Abstract
One of the biggest threats to coffee growers in East Africa are emerging vascular wilt diseases (tracheomycosis) caused by Fusarium spp. Many Trichoderma species are known to be natural antagonists of these pathogens and are widely used in biological control of fungal plant diseases. More recently, several Trichoderma spp., which exhibited high antifungal activity have been isolated as endophytes. Consequently, we have investigated the presence and the antagonistic activity of endophytic Trichoderma isolated from roots of healthy coffee plants (Coffea arabica) from the major coffee growing regions of Ethiopia. Our results showed that community of Trichoderma spp. in roots of C. arabica contains fungi from coffee rhizosphere, as well as putatively obligate endophytic fungi. The putatively “true” endophytic species, until now, isolated only from coffee plant ecosystems in Ethiopia and recently described as T. flagellatum and novel T. sp. C.P.K. 1812 were able to antagonize Fusarium spp., which cause coffee tracheomycosis. Moreover, we found that strains of these species are also highly antagonistic against other phytopathogenic fungi, such as Alternaria alternata, Botryotinia fuckeliana (anamorph: Botrytis cinerea), and Sclerotinia sclerotiorum.
1. Introduction
Endophytic fungi are defined as organisms colonizing healthy plant tissue without causing overt symptoms of apparent injury to the host [1]. They have been isolated from nearly all plants families growing in different climatic regions of the world [2]. Many commercially relevant arboreous plants, crops and officinal herbs support communities of endophytic fungi [3,4,5]. Although being biotrophic and consume plants nutrients, endophytic fungi may exhibit a beneficial role for plant health antagonizing pests via mycoparasitism, competition, and/or antibiosis. In addition they may directly stimulate plant growth and immune response inducing resistance to diseases [6,7,8].
Coffea arabica is considered a native plant in Ethiopia and coffee beans are the country’s largest export commodity [9,10]. One of the biggest threats to coffee growers world-wide are emerging fungal wilt diseases, in particular tracheomycosis caused by Gibberella xylarioides (anamorph Fusarium xylarioides) [11]. In Ethiopia incidences of tracheomycosis are reported to concern 60% of total crops, and are accompanied by significant yield losses due to severe damage and ultimate death of millions of coffee bushes [12]. Integrated Disease Management programs would be of a great potential to combat Fusarium pests in Ethiopia.
The fungal genus Trichoderma (teleomorph Hypocrea, Ascomycota, Dikarya) contains some of the most potent biocontrol agents used today [13,14]. In addition, it has been shown that several of its taxa occur as endophytes particularly in the tropical arboreous plants, and that the strains often exhibit high antagonistic activity against pathogens of these plants [15,16,17,18]. However, the importance of Trichoderma or other endophytic fungi as a part of endogenous microbial diversity and their application in the biological control of tracheomycosis on the coffee plants has not been investigated yet.
The objective of this study was to determine the endophytic fungal community of healthy roots of C. arabica with particular emphasis on the presence of Trichoderma and its mycoparasitic potential against Fusarium causing tracheomycosis.
2. Methods
2.1. Root Samples and Isolation of Endophytic Fungi
Roots of C. arabica were collected from different sites of four major coffee growing agroecological systems of Ethiopia (for details see [19]) during the wet season (August 2006) and were kept in paper bags at +4 °C until the examination. Five samples were randomly selected from the mixed sample set and processed for the isolation of endophytic fungi according to the protocol of Mysore et al. [20]. Briefly, roots were cleaned under running tap water to remove debris and were then air-dried on a sterile filter paper. From each sample, 1 cm segments of root slices were surface-sterilized by immersing them in 95% ethanol (2 min), followed by sodium hypochlorite (4% available chlorine; 5 min), and were then washed three times with sterile water. Samples were then allowed to dry on a sterile filter paper for 10 min in a sterile laminar flow chamber. Segments were placed horizontally on potato dextrose agar (PDA) supplemented with streptomycin sulphate (0.4 mg/mL). In a course of 15 days incubation at 25 °C, individual hyphal tips of the developing colonies were re-plated onto PDA plates and were further incubated for 8–10 days. Eventually, single spore cultures for each strain were prepared to assure sample purity. Isolates are stored in 50% glycerol at −80 °C in the Collection of Industrial Microorganisms of Vienna University of Technology (TUCIM, Vienna, Austria).
2.2. DNA Extraction and PCR Amplifications
For DNA extraction mycelium was cultivated three days on 3% MEA (malt extract agar). Genomic DNA was extracted using Qiagen DNeasy Plant Minikit (Hilden, Germany) according to the manufacturer’s instructions. Internal transcribed spacer region (ITS1 and 2) of the rRNA gene cluster was amplified using the primers SR6R and LR1 [21] as described by Kullnig-Gradinger et al. [22]. Additionally, the amplification of approximately1.4 kb fragment of translation elongation factor 1-alpha (tef1) using primers EF1728F [23] and TEF1LLErev (5′-AAC TTG CAG GCA ATG TGG-3′) was performed as described previously (see e.g., [24]). Template DNA was purified with the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and was subjected to automatic sequencing at Eurofins MWG Operon (Ebersberg, Germany).
2.3. DNA Barcoding
Analysis of ITS1 and 2 sequences was performed using sequence similarity search tool (blastn) against the NCBI GenBank database [25]. Trichoderma ITS1 and 2 sequences were subjected to the oligonucleotide barcode program TrichOKey [26]. Still, ambiguous cases were verified by the analysis of the 4th intron of the translation elongation factor 1-alpha-encoding gene (tef1) using sequence similarity search against NCBI GeneBank and TrichoBLAST [27] databases. Identification of Fusarium strains was done based on the analysis of 690 bp fragment of translation elongation factor 1-alpha (EF-1a) gene amplified using EF1 and EF2 primer pairs as described by O’Donnell et al. [28]. Sequences were submitted to similarity search tools against FUSARIUM-ID [29] and NCBI GenBank databases. The NCBI accession numbers for sequences obtained in this study are listed in Table 1 and Table 2.

Table 1.
List of endophytic fungi (without Trichoderma spp.) isolated from the roots of C. arabica and their identification based on tef1 barcode.
| TUCIM No. (C.P.K) | GenBank Acc. No | Identification | ||||
|---|---|---|---|---|---|---|
| % similarity/% query coverage | Phylum | Putative identification | The most similar NCBI GenBank entry | |||
| taxon | Acc. No | |||||
| 3484 | FJ827622 | 100/100 | Ascomyc. | Aspergillus | Aspergillus flavus | AM745114 |
| 3485 | FJ827626 | 100/94 | Cladosporum | Cladosporium cladosporioides | EF577236 | |
| 3478 | FJ827623 | 100/98 | Rhizopycins | Rhizopycnis sp. IBL 03177 | DQ682600 | |
| 3078 | FJ827619 | 99/94 | Penicillium | Penicillium sp. RCEF3398 | EF570358 | |
| 3084 | FJ827620 | 99/95 | ||||
| 3468 | FJ827620 | 99/92 | Phomopsis | Phomopsis columnaris | AF439625 | |
| 3471 | FJ827629 | 99/92 | ||||
| 3473 | FJ827628 | 99/93 | ||||
| 3512 | FJ827630 | 94/98 | Dipodascaceae | Dipodascus australiensis | AF157596 | |
| 3474 | FJ827625 | 99/98 | Macrophomina | Macrophomina phaseolina | EU250575 | |
| 3486 | FJ827624 | 100/97 | Pleosporales | Pleosporales sp. IBL 03175 | DQ682598 | |
| 3337 | FJ827616 | 99/99 | Fusarium equiseti | F. equiseti isolate 45/1.2.1 | DQ854855 | |
| 3469 | FJ827615 | 99/ 98 | ||||
| 3514 | FJ840530 | 99/98 | ||||
| 3466 | FJ827613 | 99/98 | ||||
| 3472 | FJ827614 | 99/98 | ||||
| 3513 | n/a | 99/98 | ||||
| 3465 | FJ840527 | 99/98 | ||||
| 3480 | FJ827612 | 93/99 | ||||
| 3470 | FJ840525 | 93/98 | F. equiseti isolate SAT73 | DQ465946 | ||
| 3479 | FJ827610 | 93/99 | F. equiseti isolate 21/1.2 | DQ854851 | ||
| 3332 | FJ840526 | 99/98 | ||||
| 3509 | FJ827611 | 99/98 | F. solani | Fusarium sp. NRRL 43704 FSSC | EF453029 | |
| 3510 | FJ840531 | 99/98 | ||||
| 3511 | FJ840532 | 99/98 | ||||
| 3330 | FJ840533 | 99/98 | ||||
| 3515 | FJ840529 | 99/99 | F. oxysporum | F. oxysporum isolate SAT77 | DQ465933 | |
| 3476 | FJ827617 | 99/99 | ||||
| 3475 | FJ827618 | 98/100 | Basidiomyc. | Trametes | Trametes versicolor | FJ608587 |
Table 2.
Endophytic Trichoderma strains isolated from roots of C. arabica.

| TUCIM No. (C.P.K) | NCBI GenBank | TUCIM No. (C.P.K) | NCBI GenBank | TUCIM No. (C.P.K) | NCBI GenBank | ||||
|---|---|---|---|---|---|---|---|---|---|
| ITS1 and 2 | tef1 | ITS1 and 2 | tef1 | ITS1 and 2 | tef1 | ||||
| Section Trichoderma | Harzianum-Catoptron Clade | Section Longibrachiatum | |||||||
| Trichoderma hamatum | Hypocrea ‘pseudoharzianumʼ nom. prov. | T. flagellatum | |||||||
| 3331 | FJ461539 | n/a | 3440 | FJ461557 | FJ763164 | 3334 | FJ461542 | FJ763149 | |
| 3333 | FJ461540 | FJ763148 | 3441 | FJ461558 | FJ763165 | 3345 | FJ461551 | FJ763158 | |
| 3335 | FJ461541 | n/a | 3442 | FJ461559 | FJ763166 | 3350 | FJ461556 | FJ763163 | |
| 3336 | FJ461543 | FJ763150 | 3443 | FJ461560 | FJ763167 | 3496 | FJ461568 | FJ763174 | |
| 3338 | FJ461544 | FJ763151 | 3501 | FJ461573 | FJ763177 | 3503 | FJ461575 | FJ763179 | |
| 3342 | FJ461548 | FJ763155 | 3502 | FJ461574 | FJ763178 | 3504 | FJ461576 | FJ763180 | |
| 3343 | FJ461549 | FJ763156 | 3506 | FJ461578 | FJ763181 | 3517 | FJ461580 | FJ763182 | |
| 3346 | FJ461552 | FJ763159 | T. sp. C.P.K. 1812 | 3522 | FJ461585 | n/a | |||
| 3347 | FJ461553 | FJ763160 | 3328 | FJ461537 | FJ763146 | 3523 | FJ461586 | n/a | |
| 3348 | FJ461554 | FJ763161 | 3329 | FJ461538 | FJ763147 | 3524 | FJ461587 | FJ763183 | |

| TUCIM No. (C.P.K) | NCBI GenBank | TUCIM No. (C.P.K) | NCBI GenBank | ||
|---|---|---|---|---|---|
| ITS1 and 2 | tef1 | ITS1 and 2 | tef1 | ||
| Section Trichoderma | Harzianum-Catoptron Clade | ||||
| Trichoderma hamatum | Hypocrea ‘pseudoharzianumʼ nom. prov. | ||||
| 3491 | FJ461563 | n/a | 3444 | FJ461561 | FJ763168 |
| 3492 | FJ461564 | FJ763170 | 3490 | FJ461562 | FJ763169 |
| 3493 | FJ461565 | FJ763171 | 3494 | FJ461566 | FJ763172 |
| 3497 | FJ461569 | n/a | 3495 | FJ461567 | FJ763173 |
| 3505 | FJ461577 | n/a | 3498 | FJ461570 | n/a |
| 3507 | FJ461579 | n/a | 3499 | FJ461571 | FJ763175 |
| 3518 | FJ461581 | n/a | 3500 | FJ461572 | FJ763176 |
| 3519 | FJ461582 | n/a | T. sp. C.P.K. 1833 | ||
| 3520 | FJ461583 | n/a | 3339 | FJ461545 | FJ763152 |
| 3521 | FJ461584 | n/a | 3341 | FJ461547 | FJ763154 |
2.4. Phylogenetic Analyses
For phylogenetic analysis DNA sequences were aligned with the CLUSTAL X program (version 1.81) [30] and visually verified with GeneDoc software (version 2.6) [31]. Phylogenetic analysis was performed using the maximum-parsimony method (observed p-distance, no evolutionary modeling required) implemented in PAUP*4.0b10 applying a heuristic search (n = 1000) with the random addition of sequences and the TBR tree-swapping algorithm. The reliability of the obtained clades was tested by 500 bootstrap replications. Bootstrap values of >74% were considered significant. The phylogenetic position of the isolates attributed to the Harzianum-Catoptron Clade was inferred using the Bayesian phylogenetic method implemented in MrBayes v.3 [32] with 3,000,000 mcmc generations and a general time-reversible model for nucleotide substitutions [33], as described previously [34]. Posterior probability above 0.94 was considered significant [35].
2.5. Screening for Antifungal Activity
Antifungal activity of endophytic Trichoderma isolates was tested against the causative agents of coffee tracheomycosis, G. xylarioides, and G. jujikuroi (anamorph Fusarium oxysporum or FOX later on in the ms), and also against three other plant pathogenic fungi, Alternaria alternata (TUCIM 199), Botryotinia fuckeliana (anamorph: Botrytis cinerea, TUCIM 200) and Sclerotinia sclerotiorum (TUCIM 201). The corresponding isolates of Fusarium spp. were obtained from infected coffee roots collected in the same region. Antagonistic ability of Trichoderma strains against G. xylarioides and FOX was tested, as described by Ortiz and Orduz [36]: a 5 mm diameter agar plug was removed from a freshly prepared PDA plate and replaced by an equally sized plug from a seven days old culture of G. xylarioides, or FOX pre-grown on PDA. After incubation for a further four days an agar plug containing mycelium of the respective Trichoderma isolate (5 mm diameter) was placed on the plate, 3 cm distant from the Fusarium colony. The plates were then further incubated for five days. Six replicates were set for each confrontation and plates containing two plugs of Fusarium culture were used as a control. Center-wise growth of Trichoderma and Fusarium, indicative by an internal mycelial growth radius (internal halo) and edge-wise growth indicated by a free mycelial growth radius (external halo), was also measured.
Dual confrontation assays with A. alternata, B. cinerea and S. sclerotiorum were made on PDA plates incubated for 10 days in a 12 h illumination cycle at 25 °C. Fresh culture agar plugs of prey and predator fungi were placed on a PDA plate located on opposite poles, each, 1 cm from the edge of the plate. Antagonistic potential was estimated as a reduction of the prey growth rate corrected for the growth rate of a pray when confronted with itself. To reveal the differences in mycoparasitic potential between endophytic and saprophytic Trichoderma sp. the results of endophytic strains isolated in this study were compared to the dual confrontation tests of saprophytic Trichoderma strains from TUCIM database (TU Collection of Industrial Microorganisms, TU Vienna). Antagonistic potential was presented as averaged growth inhibition or overgrowth of each plant pathogen (prey) by Trichoderma strains expressed in percentage. In both cases 100% represented unrestricted growth of the prey corrected for the confrontation with itself. Data were analyzed using Bartlett’s test to verify the homogeneity of variances, and then subjected to one-way ANOVA using SPSS 17.0 software. Least significant difference (LSD) was used to compare treatment means.
3. Results
3.1. The Endophytic Fungal Community in Roots of C. arabica
A total of 300 segments from five healthy C. arabica roots were used for this study, of which 80 fungal isolates were recovered and tentatively identified at the generic or higher taxonomic level (Table 1) with aid of ITS1 and 2 rRNA barcodes. Eight genera and two families were assigned to the phylum Ascomycota, while a single Basidiomycota strain was identified as Trametes spp. Members of the genera Trichoderma (n = 50) and Fusarium (n = 18) were most frequently isolated.
3.2. Diversity of Endophytic Trichoderma
ITS1 and 2 barcoding identified 21 strains as T. hamatum. In order to test whether they are identical to the T. hamatum populations which occur in coffee rhizosphere [19], we also sequenced a fragment of the elongation factor 1-alpha gene (tef1) containing the long (4th) intron. Phylogenetic analyses showed that T. hamatum isolates exhibited the same tef1 allele as the strains previously found in rhizosphere of C. arabica [19] (Figure 1A). Genetic distances of Ethiopian T. hamatum strains to the type strain and other African isolates (from Cameroon, indicated by * on Figure 1A) suggest biogeographical isolation of the Ethiopian population associated with C. arabica.
The next majority of strains (18) were identified as members of the Harzianum-Catoptron Clade [37]. The phylogenetic analysis of tef1 locus revealed that seven strains formed a separated clade inside of “T. harzianum species complex” (H. ‘pseudoharzianum’ sensu Druzhinina et al. [36]), possessing a unique tef1 allele (Figure 1B), that was not detected in rhizosphere of C. arabica [19]. Nine other isolates were identified as further members of a putative new species T. sp. C.P.K. 1812 (Figure 1B), which was so far only known from a single isolate from coffee rhizosphere [19]. The remaining two isolates C.P.K. 3339 and 3341 belonged to the putative new phylogenetic species T. sp. C.P.K. 1833 (Figure 1B), which also has been abundantly found in the rhizosphere of C. arabica in Ethiopia [19]; its formal taxonomic description based on multiloci phylogeny will be published elsewhere.
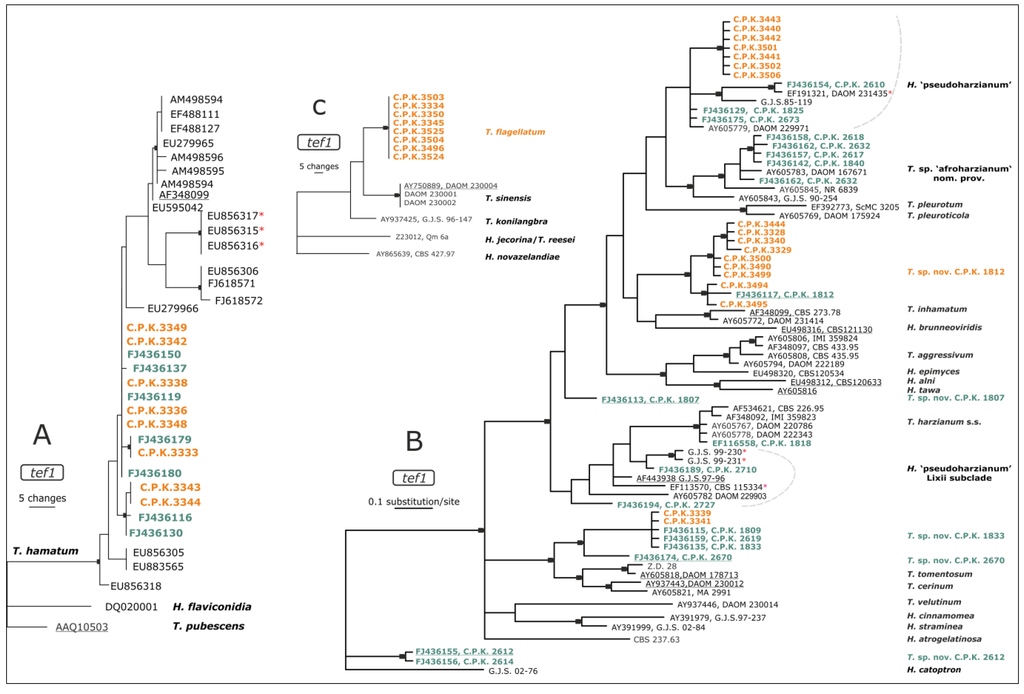
Figure 1.
Phylogeny of endophytic Trichoderma strains isolated from coffee (Coffea arabica) roots inferred by maximum parsimony (MP) (A,C) and Bayesian (MB) analyses (B). Nodes supported by bootstrap values for MP of >74% or posterior probabilities for MB>0.94 respectively are indicated by black arrows. The color code indicates isolates associated with C. arabica as green for strains that were isolated from the coffee rhizosphere [19] and orange for endophytic strains isolated in this study. Sequences of established type cultures are underlined; asterisks mark strains of African origin. (A) T. hamatum, (B) Harzianum-Catoptron Clade, and (C) section Longibrachiatum.
By the means of ITS1 and 2 barcode, 11 strains could only be identified to the section level, and were attributed to the section Longibrachiatum. Furthermore, no polymorphism was found among their ITS1 and 2 and tef1 sequences. A phylogenetic analysis of the tef1 locus grouped them in a sister clade next to T. sinensis (Figure 1C), which was described based on three isolates from Taiwan [38]. Based on our isolates Samuels et al. [39] recently described this species as T. flagellatum Mulaw, Kubicek et Samuels as a common endophyte in roots of coffee in Ethiopia. It forms a clade with T. sinense, T. konilangbra, and T. gillesii [39,40].
3.3. Diversity of Endophytic Fusarium Isolates
We assessed the diversity and species composition of endophytic Fusarium spp. from C. arabica. To reveal the endophytic community of Fusarium isolates we sequenced and analysed the gene encoding for elongation factor 1-alpha (abbreviated as EF-1α in Fusarium-related publications). The sequences were subjected to the NCBI GeneBank similarity search (blastn; [25]), and the 20 best hits were retrieved for the evaluation. By these means, the 18 Fusarium isolates were identified as four species with identical EF-1α alleles. One of them appeared to be a member of the Haematonectria haematococca (anamorph F. solani) species complex, yet differing by one SNP at a position not found in any other query sequence NCBI GenBank for June 2011. The most similar hits were G. jujikuroi and F. falciforme strains from soil in South Africa, Tanzania, Mexico, and the Philippines (Table 1). Two other groups were identified as members of the Gibberella intricans (anamorph F. equiseti) species complex by means of 99 and 93% similarity (Table 1), whereas the fourth group with two isolates belonged to FOX species complex (Table 1). Based on the result of Bayesian phylogenetic analysis (data not shown) their sequences were cospecific with the most abundant EF-1a phylotype also found in the rhizosphere of C. arabica in Ethiopia (T.B. Mulaw, I.S. Druzhinina, ms in preparation).
3.4. Antifungal Activity of Trichoderma Species
The antifungal activities of selected endophytic Trichoderma strains were first tested against the two agents of coffee tracheomycosis, FOX, and G. xylarioides respectively (Figure 2 and Figure 3). All of the isolates strongly inhibited growth of both Fusarium spp., yet the antagonistic activity was strain specific. The inhibition of G. xylarioides growth by endophytic Trichoderma isolates appeared to vary as antibiosis (Figure 2) and coiling were not observed for all strains. In contrast, inhibition of FOX growth was consistent for all tested Trichoderma strains, being considerably weaker, mostly moderate, if compared to the inhibition of G. xylarioides. Strain C.P.K. 3340, belonging to the putative new species T. sp. C.P.K. 1812, exhibited the strongest antagonistic potential against both Fusarium species tested (Figure 2 and Figure 3).
In order to test whether the mycoparasitic activity of endophytic Trichoderma strains is prey-specific or the mycoparasitism is generally increased for these strains, we made the dual confrontation assays with the three model plant pathogenic fungi A. alternata, B. cinerea, and S. sclerotiorum (Table 3). The data were evaluated in respect to 53 and 250 saprotrophic strains of T. hamatum and H. ‘pseudoharzianum’ (≡ former T. harzianum sensu lato; see Druzhinina et al. [37]), respectively, stored in TUCIM collection. Our data showed that the mycoparasitic potential of the endophytic strains is either equal or higher to the potential of saprotrophic T. hamatum and H. ‘pseudoharzianum’. Interestingly, the mode of antagonistic action in novel putative species seems to be species specific; T. sp. C.P.K. 1812 showed superior ability to overgrow B. cinera and S. sclerotiorum, while more than 50% of T. flagellatum strains failed to overgrow any of the three plant pathogens (Table 3). The endophytic strains of T. hamatum are essentially more active against B. cinerea than saprotrophic strains used in this study. Furthermore, all T. hamatum strains exhibited significantly higher potential to overgrow S. sclerotiorum compared to other species. However, all endophytic Trichoderma species found in this survey revealed high antagonistic potential and can well inhibit pray’s growth.
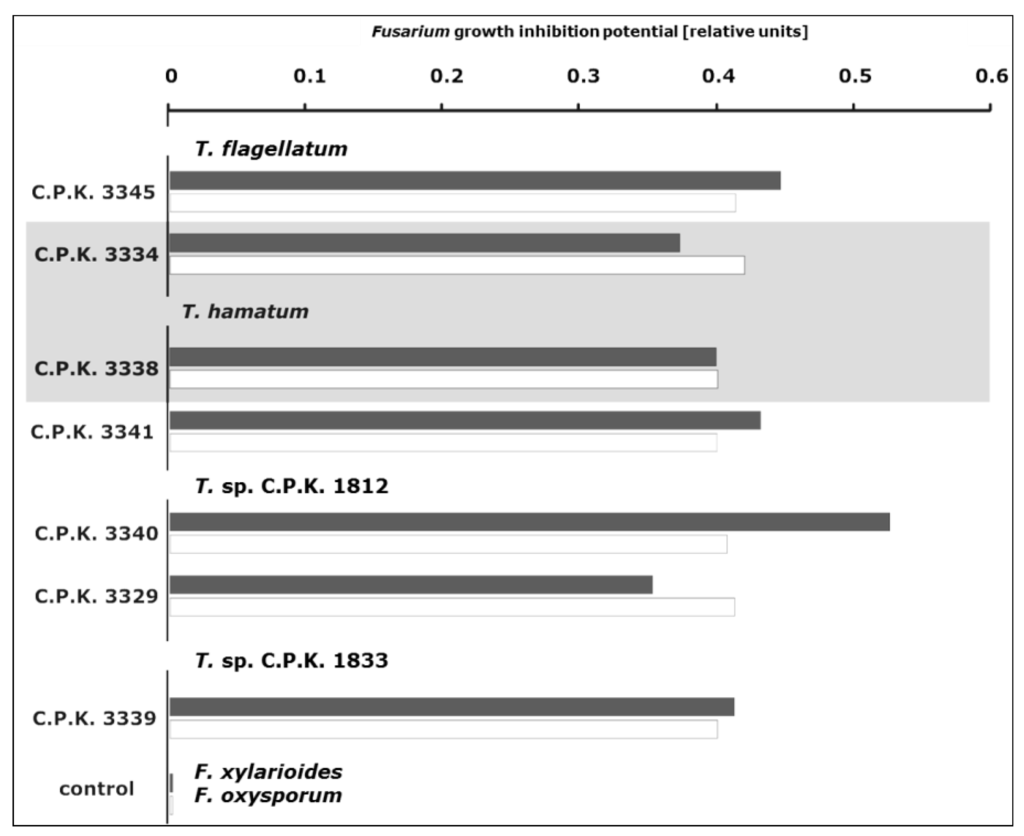
Figure 2.
Antagonistic potential of selected Trichoderma strains against G. xylarioides and G. jujikuroi. The shadowing indicates the two isolates capable of producing a diffusible inhibitor against Fusarium (antibiosis). Black and white bars indicate the inhibition of radial growth for G. xylaroides and G. jujikuroi respectively.
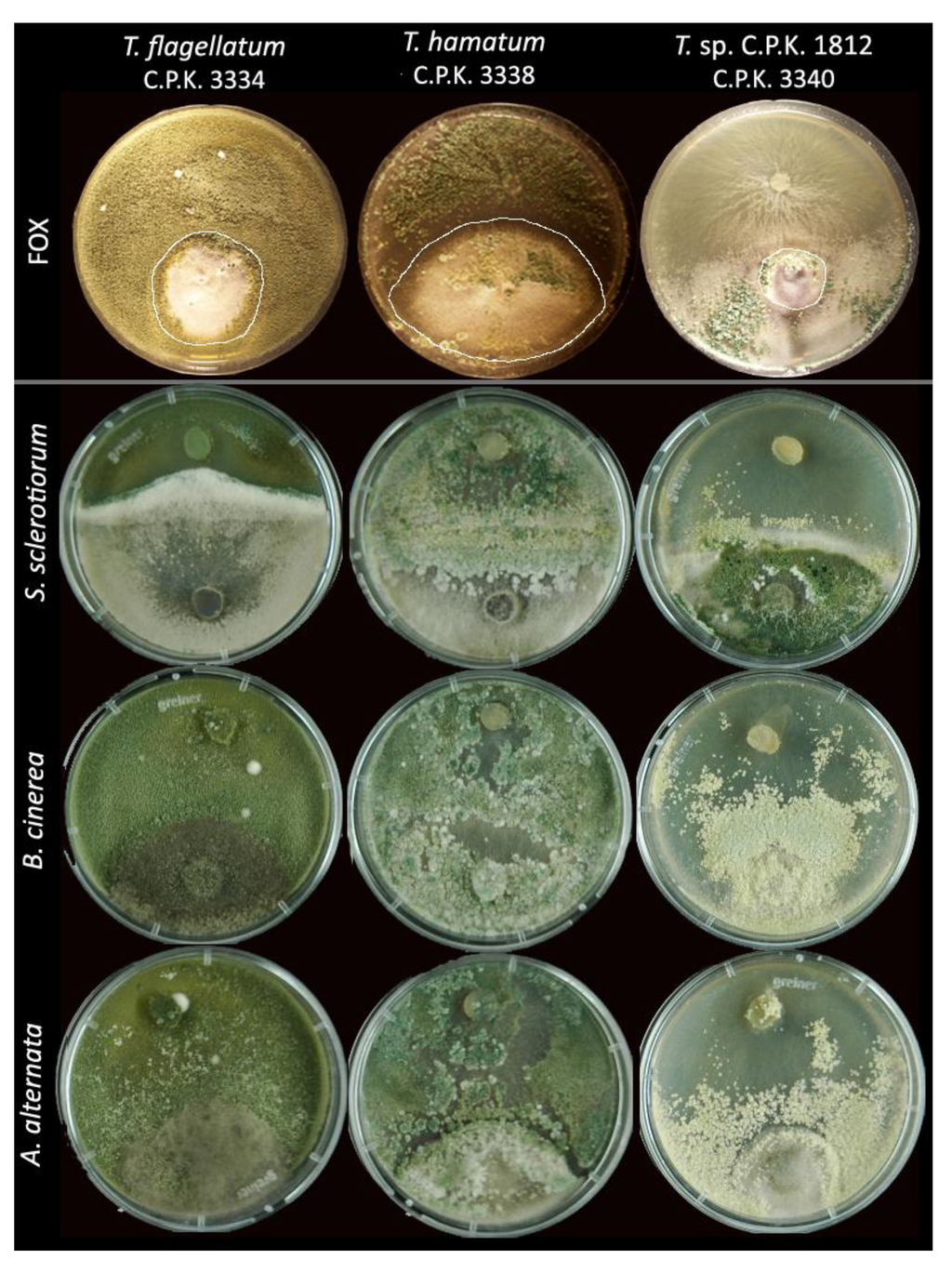
Figure 3.
Dual confrontation plates with Trichoderma strains (always above) against four plant pathogens (always below). White lines show the extension of FOX colony.

Table 3.
Antagonistic potential (plant pathogen’s growth inhibition and overgrowth) of endophytic Trichoderma strains isolated from C. arabica roots and their comparison to saprotrophic T. hamatum and H. ‘pseudoharzianum’ nom. prov.
 | Inhibition of preys (% *) | % of strains capable to overgrow the prey | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Per prey species | For 3 preys | For 3 preys | ||||||||||||||||||
| Ss | SD | Ss | ||||||||||||||||||
| SD | ||||||||||||||||||||
| Endophytic strains from roots of C. arabica | ||||||||||||||||||||
| T. hamatum | 0.65 | 17.66 | 89.5 | 84.2 | ||||||||||||||||
| Control (saprotrophic strains) | ||||||||||||||||||||
| T. hamatum | 8.47 | 0.95 | 17.12 | 75.5 | 82.3 | |||||||||||||||
* 100% is unrestricted growth of the prey corrected for the confrontation with itself (on the plate marked with red line, whereas % inhibition is marked with green arrow); conditional color formatting (the gradient from green to red marks the trend from lowest to the highest value) is applied for both panels independently. Aa, Bc and Ss indicate A. alternata, B. cinerea, and S. sclerotiorum, respectively. N means number of strains, av average value, sd standard deviation.
4. Discussion
In this study we extended our earlier findings [19] of Trichoderma species diversity in the rhizosphere of C. arabica in Ethiopia to coffee endophytes. Despite the potential benefit for C. arabica, the community of its endophytic microorganisms has not received much attention. In fact, the only studies so far have been focusing on endophytic bacteria [41], fungal foliar endosymbionts [8] and the presence of ochratoxin producing Penicillium spp. [42] in C. arabica. In contrast, in this study we focused on the diversity of fungi isolated directly from the healthy coffee roots and on screening for endophytic Trichoderma strains that can effectively combat two major pathogens of tracheomycosis, G. xylarioides, and FOX. We also tested the general ability of endophytic Trichoderma to inhibit further ascomyceteous plant pathogens and compared it to those of the saprophytic Trichoderma strains.
4.1. The Narrow Ecological Niche inside Coffee Roots Favors Diverse Community of Endophytic Fungi
C. arabica and other plants rich in xanthine alkaloids such as caffeine represent a special ecological and evolutionary challenge for microorganisms that colonize them. However, the recent studies report on a diversity of fungal endophytes in Theobroma cacao (cacao tree, [18]) and C. arabica (coffee plant, [8]). The surveys on endophytic fungi in coffee performed in Colombia, Hawaii, and Puerto Rico showed the presence of Colletotrichum, Fusarium, Penicillium, Xylaria as the most common genera but also included several entomopathogenic fungi, such as Acremonium spp., Beauveria spp., Cladosporium spp., Clonostachys spp., and Paecilomyces spp. [8]. However, in this study 63% of fungi isolated from healthy coffee roots represented Trichoderma and Fusarium species. Thus, our results show that there are different endopytic fungal communities colonizing leaves (see [8]) and roots of C. arabica. The fact that Trichoderma was isolated from C. arabica roots but not from leaves support the suggestion that Trichoderma dominates in the rhizosphere community of coffee plants [19], where it plays important role for the plant protection founding special external and also endophytic association with the plant. Understanding endophytic communities is of great importance for improvement of plant health and it is one of the most poorly explored areas in agricultural biotechnology [8].
4.2. Diversity of Endophytic Trichoderma in Roots of C. arabica is Unique
Only two of 50 Trichoderma isolates (both belonging to T. sp. C.P.K. 1833) had tef1 allele which has been found before in about 5000 Trichoderma isolates collected world-wide while all others possessed alleles either unique or previously seen in the rhizosphere of C. arabica in Ethiopia (see [19]).
The diversity of endophytic Trichoderma strains in C. arabica was essentially lower compared to the corresponding rhizosphere [19] what indicates that not all Trichoderma strains present in the rhizosphere are capable to colonize the roots. For this opportunistic avirulent symbiotic relationship it is crucial Trichoderma to penetrate into the plant’s root system and persistently survive within living plant tissues [13,43]
Consistent with our results on rhizosphere fungal community in C. arabica [19], T. hamatum was the most abundant species inhabiting the roots of coffee plant. As expected, T. hamatum strains from both ecological niches were sharing the same tef1 allele, different from any other, also African strains of this species.
Strains belonging to Harzianum-Catoptron Clade [37] were abundantly isolated, yet the detailed phylogeny revealed that they are members of three different species. The majority of this strains were attributed to the putative new species T. sp. C.P.K. 1812, so far only known by a single strain found by Mulaw et al. [19] in the rhizosphere of C. arabica. The second numerous group revealed a novel subclade within H. ‘pseudoharzianum’ sensu Druzhinina et al. [37] branching closely to the putative new agamospecies T. sp. ‘afroharzianum’ nom. prov. [37]. However, due to the intensive sexual recombination inside this group the taxonomic position of this subclade must be assessed by multiloci phylogeny elsewhere. Further two isolates belonging to the Harzianum-Catoptron Clade were attributed to the new cosmopolitan species T. sp. C.P.K. 1833 (T. ‘ethiopionense’ nom. prov, ms in preparation) abundantly found in rhizosphere of C. arabica but also in soils from Siberia [22], in Eastern Europe (R. Labuda, personal communication) and in Central America [26] suggesting the cosmopolitan distribution and saprotrophic nature of this species.
The recently formally described species T. flagellatum [39] was first found in our survey and was neither detected in rhizosphere of C. arabica nor elsewhere so far.
Many Trichoderma strains which were recently isolated as endophytes from tropical Theobroma cacao were described as new species; Trichoderma ovalisporum [44], Trichoderma martiale [16], Trichoderma stromaticum [45], Trichoderma theobromicola, and Trichoderma paucisporum [17], Trichoderma koningiopsis [46], and Trichoderma evansii [18]. Furthermore, an endophytic Trichoderma from Chinese yew (Taxus mairei), T. taxi, was also recently described [47]. In this survey we isolated a novel endophytic species T. flagellatum isolated from healthy roots of C. arabica. Its formal description was recently published by Samuels et al. [1]. Continuous discovery of species novel to science, which were isolated as endophytes likely resembles the intensive speciation in such narrow ecological niches. Our data also show that the Ethiopian semi-natural coffee soil ecosystem likely underwent major isolation, which was beneficial for formation of not only distinct genotypes but also new species. Mulaw et al. [19] emphasised that C. arabica rhizosphere in Ethiopia has been a hot spot for speciation of several Trichoderma spp. as C. arabica is known to display a high genetic diversity in Ethiopia [10] and Tanzania [48], and this fact should gave rise to new Trichoderma populations and taxa capable of establishing themselves in the rhizosphere of this genetically highly variable plant. However, due to the accumulation of various alkaloids and other biologically active compounds C. arabica may not be an easy target for an opportunistic attack and the adaptation to endophytism might require a longer evolutionary interval.
4.3. G. xylarioides is not Present in the Community of Endophytic Fungi from Healthy Coffee Plants
The finding of Fusarium spp. as endophytes of healthy C. arabica is intriguing; the majority of them belonged to the G. fujikuroi and G. intricans species complexes. G. fujikuroi/F. solani is one of the most frequently isolated fungus from soil and plant debris, but it is also a host-specific pathogen of a number of agriculturally important plants, including pea, cucurbits, and sweet potato [28,49,50]. G. intricans/F. equiseti, in contrast, is considered as a rather weak pathogen, yet capable of colonizing roots and eventually causing disease symptoms in a broad range of crop plants [51]. The EF-1a allele of endophytic G. fujikuroi/F. solani species complex shared highest similarity to isolates from tropical soil assigned as F. solani and F. falciforme. One of the most similar BLAST hits (DQ247630) even has been isolated as a parasite of a Phytium sp. They likely represent a tropical opportunistic population, which have settled in the rhizosphere of C. arabica.
The second agent of coffee tracheomycosis, G. xylarioides was not detected in the community of endophytic strains of C. arabica. These findings might imply that this agent of the coffee wilt disease has been introduced to Ethiopia, as the occurrence of G. xylarioides on C. arabica in Ethiopia was first detected in the early 1970s [12], yet this should be confirmed in further investigations. Furthermore, the comparison of G. xylarioides isolates from different coffee species, also including earlier isolates, revealed host specialization of this pathogen on Coffea spp. [12].
4.4. Endophytic Trichoderma Strains Exhibit High Antifungal Potential
Several epiphytic and endophytic Trichoderma species and strains are already being well studied and further recommended as potential biological control agents against phytophatogenic fungi such as Phytophthora capsici in hot pepper [52], Moniliophthora roreri, M. fide, M. perniciosa, and Phytophthora species in cacao trees [43,53]. Trichoderma strains being studied for their potential to control cacao diseases and ameliorate damage caused by abiotic stresses also have an ability to alter cacao gene expression during colonization [43].
Our data revealed high antagonistic potential against the two agents of coffee tracheomycosis, G. xylarioides and FOX. However, the endophytic strains less successfully combated FOX, which was also isolated from roots of C. arabica, then G. xylarioides, which was not detected in roots of healthy plants. This likely reflects a degree of adaptation between endophytic FOX and Trichoderma in this specialized native environment.
Furthermore, the endophytic Trichoderma strains expressed high mycoparasitic potential against three tested plant pathogens (A. alternata, B. cinerea, and S. sclerotiorum). Their antagonistic activity was in range or even higher with those from saprotrophic strains of T. hamatum and H. ‘pseudoharzianum’. The growth of Fusarium pathogens was efficiently inhibited by the endogenous Trichoderma strains, therefore these endophytic Trichoderma spp. may represent attractive candidates for biocontrol of coffee tracheomycosis and eventually also for other fungal plant diseases. Particularly the putatively exclusive endophytic species (T. flagellatum and T. sp. C.P.K. 1812) are able to antagonize G. xylarioides and FOX which cause coffee tracheomycosis, and additionally also successfully inhibit growth of other plant pathogens. The strain C.P.K. 3340, belonging to the putative new species T. sp. C.P.K. 1812, exhibited the strongest antagonistic potential against both Fusarium species tested and possessed a great ability to overgrow other three pathogens, thus rendering a potential biocontrol candidate that could simultaneously combat several plant diseases. Herewith, we propose in vivo experiments with novel endophytic Trichoderma species on the C. arabica cultivars to assess effective value of their application in future.
5. Conclusions
In this study we confirmed that endophytic Trichoderma spp. from C. arabica display antagonistic activity against G. xylarioides and FOX, the two Fusarium species found in wilted coffee roots. The diversity of Trichoderma inside C. arabica roots is essentially lower compared to rhizosphere and is composed of mainly endemic species which are either new to science or were previously detected in rhizosphere of this plant. Endophytic strains of the common opportunistic species T. hamatum and H. ‘pseudoharzianum’ possess unique DNA barcodes suggesting emerging genetic isolation. Strains of the putative new species T. sp. C.P.K. 1812 and recently described T. flagellatum so far isolated only from C. arabica roots from Ethiopia have shown high mycoparasitic capacity against the studied set of pathogens and therefore may be recommended for the integrated disease management of coffee plants in Ethiopia.
Acknowledgments
This work was partly supported by the Austrian Science Fund grants FWF P17859 to I.S.D and by OeAD 894/07 North-South Dialogue Program, doctorate scholarship to T. B. Mulaw. The authors are grateful to Liliana Espino de Rammer, Monika Komon-Zelazowska, and Benigno Aquino (Vienna University of Technology, Austria) for laboratory assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bills, G.F. Isolation and Analysis of Endophytic Fungal Communities from Woody Plants. In Systematics, Ecology and Evolution of Endophytic Fungi in Grasses and Woody Plants; Redlin, S., Carris, L.M., Eds.; APS Press: St. Paul, MN, USA, 1996; pp. 31–65. [Google Scholar]
- Larran, S.; Perello, A.; Simon, M.R.; Moreno, V. Isolation and analysis of endophytic microorganisms in wheat (Triticum aestivum L.) leaves. World J. Microb. Biot. 2002, 18, 683–686. [Google Scholar] [CrossRef]
- Redlin, S.C.; Carris, L.M. Endophytic Fungi in Grasses and Woody Plants. In Systematics, Ecology and Evolution of Endophytic Fungi in Grasses and Woody Plants; Redlin, S., Carris, L.M., Eds.; APS press: St. Paul, MN, USA, 1996; p. 216. [Google Scholar]
- Marshall, D.; Tunali, B.; Nelson, L.R. Occurrence of fungal endophytes in species of wild Triticum. Crop Sci. 1996, 39, 1507–1512. [Google Scholar] [CrossRef]
- Huang, Y.; Jianfeng, W.G.L.; Zhonghui, Z.; Wenjin, S. Anti tumor and antifungalactivities in endophytic fungi isolated from pharmaceutical plants Taxus mairei, Cephalataxus fortunei and Torreya grandis. FEMS Immunol. Med. Microbiol. 2001, 31, 163–167. [Google Scholar] [CrossRef]
- Arnold, A.E.; Mejía, L.C.; Kyllo, D.; Rojas, E.I.; Maynard, Z.; Robbins, N.; Herre, E.A. Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. USA 2003, 100, 15649–15654. [Google Scholar]
- Niere, B.; Gold, C.S.; Coyne, D. Can fungal endophytes control soilborne pests in banana? Bull. OILB/SROP 2004, 27, 203–209. [Google Scholar]
- Santamaría, J.; Bayman, P. Fungal epiphytes and endophytes of Coffee Leaves (Coffea arabica). Microb. Ecol. 2005, 50, 1–8. [Google Scholar] [CrossRef]
- Sylvain, P. Ethiopian coffee—its significance to world coffee problems. Econ. Bot. 1958, 12, 111–139. [Google Scholar] [CrossRef]
- Aga, E.; Bryngelsson, T.; Bekele, E.; Salomon, B. Genetic diversity of forest arabica coffee (Coffea arabica L.) in Ethiopia as revealed by random amplified polymorphic DNA (RAPD) analysis. Hereditas 2003, 138, 36–46. [Google Scholar] [CrossRef]
- Geiser, D.M.; Ivey, M.L.L.; Hakiza, G.; Juba, J.H.; Miller, S.A. Gibberella xylarioides (anamorph: Fusarium xylarioides), a causative agent of coffee wilt disease in Africa, is a previously unrecognized member of the G. fujikuroi species complex. Mycologia 2005, 97, 191–201. [Google Scholar] [CrossRef]
- Girma, A.; Hulluka, M.; Hindorf, H. Incidence of tracheomycosis, Gibberella xylarioides (Fusarium xylarioides), on Arabica coffee in Ethiopia. Zeitschrift fur Pflanzenkrankheiten und Pflanzenschutz 2001, 108, 136–142. [Google Scholar]
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma species: Opportunistic avirulent plant symbionts. Nat. Rev. Microbiol. 2004, 2, 43–56. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Seidl-Seiboth, V.; Herrera-Estrella, A.; Horwitz, B.A.; Kenerley, C.M.; Monte, E.; Mukherjee, P.K.; Zeilinger, S.; Grigoriev, I.V.; Kubicek, C.P. Trichoderma: The genomics of opportunistic success. Nat. Rev. Microbiol. 2011, 9, 749–759. [Google Scholar] [CrossRef]
- Zhang, C.L.; Druzhinina, I.S.; Kubicek, C.P.; Xu, T. Biodiversity of Trichoderma in China: Evidence for a North to South difference of species distribution in East Asia. FEMS Microbiol. Letts. 2005, 251, 251–257. [Google Scholar] [CrossRef]
- Hanada, R.E.; de Jorge Souza, T.; Pomella, A.W.; Hebbar, K.P.; Pereira, J.O.; Ismaiel, A.; Samuels, G.J. Trichoderma martiale sp. nov., a new endophyte from sapwood of Theobroma cacao with a potential for biological control. Mycol. Res. 2008, 112, 1335–1343. [Google Scholar] [CrossRef]
- Samuels, G.J.; Suarez, C.; Solis, K.; Holmes, K.A.; Thomas, S.E.; Ismaiel, A.; Evans, H.C. Trichoderma theobromicola and T. paucisporum: Two new species isolated from cacao in South America. Mycol. Res. 2006, 110, 381–392. [Google Scholar] [CrossRef]
- Samuels, G.J.; Ismaiel, A. Trichoderma evansii and T. lieckfeldtiae: Two new T. hamatum-like species. Mycologia 2009, 101, 142–156. [Google Scholar] [CrossRef]
- Mulaw, T.B.; Kubicek, C.P.; Druzhinina, I.S. The distinguished diversity of Trichoderma is associated with rhizosphere of Coffea arabica in highland forests of Ethiopia. Diversity 2010, 2, 527–549. [Google Scholar] [CrossRef]
- Mysore, V.T.; Basavanna, M.; Monnanda, S.N.; Harishchandra, S.P.; Kukkundoor, R.K.; Ven, S.; Hunthrike, S.S. Endophytic fungal assemblages from inner and twig of Terminalia arjuna W. and A. (Combretaceae). World J. Microbiol. Biotechnol. 2005, 21, 1535–1540. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Kullnig-Gradinger, C.M.; Szakacs, G.; Kubicek, C.P. Phylogeny and evolution of the fungal genus Trichoderma—a multigene approach. Mycol. Res. 2002, 106, 757–767. [Google Scholar] [CrossRef]
- Chaverri, P.; Castlebury, L.A.; Samuels, G.J.; Geiser, D. Multilocus phylogenetic structure within the Trichoderma/Hypocrea lixii complex. Mol. Phylogenet. Evol. 2003, 27, 302–313. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Komoń-Zelazowska, M.; Kredics, L.; Hatvani, L.; Antal, Z.; Belayneh, T.; Kubicek, C.P. Alternative reproductive strategies of Hypocrea orientalis and genetically close but clonal Trichoderma longibrachiatum, both capable of causing invasive mycoses of humans. Microbiology 2008, 154, 3447–3459. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar]
- Druzhinina, S.I.; Kopchinskiy, G.A.; Komon, M.; Bissett, J.; Szakacs, G.; Kubicek, P.C. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet. Biol. 2005, 42, 813–828. [Google Scholar] [CrossRef]
- Kopchinskiy, A.; Komon, M.; Kubicek, C.P.; Druzhinina, I.S. TRICHOBLAST: A multilocus database for Trichoderma and Hypocrea identifications. Mycol. Res. 2005, 109, 658–660. [Google Scholar] [CrossRef]
- O’Donnell, K. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 2000, 92, 919–938. [Google Scholar] [CrossRef]
- Geiser, D.M.; Jiménez-Gasco, M.; Kang, S.; Makalowska, I.; Veeraraghavan, N.; Ward, T.J.; Zhang, N.; Kuldau, G.A.; O’Donnell, K. FUSARIUM-ID v.1.0: A DNA Sequence Database for Identifying Fusarium. Eur. J. Plant Pathol. 2004, 110, 473–479. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Nicholas, K.B.; Nicholas, H.B., Jr.; Deerfield, D.W., II. GeneDoc: Analysis and Visualization of Genetic Variation, EMBNEW.NEWS 1997, 4:14. Available online: http://www.psc.edu/biomed/genedoc (accessed on 28 August 2013).
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Tavaré, S. Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences. In Some Mathematical Questions in Biology—DNA Sequence Analysis; Miura, R.M., Ed.; American Mathematical Society: Providence, RI, USA, 1986; pp. 57–86. [Google Scholar]
- Atanasova, L.; Jaklitsch, W.M.; Komon-Zelazowska, M.; Kubicek, C.P.; Druzhinina, I.S. Clonal species Trichoderma parareesei sp. nov. likely resembles the ancestor of the cellulase producer Hypocrea jecorina/T. reesei. Appl. Environ. Microbiol. 2010, 76, 7259–7267. [Google Scholar] [CrossRef]
- Leaché, A.D.; Reeder, T.W. Molecular systematics of the Eastern Fence lizard (Sceloporus undulatus): A comparison of parsimony, likelihood and Bayesian approaches. Syst. Biol. 2002, 51, 44–68. [Google Scholar] [CrossRef]
- Ortiz, A.; Orduz, S. In vitro evaluation of Trichoderma and Gliocladium antagonism against the symbiotic fungus of the leaf-cutting ant Atta cephalotes. Mycopathologia 2001, 150, 53–60. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Kubicek, C.P.; Komon-Zelazowska, M.; Mulaw, T.B.; Bissett, J. The Trichoderma harzianum demon: Complex speciation history resulting in coexistence of hypothetical biological species, recent agamospecies and numerous relict lineages. BMC Evol. Biol. 2010. [Google Scholar] [CrossRef]
- Bisset, J.; Szakacs, G.; Nolan, C.A.; Druzhinina, S.I.; Gradinger, C.M.; Kubicek, C.P. New species of Trichoderma from Asia. Can. J. Bot. 2003, 81, 570–586. [Google Scholar] [CrossRef]
- Samuels, G.J.; Ismaiel, A.; Mulaw, T.B.; Szakacs, G.; Druzhinina, I.S.; Kubicek, C.P.; Jaklitsch, W.M. The Longibrachiatum Clade of Trichoderma: A revision with new species. Fungal Divers. 2012, 55, 77–108. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Komoń-Zelazowska, M.; Ismaiel, A.; Jaklitsch, W.; Mullaw, T.; Samuels, G.J.; Kubicek, C.P. Molecular phylogeny and species delimitation in the section Longibrachiatum of Trichoderma. Fungal Genet. Biol. 2012, 49, 358–368. [Google Scholar] [CrossRef]
- Vega, F.E.; Pava-Ripoll, M.; Posada, F.; Buyer, J.S. Endophytic bacteria in Coffea arabica L. J. Basic Microbiol. 2005, 45, 371–380. [Google Scholar] [CrossRef]
- Vega, F.E.; Posada, F.; Peterson, S.W.; Gianfagna, T.J.; Chaves, F. Penicillium species endophytic in coffee plants and ochratoxin A production. Mycologia 2006, 98, 31–42. [Google Scholar] [CrossRef]
- Bailey, B.A.; Strem, M.D.; Wood, D. Trichoderma species form endophytic associations within Theobroma cacao trichomes. Mycol. Res. 2009, 113, 1365–1376. [Google Scholar] [CrossRef]
- Holmes, K.A.; Schroers, H.-J.; Thomas, S.E.; Evans, H.C.; Samuels, G.J. Taxonomy and biocontrol potential of a new species of Trichoderma from the Amazon basin of South America. Mycol. Prog. 2004, 3, 199–210. [Google Scholar] [CrossRef]
- Samuels, G.J.; Pardo-Schultheiss, R.; Hebbar, K.P.; Lumsden, R.D.; Bastos, C.N.; Costa, J.C.; Bezerra, J.L. Trichoderma stromaticum sp. nov., a parasite of the cacao witches broom pathogen. Mycol. Res. 2000, 104, 760–764. [Google Scholar] [CrossRef]
- Samuels, G.J.; Dodd, S.L.; Lu, B.-S.; Petrini, O.; Schroers, H.-J.; Druzhinina, I.S. The Trichoderma koningii aggregate species. Stud. Mycol. 2006, 56, 67–133. [Google Scholar] [CrossRef]
- Zhang, C.L.; Liu, S.P.; Lin, F.C.; Kubicek, C.P.; Druzhinina, I.S. Trichoderma taxi sp. nov., an endophytic fungus from Chinese yew Taxus mairei. FEMS Microbiol. Lett. 2007, 270, 90–96. [Google Scholar] [CrossRef]
- Masumbuko, L.I.; Bryngelsson, T.; Mneney, E.E.; Salomon, B. Genetic diversity in Tanzanian Arabica coffee using random amplified polymorphic DNA (RAPD) markers. Hereditas 2003, 139, 56–63. [Google Scholar] [CrossRef]
- Booth, C. The Genus Fusarium; Commonwealth Mycological Institute: Surrey, UK, 1971. [Google Scholar]
- Zhang, N.; O’Donnell, K.; Sutton, D.A.; Nalim, F.A.; Summerbell, R.C.; Padhye, A.A.; Geiser, D.M. Members of the Fusariuim solani species complex that cause infections in both humans and plants are common in the environment. J. Clin. Microbiol. 2006, 44, 2186–2190. [Google Scholar] [CrossRef]
- Goswami, R.S.; Dong, Y.; Punja, Z.K. Host rang and mycotoxin production by Fusarium equiseti isolates originating from Ginseng fields. Can. J. Plant. Pathol. 2008, 30, 155–160. [Google Scholar] [CrossRef]
- Bae, H.; Sicher, R.C.; Kim, M.S.; Kim, S.H.; Strem, M.D.; Melnick, R.L.; Bailey, B.A. The beneficial endophyte Trichoderma hamatum isolate DIS 219b promotes growth and delays the onset of the drought response in Theobroma cacao. J. Exp. Bot. 2009, 60, 3279–3295. [Google Scholar] [CrossRef]
- Bae, H.; Roberts, D.P.; Lim, H.S.; Strem, M.D.; Park, S.C.; Ryu, C.M.; Melnick, R.L.; Bailey, B.A. Endophytic Trichoderma isolates from tropical environments delay disease onset and induce resistance against Phytophthora capsici in hot pepper using multiple mechanisms. Mol. Plant Microbe. Interact. 2011, 24, 336–351. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).