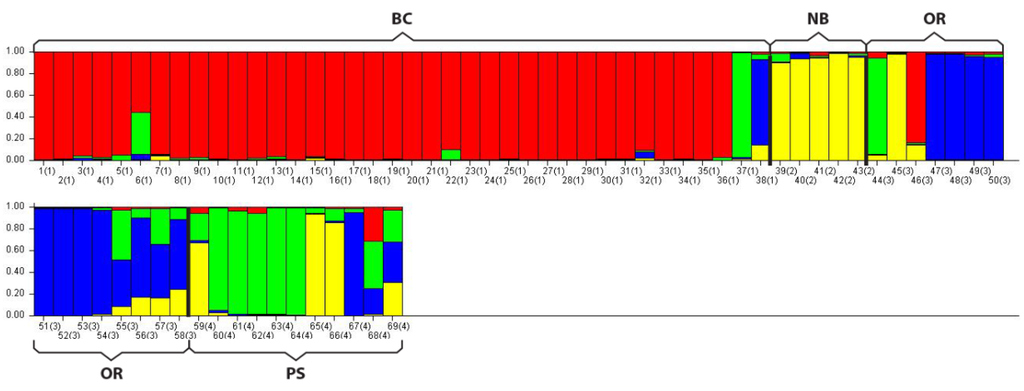
Figure 2.
STRUCTURE analysis bar plot. This program assigns unique genetic variation a color code. Populations with one solid color that is not shared by another group are genetically distinct. Populations that share colors are more similar. Note dominant colors in sampled populations: BC = Red; NB = Yellow; OR = Blue; PS = Green. Within PS two individuals were similar to NB (yellow), one was similar to Oregon (blue) and two were a mixture of signatures suggesting geneflow between PS and the other groups.
The evidence of significant population structuring among the sampled populations reported here is not unique for octopuses. Most studies of population genetics within octopuses report significant population structure among groups [,,]. This is not surprising due to the relatively sedentary nature of adult octopuses. Sampling areas in this study were separated by 144 km (PS-BC), 200 km (PS-NB) and up to 550 km (PS-OC). Even though these distances are rather small in the oceanic realm, the structure observed here may be explained by the behavior of adult giant Pacific octopuses found to move less than 10 km throughout their lifetimes []. A study on den utilization and movements of tagged octopuses discovered they moved an average of only 13.2 m between den sites and were sedentary most of the time showing high preferences to small home ranges []. Indeed the greatest dispersal of octopuses is thought to occur during their planktonic phase which may last between one to six months during which they drift with oceanic currents and may move long distances ([,,] and Cosgrove unpublished data).
It is unknown how far larval planktonic giant Pacific octopuses disperse throughout the sampled study region. However the oceanic currents along the outer west coast of Oregon and Washington are known to be dominated by the California Current System (CCS) which changes direction based in part on wind and atmospheric pressures. During the winter when the coasts are dominated by low atmospheric pressure the currents primarily run northward, while during the summer when the weather is dominated by high atmospheric pressure the currents primarily run southward []. In addition just off the northwest coast of Washington the Juan de Fuca eddy runs counterclockwise and is the dominant current during the summer []. Unlike the open ocean currents, the currents in the inland waterways of the Strait of Juan de Fuca, the Strait of Georgia and Puget Sound are dominated by daily tidal currents with flow changing direction (incoming flood currents and outgoing ebb currents) four times daily []. Current flows throughout these straits and estuaries are known to be complex with multiple nearshore eddies occurring throughout. Drift cards released at many points throughout this system have been found beached throughout the waterways as well as in the Strait of Juan de Fuca moving westward towards the open ocean []. Thus the dispersal of planktonic larval giant Pacific octopuses throughout this system is likely complex and driven by many factors such as season, weather and tides. More work should be done to track movements of early life stages of this species.
Even with minimal movements as adults and unknown dispersal rates and distances as juveniles one study by Cabranes et al. (2008) [] found no significant structuring among octopus groups separated by as much as 200 km. In contrast this study found structure between the two PS and BC octopus populations separated by only 144 km. However we also found similarities between groups sampled such as the smaller FST between BC and PS (Table 2), the overlap in samples between groups such as NB on the PCoA plot (Figure 1), and the genetic similarities found in the STRUCTURE analysis particularly within PS and the other groups (Figure 2). How geneflow between the populations sampled here occurs is unknown. Migration is most likely due to distribution of larvae in ocean currents and/or mixing between adjacent populations over time allowing for some genetic connectivity along the northeast pacific coast. More sampling is needed at a finer geographic scale not only to increase sample sizes but also to determine boundaries between adjacent populations and document geneflow among groups.
Finally this study revealed multiple paternity or polyandry as a reproductive strategy in giant Pacific octopuses. MLRelate and Colony analyses suggest a mixture of full sibings (FS) and half siblings (HS) within each female octopuses egg clutch with an average of 64% of fertilized embryos full siblings and 38% half siblings (Table 3). At the individual egg string level Gerud analysis suggest polyandry of between two to three males fathering a female’s eggs within a string and up to four males in total contributing to the progeny within the sampled eggs from a single females clutch or den. In addition Colony analysis suggested that not all males were equally successful with one male thought to have sired progeny in all three dens sampled (male 4), two males siring progeny in two dens (males 1 and 3), and two other males represented in only one of the dens (males 2 and 9). While polyandry was found in all three dens only one female’s clutch was sired by four males (Den 9 was sired by males 1, 3, 4 and male 9, the one resident adult male sampled).

Table 3.
Proportional relatedness among eggs sampled at three octopus dens (1 = 100%).
| MLRelate | FS | HS | Pot. Males |
| Den 1 | 0.46 | 0.64 | |
| Den 7 | 0.67 | 0.33 | |
| Den 9 | 0.69 | 0.31 | |
| Ave | 0.61 | 0.43 | |
| Colony | FS | HS | |
| Den 1 | 0.72 | 0.28 | 1 and 4 |
| Den 7 | 0.67 | 0.33 | 2, 3, and 4 |
| Den 9 | 0.63 | 0.37 | 1, 3, 4 and 9 * |
| Ave | 0.67 | 0.33 | |
| Ave overall | 0.64 | 0.38 |
* = male samples for analysis near Den 9.
Previously it was thought that giant Pacific octopuses employed a polygynous mating system with only the males mating with more than one female []. In this study we did find polygyny with more than one male contributing to more than one female’s egg clutch and also polyandry, females mating with more than one male, or multiple paternity within each female’s egg clutch. Giant Pacific octopuses lay their eggs in strings of connected eggs. Our collection procedure allowed us to determine that multiple paternity is present even within individual strings, as opposed to a string representing fertilization by a single male. Polyandry as a mating strategy is not a surprising tactic given the large numbers of eggs laid, the fact that females are known to store spermatophores after mating, and given that octopuses are terminal phase spawners allowing just one chance to produce viable offspring [,]. Thus females that allow more than one male’s sperm to sire their progeny may increase the chance that at least one of the pairings will produce viable offspring. Finally multiple paternity is not unique within cephalopods has been documented in other octopuses and squid [,,].
4. Conclusions
Giant Pacific octopuses sampled in Oregon, Neah Bay, Puget Sound and Vancouver Island have moderate genetic diversity; moderate population structure suggesting limited geneflow between groups; and three females egg clutches showed evidence of multiple paternity and suggest the females employ a polyandrous mating system. Due to the significant local population structuring found here, we believe conservation measures and fishery management policies should incorporate geographic and population specific goals to ensure the long term preservation of local giant Pacific octopus populations throughout their range.
Acknowledgments
We wish like to thank Keith Chandler from the Seaside Aquarium in Oregon for providing Oregon Coast samples, the many biologists at the Seattle Aquarium who provided the Puget Sound and Neah Bay samples, and divers Jeannie Cosgrove, Leah Saville, David Gagliardi and Fred Demchuk for their assistance in collecting eggs and tissues in Saanich Inlet, BC, Canada. Thanks to Angela Smith, Satya Advani and Lauren Ball for assistance in processing samples and lab analyses. Thanks to CJ Casson, John Braden and Bob Davidson for ongoing support. This work is dedicated to the memory of the late Roland Anderson.
Author Contributions
Shawn Larson collected samples; designed, conducted and analyzed experiments; and prepared manuscript. Catherine Ramsey conducted and analyzed experiments and assisted in preparation of manuscript. James Cosgrove collected samples and assisted in preparation of manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Norman, M. Cephalopods: A World Guide; Conch Books: Hackenheim, Germany, 2000; p. 320. [Google Scholar]
- Hanlon, R.T.; Messenger, J.B. Cephalopod Behaviour. Cambridge University Press: Cambridge, UK, 1996; p. 232. [Google Scholar]
- Mather, J.A.; Kuba, M.J. The cephalopod specialties: Complex nervous system, learning and cognition. Can. J. Zool. 2013, 91, 431–449. [Google Scholar] [CrossRef]
- High, W.L. The giant Pacific octopus. U.S. National Marine Fisheries Service. Mar. Fish. Rev. 1976, 38, 17–22. [Google Scholar]
- Hochberg, F.G. Class Cephalopoda volume 8, the Mollusca, part 1. In Taxonomic Atlas of the Benthic Fauna of the Santa Maria Basin and the Western Santa Barbara Channel; Scott, P.V., Blake, J.A., Eds.; Santa Barbara Museum of Natural History: Santa Barbara, CA, USA, 1998; pp. 175–236. [Google Scholar]
- Cosgrove, J.A. Aspects of the Natural History of, Octopus dofleini, the Giant Pacific Octopus. Master Thesis, Department of Biology, University of Victoria, Victoria, BC, Canada, 1987; p. 101. [Google Scholar]
- Carlson, B.A.; Delbeek, J.C. Cephalopod husbandry: Progress and problems. In Annual Conference Proceedings 1999; American Zoo and Aquarium Association: Silver Spring, MD, USA, 1999; pp. 28–36. [Google Scholar]
- Anderson, R.C. Octopus dofleini and O. rubescens: Animal husbandry. In Proceedings of the Workshop on Fishery and Market Potential of Octopus in California; Lang, M.A., Hochberg, F.G., Eds.; Smithsonian Institution: Washington, DC, USA, 1997; pp. 141–149. [Google Scholar]
- Anderson, R.C.; Mather, J.A. The packaging problem: Bivalve prey selection and prey entry techniques of the octopus Enteroctopus dofleini. J. Comp. Psychol. 2007, 121, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Mather, J.A.; Anderson, R.C. Exploration, play, and habituation in Octopuses (Octopus dofleini). J. Comp. Psychol. 1999, 113, 333–338. [Google Scholar] [CrossRef]
- Greatorex, E.C.; Jones, C.S.; Murphy, J.; Key, L.N.; Emery, A.M.; Boyle, P.R. Microsatellite markers for investigating population structure in Octopus vulgaris (Mollusca: Cephalopoda). Mol. Ecol. 2000, 9, 641–642. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.M.; Balguerías, E.; Key, L.N.; Boyle, P.R. Microsatellite DNA markers discriminate between two Octopus vulgaris (Cephalopoda: Octopoda) fisheries along the North-west African Coast. Bull. Mar. Sci. 2002, 71, 545–553. [Google Scholar]
- Casu, M.; Maltagliati, F.; Meloni, M.; Casu, D.; Cossu, P.; Binelli, G.; Curini-Galletti, M.; Castelli, A. Genetic structure of O. vulgaris (Mollusca, Cephalopoda) from the Mediterranean Sea as revealed by a microsatellite locus. Ital. J. Zool. 2002, 71, 473–486. [Google Scholar]
- Cabranes, C.; Fernandez-Rueda, P.; Martinez, J.L. Genetic structure of Octopus vulgaris around the Iberian Peninsula and Canary Islands as indicated by microsatellite DNA variation. ICES J. Mar. Sci. 2008, 65, 12–16. [Google Scholar] [CrossRef]
- Moreiraa, A.A.; Tomásc, A.R.G.; Hilsdorf, A.W.S. Evidence for genetic differentiation of Octopus vulgaris (Mollusca, Cephalopoda) fishery populations from the southern coast of Brazil as revealed by microsatellites. J. Exper Mar. Biol. Ecol. 2011, 407, 34–40. [Google Scholar] [CrossRef]
- Kang, J.H.; Kim, Y.K.; Park, J.Y.; An, C.M.; Jun, J.C. Development of microsatellite markers to genetically differentiate populations of Octopus minor from Korea and China. Mol. Biol. Rep. 2012, 39, 8277–8286. [Google Scholar] [CrossRef] [PubMed]
- Juárez, O.E.; Rosas, C.; Arena, L. Heterologous microsatellites reveal moderate genetic structure in the Octopus maya population. Fish. Res. 2010, 106, 117–248. [Google Scholar] [CrossRef]
- Quinteiro, J.; Baibai, T.; Oukhattar, L.; Soukri, A.; Seixas, P.; Rey-Mendez, M. Multiple paternity in the common octopus Octopus vulgaris (Cuvier, 1797), as revealed by microsatellite DNA analysis. Mol. Res. 2011, 31, 15–20. [Google Scholar]
- Voight, J.R.; Feldheim, K.A. Microsatellite inheritance and multiple paternity in the deep-sea octopus Graneledone boreopacifica (Mollusca: Cephalopoda). Invertebr. Biol. 2009, 128, 26–30. [Google Scholar] [CrossRef]
- Squires, Z.E.; Wong, B.B.M.; Norman, M.D.; Stuart-Fox, D. Multiple paternity but no evidence of biased sperm use in female dumpling squid Euprymna tasmanica. Mar. Ecol. Prog. Ser. 2014, 511, 93–103. [Google Scholar] [CrossRef]
- Toussaint, R.K.; Sage, G.K.; Talbot, S.L.; Scheel, D. Microsatellite markers isolation and development for the giant Pacific octopus (Enteroctopus dofleini). Conserv. Genet. Resour. 2012, 4, 545–548. [Google Scholar] [CrossRef]
- Raymond, M.; Roussett, F. Genepop (Version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT (Version 1.2): A Computer Program to Calculate F-Statistics. J. Hered. 1995, 86, 485–486. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Wang, J. Sibship reconstruction from genetic data with typing errors. Genetics 2004, 166, 1963–1979. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.G. GERUD 2.0: A computer program for the reconstruction of parental genotypes from half-sib progeny arrays with known or unknown parents. Mol. Ecol. Notes 2005, 5, 708–711. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Wagner, A.P.; Taper, M.L. ML-Relate: A computer program for maximum likelihood estimation of relatedness and relationship. Mol. Ecol. Notes 2006, 6, 576–579. [Google Scholar] [CrossRef]
- Scheel, D.; Bisson, L. Movement patterns of giant Pacific octopuses, Enteroctopus dofleini (Wülker, 1910). J. Exper Mar. Biol. Ecol. 2012, 416–417, 21–31. [Google Scholar] [CrossRef]
- Hartwick, E.B.; Ambrose, R.F.; Robinson, S.M.C. Den utilization and the movements of tagged Octopus dofleini. Mar. Behav. Physiol. 1984, 11, 95–110. [Google Scholar] [CrossRef]
- Mottet, M.G. The Fishery Biology of Octopus dofleini (Wülker); Washington State Department of Fisheries Technical Report; Washington State Department of Fisheries: Washington, DC, USA, 1975; pp. 1–39.
- Snyder, S. Successful rearing of Octopus dofleini from hatching to settlement. In Proceedings of the American Association of Zoological Parks and Aquariums; American Zoo and Aquarium Association: Silver Spring, MD, USA, 1986; pp. 781–783. [Google Scholar]
- Kubodera, T. Distribution and abundance of the early stages of octopus, Octopus dofleini Wulker, 1910 in the north Pacific. Bull. Mar. Sci. 1991, 49, 235–243. [Google Scholar]
- Banas, N.S.; Hickey, B.M. Oceanography of the U.S. Pacific Northwest Coastal Ocean and Estuaries with Application to Coastal Ecology. Estuaries 2003, 26, 1010–1031. [Google Scholar]
- Cannon, G.A. Circulation in the Strait of Juan de Fuca; Some Recent Oceanographic Observations (1978); NOAA Technical Report; Pacific Marine Environmental Laboratory Seattle: Washington, DC, USA, 1978. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).