Abstract
Fish play a critical role in nutrient cycling and organic matter flow in aquatic environments. However, little is known about the microbial diversity within the gastrointestinal tracts that may be essential in these degradation activities. Panaque nigrolineatus is a loricariid catfish found in the Neotropics that have a rare dietary strategy of consuming large amounts of woody material in its natural environment. As a consequence, the gastrointestinal (GI) tract of P. nigrolineatus is continually exposed to high levels of cellulose and other recalcitrant wood compounds and is, therefore, an attractive, uncharacterized system to study microbial community diversity. Our previous 16S rRNA gene surveys demonstrated that the GI tract microbial community includes phylotypes having the capacity to degrade cellulose and fix molecular nitrogen. In the present study we verify the presence of a resident microbial community by fluorescence microscopy and focus on the cellulose-degrading members by culture-based and 13C-labeled cellulose DNA stable-isotope probing (SIP) approaches. Analysis of GI tract communities generated from anaerobic microcrystalline cellulose enrichment cultures by 16S rRNA gene analysis revealed phylotypes sharing high sequence similarity to known cellulolytic bacteria including Clostridium, Cellulomonas, Bacteroides, Eubacterium and Aeromonas spp. Related bacteria were identified in the SIP community, which also included nitrogen-fixing Azospirillum spp. Our ability to enrich for specialized cellulose-degrading communities suggests that the P. nigrolineatus GI tract provides a favorable environment for this activity and these communities may be involved in providing assimilable carbon under challenging dietary conditions.
1. Introduction
Fish are a critical component of organic matter flow and nutrient cycling in the aquatic environment due to their trophic interactions and widespread distribution [1,2,3,4,5]. Using extinction simulations it has been estimated that fish could supply up to 75% of dissolved inorganic nitrogen in Neotropical river systems [5]. A number of dietary strategies—carnivory, insectivory, herbivory and omnivory—are employed by fish across the classes Agnatha and Gnathostomata. Each dietary regime will affect the abiotic and biotic environment and alter the resident GI tract microbial community of the fish [6,7]. It has been determined that the vertebrate GI tract of the host has a critical role in determining the diversity of the GI tract community [6]. However, the microbiome of the fish GI tract can play a number of roles in the overall health and performance of the fish, including development of the mucosal immune system [8], epithelial differentiation [9], vitamin production (e.g., B12 [10]), and competition and niche exclusion of enteric pathogens [11]. Little is known about herbivorous fish GI tract microbial communities, especially those from freshwater environments [12,13,14], which play a major role in carbon and nutrient cycling in these environments.
The Loricariidae family of Neotropical catfish consists of 80 genera with approximately 830 characterized species [15,16]. Reports of algae, detritus, vegetal matter, seed, and benthic matter consumption by loricariids are common [17,18,19] and they are generally classed as omnivorous [20]. Furthermore, consumption of wood is unique to the genus Panaque (Eigenmann and Eigenmann) (Heckel) [20] and Hypostomus cochliodon [21]. These xylivorous Loricariidae have distinctive musculature around the suckermouth and robust, fully mineralized, spoon-shaped teeth [20,22,23]; both adaptations are believed to allow the fish to adhere to and ingest submerged woody materials. Field observations and gut content analysis confirm that Panaque spp. consume allocthonous sources of carbon in the environment, e.g., terrestrial woody debris [20,24], and wild caught Panaque cf. nigrolineatus “Maranon” contained up to 75% wood in the GI tract [25]. Studies on other herbivorous fish have revealed the presence of a diverse hindgut microflora responsible for producing short chain fatty acids via fermentation, which are absorbed and metabolized by the fish [12,26]. However, comprehensive studies examining digestive enzyme activity levels, concentration of fermentative end-products, and gut transit time indicate that Panaque are detritivores and do not obtain energy directly from the digestion of wood [25,27], although the fish do swallow microbes associated with the wood and by-products generated by microbial wood degradation within the GI tract [28]. Thus, the Panaque GI tract provides an interesting environment—a vertebrate host GI tract enriched in cellulose and other recalcitrant wood compounds—offering an attractive system for discovery of new microbial species and novel enzymes.
Degradation of cellulose requires a complex system of enzymes, often working synergistically [29]. In a recent study comparing different fish species from Brazil, microbial functional diversity assessed with BIOLOG, indicated that adaption for organic matter degradation was high [30]. Cellulose degradation can occur either aerobically or anaerobically [31] and is a widely distributed activity across bacterial genera [32]. Invertebrate model systems (termites and wood-boring shipworm) are well characterized and have provided insight into unique and complex degradation pathways [33,34,35,36,37]. Although cellulose degradation activities have been detected in a wide range of microbes, it requires specialized enzymes due to the presence of β-1,4 glycosidic linkages that join the repeating glucose monomers [38]. Other biopolymers present in wood (e.g., lignin) increase cellulose recalcitrance to microbial attack [39,40]. Increasingly, enzymes involved in cellulose degradation are an area of great interest due to their role in the global carbon cycle [31], potential in biofuel production [41] and other commercial applications.
In a previous study, we examined 16S rRNA gene clone libraries created from the microbial communities associated with the foregut, midgut, hindgut, and auxiliary lobe (AL) of P. nigrolineatus [42]. Our results revealed the presence of diverse and different communities in these GI tract regions, with phylotypes having high levels of 16S rRNA gene sequence similarity to Clostridium xylanovorans and Clostridium saccharolyticum, dominating the midgut community. However, the hindgut was dominated almost exclusively by phylotypes with the highest 16S rRNA sequence similarity to the Bacteroidetes phylum. In the present study, we focused on culturable microbial communities associated with cellulose-degrading microbial species from different areas of the GI tract of P. nigrolineatus to begin understanding functional roles and activity in the GI tract. By utilizing an enrichment approach, microbial populations involved in cellulose degradation were examined by molecular cloning and DNA stable-isotope probing (DNA-SIP) techniques. Our results show that the Panaque GI tract possesses a specialized group of microorganisms that may play a role in providing assimilable carbon under challenging dietary conditions.
2. Experimental Section
2.1. Fish Maintenance and Sacrifice
P. nigrolineatus were imported from Rio Xingu, Peru, through commercial wholesale distributors (AquaScape Online, Belleville, NJ, USA). The fish were wild caught and not treated with antibiotics and ranged in size from 5.1 to 8.3 cm standard length. Upon receipt, individuals were randomly assigned to 40 L glass aquaria where they were acclimated to 29 °C aerated, filtered tap water. Fish were fed a mixed diet of hearts of palm (Euterpe precatoria) (Edward and Sons, Carpinteria, CA, USA), algae pellets (Hikari Tropical Sinking Algae Wafers, Hayward, CA, USA), and date palm wood (Phoenix dactylifera) during an acclimation period of three weeks. Following acclimation, fish were anaesthetized using 3-aminobenzoic acid ethyl ester (MS-222, Sigma Chemical Co., St. Louis, MO, USA), (25 mg/L) so that initial body mass and standard length could be recorded. Fish were immediately transferred to individual 19 L plastic jugs containing aerated tap water kept at 29 °C inside a heat-controlled room and fed only autoclaved palm wood for the following 28 days. A 30%–40% water change was performed every four days. Room lights remained off approximately 23 h per day to encourage feeding and limit algae growth as a potential uncontrolled source of food. During maintenance periods, burlap sheets were used to dim room lights and tanks were covered with black plastic. Fish were sacrificed by anesthetic overdose in MS-222 (200 mg/L), and immediately placed in an anaerobic glove bag (Cole Parmer, Vernon Hills, IL, USA) under a N2 atmosphere (zero grade, 99.998% purity) for dissection. After removing the ventral body plate, sterile ice-cold phosphate buffered saline (PBS) was added to the abdominal cavity. The intestine was separated immediately distal to the stomach, removed from the body cavity, uncoiled, and measured rapidly in cold PBS. The auxiliary lobe an organ vascularly connected to the midgut region and whose function is unknown, was separated from the intestine which was divided into three parts of equal length from anterior to posterior ends.
2.2. Fluorescence Microscopy
Following dissection on ice, GI tract tissue was immediately fixed in either Carnoy’s solution (60% absolute ethanol, 30% chloroform, 10% acetic acid) for 2 h at 4 °C or 4% phosphate buffered formalin (pH 7.0) for 2 h at room temperature. The Carnoy’s fixed samples were placed directly in cold 100% ethanol and stored at −20 °C for later processing. Formalin-fixed tissues were washed twice with cold PBS for 15 min. followed by dehydration in a graded ethanol series. Tissue samples were equilibrated in a 30% sucrose solution overnight at 4 °C before being embedded in Tissue-Tek O.C.T. Compound (Torrance, CA, USA). Samples were immediately frozen in a cold ethanol bath and stored at −20 °C until sectioning. Tissues were serially sectioned at 10 µm at −25 °C using a Sakura Tissue-Tek Cryo3 (Torrance, CA, USA). Sections were immobilized on Superfrost Plus slides (VWR International, Radnor, PA, USA) and stored at 4 °C until staining. Tissues were then stained with 1× SYBR green for 2 min. in the dark and rinsed once with deionized water. Sections were imaged using a Zeiss Axioskop microscope using a BioRad Radiance 2100 Laser Scanning System at 488 nm.
2.3. Enrichment Cultures
Anaerobic enrichment cultures were established for all three regions of the GI tract as well as the tank water and wood. The media was prepared with microcrystalline cellulose as the sole carbon source (Acros Organics, Fairlawn, NJ, USA) and included 15 mg resazurin, 750 mg sodium thioglycolate, 15 g microcrystalline cellulose, 1.0 mL 2× vitamin solution (DSM Medium 141), 150 mL 5× M9 salt solution (64 g Na2HPO4, 15 g KH2HPO4, 2.5 g NaCl, 5.0 g NH4Cl per 1.0 L), pH 7.4, made up to 750 mL with dH2O, anaerobically as described previously [43]. Cultures were generated in duplicate by placing half of an intact GI tract region (fore-, mid-, and hindgut) into a 20 mL Hungate tube with 10 ml of anaerobic media. Cultures were also generated from the tank water and wood used to feed the fish. For the tank water cultures, 500 µL of water was injected directly into sealed Hungate tubes. Wood samples were prepared by cutting several small (~0.5 cm3) sections from the wood’s surface using a sterile scalpel. Approximately 10 sections were used to inoculate each enrichment culture. Tubes were sealed and cultures were incubated at 22 °C on a rocking platform. After 5 days of incubation, 250 µL of culture were removed and used to inoculate 10 mL of fresh media. Cultures were incubated for an additional 5 days at 22 °C on a rocking platform prior to DNA extraction.
2.4. Extraction of Microbial DNA
Total DNA was extracted from all second-passage enrichment cultures. Briefly, cultures were vortexed vigorously and inverted for one minute to allow insoluble cellulose to settle. Using a syringe, 1.0 mL of supernatant was removed and DNA was extracted using the Wizard genomic DNA purification kit (Promega Corp., Madison, WI) according to the manufacturer’s instructions.
2.5. DNA Sequencing and Analysis
Microbial community 16S rRNA genes were amplified using universal bacterial primers 27F.1 (5'-AGAGTTTGATCMTGGCTCAG-3') [44] and 1392R (5'-ACGGGCGGTGTGTAC-3') [45]. PCR amplification was performed using a BioRad s1000 thermal cycler with Qiagen Taq PCR master mix (Germantown, MD) and the following parameters: initial denaturation step of 3 min. at 94 °C followed by 30 cycles of denaturation for 1 min at 94 °C, annealing for 1.5 min at 58 °C, elongation for 1.5 min at 72 °C, followed by a final elongation for 10 min at 72 °C. The size and yield of PCR products were verified by gel electrophoresis. Amplified 16S rRNA genes were ligated into pCR 2.1 vector (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and ligation products were used to transform One Shot Top 10 chemically competent cells (Invitrogen, Carlsbad, CA, USA). Prior to sequencing, clones were cultured in 200 μL of a phosphate buffered terrific broth (TB) (180 μL TB; 20 μL KH2PO4 [0.17 M] + K2HPO4 [0.72 M]) in medium-binding microtiter plates at 37 °C for 19 h at 300 RPM. Plasmid preparation and sequencing was performed by the Institute of Marine and Environmental Technology BioAnalytical Services Laboratory. Sequences of the partial 16S rRNA gene of representative clones were submitted to GenBank [48]; accession numbers for enrichment library clones are KC000008-KC000102. Recovered sequences were trimmed automatically and vector sequence was removed using the NCBI VecScreen program. Trimmed sequences were identified using the NCBI Basic Local Alignment Search Tool optimized for highly similar sequences (megablast). Uncultured and environmental samples were excluded from the results. The trimmed sequences were also imported into Mega 5 and automatically aligned pairwise using MUSCLE followed by manual adjustment. Phylogenies were constructed by the neighbor joining method using the Kimura 2-parameter distance model with 10,000 bootstrap resamplings. All default parameters were used.
2.6. DNA Stable-Isotope Probing (SIP)
Enrichment media were prepared using a minimal salts media with cellobiose as the sole carbon source (Pfanstiehl Laboratories, Inc., Waukegan, IL, USA) as previously described [46]. Tissue samples (100 mg) were placed into a sterilized mortar and pestle and ground with 1 ml sterile PBS and subsequently serially diluted. Media were inoculated with GI tract samples (three replicates) and incubated in the dark at 29 °C for 30 days. Aerobic cellobiose enrichment cultures derived from each fish GI tract section were subjected to SIP as described by Neufeld et al. [47]. Briefly, samples were prepared by adding 0.65 g 13C-labeled cellulose from maize (Sigma, St. Louis, MO, USA) to 150 mL of minimal media (800 mL distilled H2O, 200 mL 5× M9 salts solution, pH 7.4) and inoculated with 1.0 mL of enrichment culture in triplicate for each GI tract section. Control cultures containing unlabeled cellulose were prepared and treated in the same manner. Samples were placed in serum vials, sealed and incubated for 36 days at 28 °C. DNA was extracted directly from these enrichments using bead-beating disruption followed by phenol-chloroform extraction. Separation was performed using a CsCl density gradient with a VTi80 rotor (Beckman Coulter, Brea, CA, USA) at 140,000 × g and 20 °C for 72 h. After centrifugation, fractions across the gradient were collected and DNA was processed by PCR, cloning, and sequencing as described above. Sequences of the partial 16S rRNA gene of representative clones were submitted to GenBank [48] with accession numbers JN169765-JN169780.
3. Results and Discussion
Wood-eating Panaque spp. provide an attractive system to examine microbial communities associated with a high cellulose-enriched vertebrate GI tract. Fluorescence microscopy of non-aqueous Carnoy’s-fixed tissue stained with DAPI (Figure 1) revealed a distinct mucus layer adhering to the intestinal wall that was extensively colonized by microbial bacillus and coccoidal cells. This mucus layer was most prevalent in the foregut, but also extended into the mid- and hindgut regions. This structure was often seen attached to the epithelial lining, but occasionally extended into the lumen contacting the digesta, which is likely a result of the mucus pulling away from the intestinal wall during tissue dehydration. Attempts to visualize a mucus-associated community in 4% paraformaldehyde-fixed tissues were unsuccessful, which may be explained by the inability of formaldehyde-based fixatives to preserve mucus [49]. Non-fixed mucus is easily destroyed by the mechanical forces of the wash steps. These findings may explain the discrepancy between results reported here and those of German (2009) who examined the GI tract of Panaque sp. using scanning electron microscopy of 4% paraformaldehyde-fixed tissues and reported the absence of any bacterial cell conglomerations. Although qualitative, the highest densities of microorganisms could be observed in the mucus membrane; however, bacteria could also be detected attached to wood particles.
Our previous study describing the microbial diversity of the GI tract using a molecular approach detected a population of putative cellulose degrading bacteria [42]. To examine this population further and gain a better understanding of activity potential, cultivation-based methods were applied in the present study. Enrichment cultures were established to gain an understanding of the bacterial species capable of cellulolytic degradation present within the GI tract. Minimal media, containing cellulose as the sole carbon source, were inoculated using different sections of GI tract, tank water, or wood under anaerobic conditions. From these cellulose enrichment cultures, 16S rRNA gene clone libraries were generated and representative clones of prevalent OTU types were examined. The closest phylogenetic relatives for each OTU type are shown in Table 1.
The microbial communities obtained from the enrichment cultures were dominated by OTUs with sequence similarity to known xylan, hemi-cellulose, cellobiose and cellulose degraders, such as Clostridium hathewayi, C. intestinale, C. xylanolyticum, Cellulomonas chitinilytica, Bacteroides xylanolyticus, Anaerosporobacter mobilis, Enterobacter soli, Acinetobacter junii, Paenibacillus illinoisensis, Paenibacillus turicensis and Bacillus circulans (Table 1). In our previous 16S rRNA analysis, the cellulolytic community was dominated by members of the Clostridiales and Bacteroidetes group. This current study has expanded this community to include several species of gammaproteobacteria and Bacilliales as well as Actinomycetales. C. chitinilytica, C. xylanolyticum, A. mobilis, and E. soli have been identified in highly cellulose-enriched environments including wood chip piles [50] forest soils [51,52], cattle waste [53] and showed high levels of activity against several plant structural polysaccharides [53,54]. Although not cellulolytic, an additional phylotype, Enterobacter ludwigii, was identified in the midgut. This microorganism was previously isolated from the rumen of goats and found to produce tannases, which may increase the nutritional value of plant digesta [55].
Several OTUs identified in the enrichment cultures displayed similarities to bacteria not known to be cellulolytic, based on the physiology of related cultured specimens. These included P. acnes (KC000009 and KC000071), D. nishinomiyanensis (KC000084), P. koreensis (KC000087), and several species of γ Proteobacteria. These species were previously isolated predominantly from municipal wastewater bioreactors and clinical samples [56,57,58]. It is unclear what role these microorganisms contribute to fish health or nutrition, and if so, whether it is through either direct degradation of plant polysaccharides, providing vitamins, or through metabolic cross-feeding.
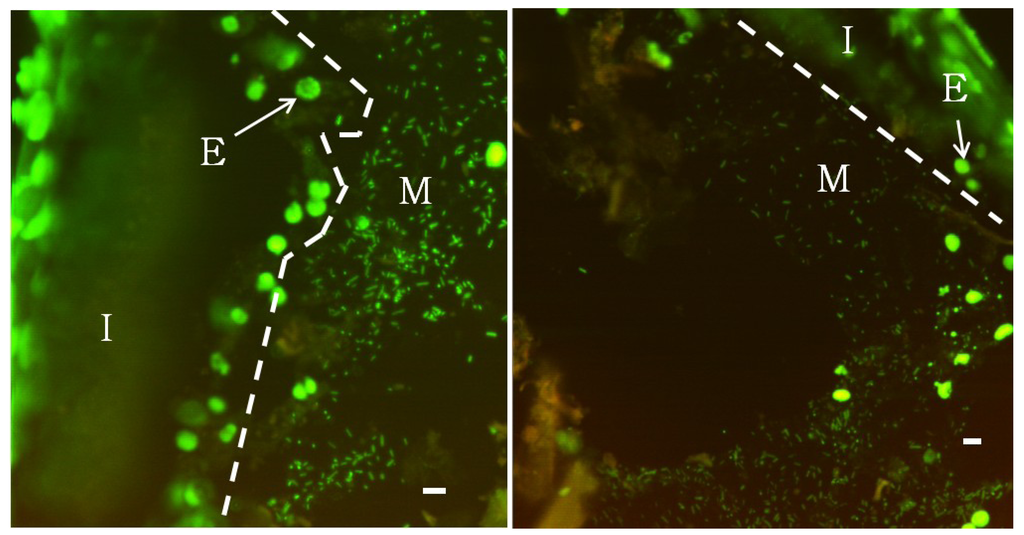
Figure 1.
Epifluorescence microscopy of SYBR Green stained semi-thin serially sectioned foregut tissue.

Table 1.
Sequence similarity of the 16S rRNA gene from isolates obtained from cellulose enrichment cultures inoculated with different GI tract regions from P. nigrolineatus.
| Class | Phylotype Detected (Source*) | Closest GenBank Species (Accession No.) | % Sequence Similarity |
|---|---|---|---|
| Actinobacteria | |||
| KC000009 (FG) | Propionibacterium acnes (GU814270) | ||
| KC000071 (W) | “ | ||
| KC000084 (WD) | Dermacoccus nishinomiyaensis (NR044872) | 99 | |
| KC000082 (W) | Cellulomonas chitinilytica (JQ659654) | 99 | |
| KC000097 (WD) | “ | ||
| Bacilli | |||
| KC000067 (W) | Paenibacillus illinoisensis (D85397) | 99 | |
| KC000081 (W) | Paenibacillus turicensis (JN378529) | 97 | |
| KC000085 (WD) | Bacillus circulans (AB680477) | 99 | |
| Bacteroides | |||
| KC000017 (FG) | Bacteroides xylanolyticus (FR850058) | 99 | |
| KC000040 (MG) | “ | ||
| Clostridia | |||
| KC000010 (FG) | Eubacterium contortum (EU980608) | 99 | |
| KC000011(FG) | Clostridium xylanolyticum (AB601097) | 99 | |
| KC000027 (FG) | Clostridium intestinale (AM158323) | 99 | |
| KC000090 (WD) | “ | ||
| KC000058 (HG) | Clostridium hathewayi (AY552788) | 99 | |
| KC000018 (FG) | Anaerosporobacter mobilis (AY534872) | 99 | |
| KC000038 (MG) | “ | ||
| KC000049 (HG) | “ | ||
| Flavobacteria | |||
| KC000048 (HG) | Elizabethkingia anophelis (EF426430) | 99 | |
| KC000079 (W) | Cloacibacterium normanense (FJ544401) | 99 | |
| α Proteobacteria | |||
| KC000087 (WD) | Pleomorphomonas koreensis (AB127971) | 99 | |
| γ Proteobacteria | |||
| KC000008 (FG), | Enterobacter soli (GU814270) | 99 | |
| KC000070 (W), | “ | ||
| KC000028 (MG) | “ | ||
| KC000030 (MG) | Enterobacter ludwigii (JN644550) | 99 | |
| KC000013 (FG) | Aeromonas allosaccharophila (GU205192) | 98 | |
| KC000016 (FG), | Aeromonas enteropelogenes (FJ940838) | 99 | |
| KC000031 (MG) | “ | ||
| KC000064 (HG) | “ | ||
| KC000068 (W) | “ | ||
| KC000032 (MG) | Aeromonas sharmana (NR_043470) | 99 | |
| KC000073 (W) | “ | ||
| KC000035 (MG) | Shewanella putrefaciens (AJ000213) | 99 | |
| KC000051 (HG) | Acinetobacter junii (JN644576) | 99 | |
| KC000098 (WD) | Moraxella osloensis (AB643595) | 99 |
*FG—foregut; MG—midgut; HG—hindgut; wood—WD, or system water—W.
Expanding our BLAST analyses to include environmental and uncultured samples we found many sequences having high similarity (>99% identity) to those recovered from cellulose-rich environments. These included phylotypes similar to Clostridium intestinale (KC000027 and KC000090), Eubacterium contortum (KC000010), and Cloacibacterium normanse (KC000079), which were identified in corn-stover bioreactors [59], cow feces cellulose enrichment cultures (Accession No. GQ920790), and termite guts (Accession No. GQ502503), respectively. Since cellulose was the only carbon source available in the enrichment culture, these OTUs might represent previously unknown cellulolytic species or arose due to extensive metabolic cross-feeding by the cellulolytic population.
The specialized cellulose-enriched community was obtained in direct response to the nature of the selective conditions imposed—cellulose was the only carbon source present and, as such, reflects a subset of the GI tract microbial community. While we note that enrichment cultures are not a reliable indicator of in situ community diversity due to the shift in population numbers, they do provide a basis for isolating new species that may provide novel pathways for cellulose degradation after further study [31]. Consistent with our previous study [42], cultivation-based analysis resulted in the identification of distinct microbial communities in each of the tissue regions with the tank water sharing many phylotypes with foregut and midgut regions. Interestingly, OTUs obtained from the wood samples were largely missing from the GI tract, indicating that the wood is likely not the source of cellulolytic microorganisms that colonize the intestines as previously suggested [25]. This difference further suggests that the intestinal microbial community is stably maintained by the fish and spatially structured, as may be seen for the hindgut enrichment, which shares no phylotypes with the wood and only a single phylotype with the tank water, Aeromonas enteropelogenes, which was ubiquitous in all tissue regions (Table 1).
In the DNA-SIP procedure, cultures grown in the presence of 13C-labeled cellulose contained a DNA band after ultracentrifugation that was distinct from the unlabeled control culture (not shown); 16S rRNA gene libraries constructed from 13C-labeled bands for foregut, midgut and hindgut enrichments were compared to GenBank databases (Table 2). A number of DNA-SIP phylotypes were similar to enrichment culture clones, e.g., Clostridium spp., but also contained phylotypes closely related to other known cellulose degraders—Cellulosimicrobium cellulans, Pseudoxanthomonas mexicana, and Bosea thiooxidans as well as nitrogen-fixing Azospirillum spp. (not detected in the enrichment cultures). The presence of phylotypes with similar phylogeny to species having the capacity for nitrogen-fixation and cellulose-degradation is not unexpected given the very low nitrogen content associated with a wood diet and consistent with our finding of nitrogenase production in GI tract samples (McDonald, Watts and Schreier, manuscript in preparation).
Unique to the foregut, phylotype SIP1A was found to be closely related to Flavobacterium aquatile, and cellulose degrading Flavobacterium spp. have been previously described [60]. Phylotypes SIP9C and SIP8D appeared to form a monophyletic clade with A. brasilense (Figure 2) and are consistent with the presence of a nitrogen-fixing GI tract community. Interestingly, SIP9D is closely related to Cellulosimicrobium cellulans, which has been identified as part of a beneficial association with A. brasilense [61] promoting degradation of wheat straw. While any relationship between phylotypes SIP9C and SIP8D with SIP9D in the GI tract is unknown, their presence together is intriguing and is consistent with other cellulose degrading systems. Phylotype SIP11D was closely related to cellobiose-degrading C. intestinale an organism capable of growing on cellobiose and producing H2 and ethanol as end products and phylotype SIP3C was most closely related to Sporomosa aerivorans. Trophic interactions between these two microorganisms have been reported previously [62]. Under anoxic conditions a syntrophic relationship appears to exist between the butyrate fermenting C. intestinale producing H2, which is subsequently utilized by an acetate-producing Sporumosa spp. Whether similar interaction between these or related bacteria occurs in the GI tract of Panaque is presently unknown. However, in two species of Panaque, and Hypostomus pyrineusi SCFA concentrations are significantly lower than measured in any fish species for which SCFA assimilation appears to be important for the hosts energy metabolism [27].

Table 2.
Sequence similarity of OTUs detected in 16S rRNA gene clone libraries generated from DNA-stable isoptope probing (SIP) using enrichments inoculated with different GI tract regions from P. nigrolineatus.
| Class | Phylotype Detected (Accession No.) and Source* | Closest GenBank Species (Accession No.) | % Sequence Similarity |
|---|---|---|---|
| Actinobacteria | |||
| SIP9D (JN169776) HG | Cellulosimicrobium cellulans (AB116667) | 99 | |
| SIP9G (JN169780) MG | Curtobacterium flaccumfaciens (EU977762) | 99 | |
| Bacilli | |||
| SIP9A (JN169768) FG | Bacillus cereus strain (HM068888) | 99 | |
| Bacteroides | |||
| SIP1A (JN169765) FG | Flavobacterium aquatile (AM230485) | 95 | |
| Clostridia | |||
| SIP4D (JN169773) MG, HG | Clostridium saccharolyticum (FJ957875) | 97 | |
| SIP11D (JN169778) HG | Clostridium intestinale (AY781385) | 99 | |
| Negativicutes | |||
| SIP3C (JN169769) MG | Sporomusa aerivorans TMAO3 (NR028991) | 96 | |
| α Proteobacteria | |||
| SIP9C (JN169771) MG, HG | Azospirillum brasilense GR2(FR667907) | 99 | |
| SIP8D (JN169775) HG | Azospirillum brasilense (AB480699) | 97 | |
| SIP12D (JN169779) HG | Bosea thiooxidans (AJ250797) | 99 | |
| β Proteobacteria | |||
| SIP7C (JN169770) MG | Achromobacter denitrificans (FJ810080) | 99 | |
| γ Proteobacteria | |||
| SIP3A (JN169766) FG, HG | Pseudoxanthomonas Mexicana (AB375392) | 99 | |
*FG—foregut; MG—midgut; HG—hindgut.
The DNA-SIP experiment has enabled the identification of putative cellulose-degrading microorganisms present in enrichment cultures and may indicate possible functional roles within the fish GI tract. However, we point out that a number of limitations with this technique have been well documented, such as cross feeding, and the dependence on initial substrate concentration and incubation time [47,63]. In addition, use of the 16S rRNA gene as an indicator of function is often inappropriate [64,65] and further analysis is underway to better understand the genetic pathways involved in the cellulose degradation.
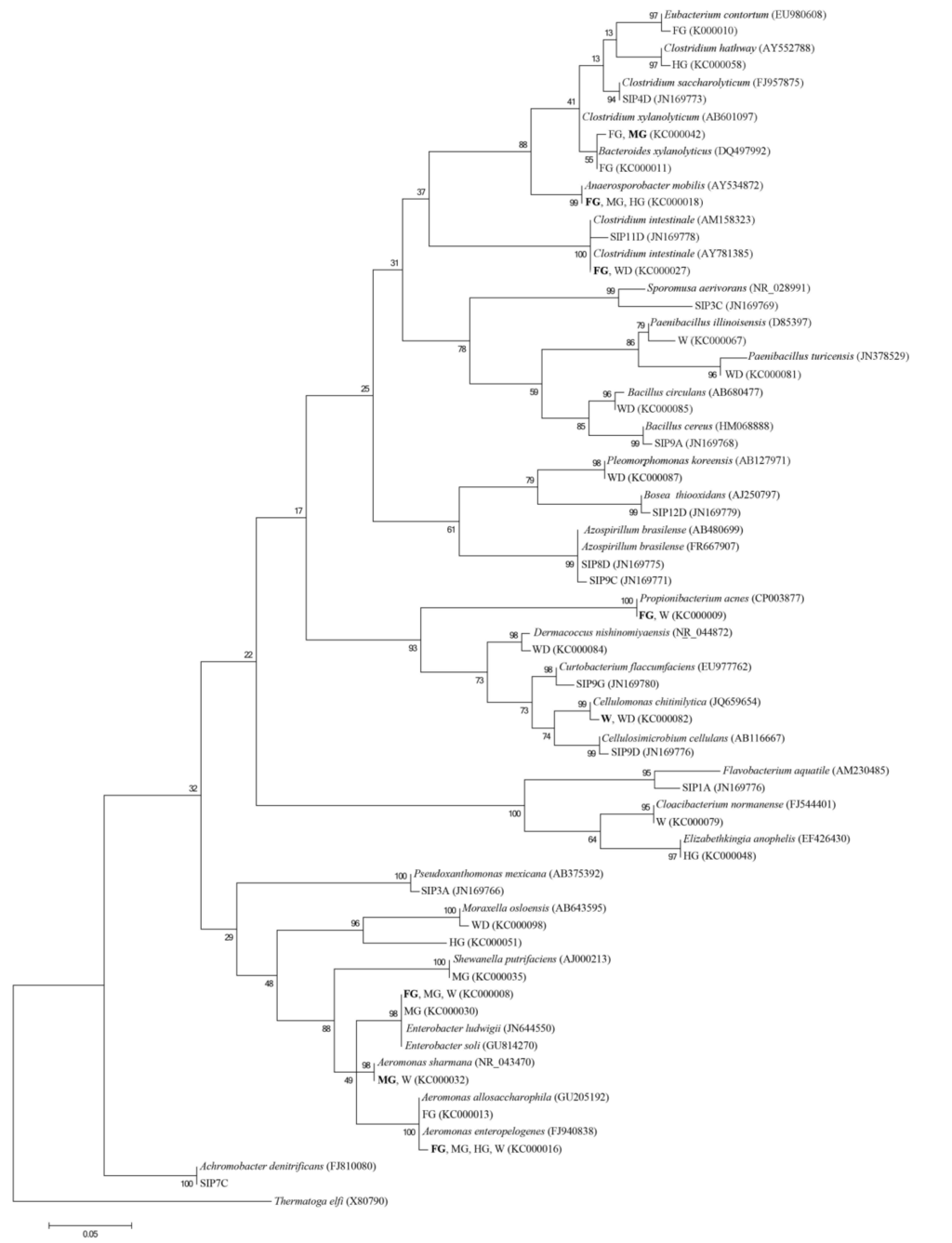
Figure 2.
Phylogenetic tree showing the relationship of 16S rRNA gene sequences (>1.2 kb) identified in the cellulose enrichment culture and DNA-SIP clone library.
Although different growth conditions were used for enrichments—cellulose and anaerobic compared to prior aerobic and cellobiose initial selection for the DNA-SIP experiments —the resulting diversity detected was similar at the genus level. However, due to the nutrient and enrichment limitations, diversity detected using these techniques was considerably reduced compared to those obtained from 16S rRNA clone libraries [42]. When examining the phylogeny of the detected OTUs it is clear that a number of key families appear to be involved in cellulose degradation in these cultures; the Clostridiales, α, γ Proteobacteria, Bacilliales, Bacteroidetes, Flavobacteria and Actinobacteria (Figure 2). However, the DNA-SIP procedure also exclusively detected β Proteobacteria-SIP7C was most closely related to Achromobacter denitrificans (Figure 2)—which was not detected in the cellulose enrichment cultures and may be a consequence of the different enrichment conditions or inocula. Diversity differences at the species level (Figure 2) are likely due to different ecotypes being present in individual fish. Since different fish were used for the inocula it is likely that multiple factors are involved in the colonization of fish GI tracts including their habitat or diet prior to being raised in the lab.
The collection, transport, and maintenance of these fish in laboratory aquaria likely play a significant role in affecting the composition of the cellulolytic microbial communities compared to wild animals. However, the detection of phylotypes having similarities to known cellulolytic bacteria in different regions of the Panaque GI tract strongly supports a view that the intestinal habitat favors a core microbiome that is capable of cellulose utilization as we have noted previously [42]. A similar study in zebrafish demonstrated that while differences could be detected in the microbial communities within wild vs. laboratory-reared fish, there was a shared core gut microbiota that was not affected by domestication [66].
4. Conclusions
Panaque spp. possess a number of morphological adaptations that could support the ability to obtain dietary supplements from a resident microbial community located in the mucus layer along the GI tract surface. These adaptions include spoon-like teeth for eating and scraping wood [22] and a long GI tract providing a high surface area for microbial colonization. Our results suggest that the P. nigrolineatus GI tract contains a microbial community that has the capacity to degrade cellulose and are likely to be involved in the breakdown of cellulolytic substrates. The fish may obtain energy by digesting this microbial population, a strategy that is consistent with recent studies using isotope tracking, which found that wood-eating loricariids are assimilating microbially-derived carbon [28]. Our study provides the first images and culture based studies providing compelling evidence for a specialized microbial community involved in cellulose digestion within the P. nigrolineatus GI tract and is the basis for ongoing studies focusing upon the molecular analysis of microbial cellulose-degrading population present in this interesting wood-consuming vertebrate.
Acknowledgments
This work was supported, in part, by the National Science Foundation, award 0801830 (J.E.M.W. and H.J.S.) and Marie Curie FP7-RG 276948 (J.E.M.W). The assistance of Damian Smoot (NSF-REU 0552654), Ashley Kurzweil, Rebecca Kulp (NSF-REU 0552654), Moses Demehin (NSF-REU 0552654), Mickey Dehn and Fan Zhang with various portions of the laboratory work was greatly appreciated. We also thank Jay Nelson for assistance with fish maintenance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Power, M.E. Effects of fish in river food webs. Science 1990, 250, 811–814. [Google Scholar]
- Schindler, D.E.; Carpenter, S.R.; Cole, J.J.; Kitchell, J.F.; Pace, M.L. Influence of food web structure on carbon exchange between lakes and the atmosphere. Science 1997, 277, 248–251. [Google Scholar] [CrossRef]
- Vanni, M.J. Nutrient cycling by animals in freshwater ecosystems. Annu. Rev. Ecol. Syst. 2002, 33, 341–370. [Google Scholar] [CrossRef]
- Taylor, B.W.; Flecker, A.S.; Hall, R.O. Loss of a harvested fish species disrupts carbon flow in a diverse tropical river. Science 2006, 313, 833–836. [Google Scholar] [CrossRef]
- McIntyre, P.B.; Jones, L.E.; Flecker, A.S.; Vanni, M.J. Fish extinctions alter nutrient recycling in tropical freshwaters. Proc. Natl. Acad. Sci. USA 2007, 104, 4461–4466. [Google Scholar] [CrossRef]
- Rawls, J.F.; Mahowald, M.A.; Ley, R.E.; Gordon, J.I. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 2006, 127, 423–433. [Google Scholar] [CrossRef]
- Sullam, K.E.; Essinger, S.D.; Lozupone, C.A.; O’Connor, M.P.; Rosen, G.L.; Knight, R.O.; Kilham, S.S.; Russell, J.A. Environmental and ecological factors that shape the gut bacterial communities of fish: A meta-analysis. Mol. Ecol. 2012, 21, 3363–3378. [Google Scholar] [CrossRef]
- Rawls, J.F.; Samuel, B.S.; Gordon, J.I. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. USA 2004, 101, 4596–4601. [Google Scholar] [CrossRef]
- Bates, J.M.; Mittge, E.; Kuhlman, J.; Baden, K.N.; Cheesman, S.E.; Guillemin, K. Distinct signals fromthe microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 2006, 297, 374–386. [Google Scholar] [CrossRef]
- Sugita, H.; Miyajima, C.; Deguchi, Y. Vitamin B12 producing ability of the intestinal microfora of fresh-water fish. Aquaculture 1991, 92, 267–276. [Google Scholar] [CrossRef]
- Sekirov, I.; Finlay, B.B. The role of the intestinal microbiota in enteric infection. J. Physiol. 2009, 587, 4159–4167. [Google Scholar] [CrossRef]
- Choat, J.H.; Clements, K.D. Vertebrate herbivores in marine and terrestrial environments: A nutritional ecology perspective. Annu. Rev. Ecol. Syst. 1998, 29, 375–403. [Google Scholar] [CrossRef]
- Choat, J.H.; Clements, K.D.; Robbins, W.D. The trophic status of herbivorous fishes on coral reefs 1: Dietary analyses. Mar. Biol. 2002, 140, 613–623. [Google Scholar] [CrossRef]
- Choat, J.H.; Robbins, W.D.; Clements, K.D. The trophic status of herbivorous fishes on coral reefs II: Food processing modes and trophodynamics. Mar. Biol. 2004, 145, 445–454. [Google Scholar]
- Armbruster, J.W. Phylogenetic relationships of the suckermouth armoured catfishes (Loricariidae) with emphasis on the Hypostominae and the Ancistrinae. Zool. J. Linn. Soc. 2004, 141, 1–80. [Google Scholar] [CrossRef]
- Lujan, N.K.; Winemiller, K.O.; Armbruster, J.W. Trophic diversity in the evolution and community assembly of loricariid catfishes. BMC Evol. Biol. 2012, 26, 124–136. [Google Scholar]
- Saul, W.G. An ecological study of fishes at a site in upper amazonian ecuador. Proc. Acad. Nat. Sci. USA 1975, 127, 93–114. [Google Scholar]
- Delariva, R.L.; Agostinho, A.A. Relationship between morphology and diets of six neotropical loricariids. J. Fish Biol. 2001, 58, 832–847. [Google Scholar] [CrossRef]
- Salvador, L.F., Jr.; Salvador, G.N.; Santos, G.B. Morphology of the digestive tract and feeding habits of Loricaria lentiginosa Isbrücker, 1979 in a Brazilian reservoir. Acta Zool. 2009, 90, 101–109. [Google Scholar] [CrossRef]
- Schaefer, S.A.; Stewart, D.J. Systematics of Panaque dentex species group (Siluriformes, Loricariidae) wood-eating armored catfishes from tropical South America. Ichthyol. Explor. Freshw. 1993, 4, 309–342. [Google Scholar]
- Armbruster, J.W. The Species of the Hypostomus Cochliodon Group (Siluriformes: Loricariidae). Zootaxa 2003, 249, 1–60. [Google Scholar]
- Geerinckx, T.; de Poorter, J.; Adriaens, D. Morphology and development of teeth and epidermal brushes in loricariid catfishes. J. Morphol. 2007, 268, 805–814. [Google Scholar] [CrossRef]
- Lujan, N.K.; Armbruster, J.W. Morphological and functional diversity of the mandible in suckermouth armored catfishes (Siluriformes: Loricariidae). J. Morphol. 2012, 273, 24–39. [Google Scholar] [CrossRef]
- Nonogaki, H.; Nelson, J.A.; Patterson, W.P. Dietary histories of herbivorous loricariid catfishes: evidence from δ13C values of otoliths. Environ. Biol. Fishes 2007, 78, 13–21. [Google Scholar] [CrossRef]
- German, D. Inside the guts of wood-eating catfishes: Can they digest wood? J. Comp. Physiol. B 2009, 179, 1011–1023. [Google Scholar] [CrossRef]
- Clements, K.D.; Gleeson, V.P.; Slaytor, M. Short-chain fatty acid metabolism in temperate marine herbivorous fish. J. Comp. Physiol. B 1994, 164, 372–377. [Google Scholar] [CrossRef]
- German, D.; Bittong, R. Digestive enzyme activities and gastrointestinal fermentation in wood-eating catfishes. J. Comp. Physiol. B 2009, 179, 1025–1042. [Google Scholar] [CrossRef]
- Lujan, N.K.; German, D.P.; Winemiller, K.O. Do wood-grazing fishes partition their niche?: morphological and isotopic evidence for trophic segregation in Neotropical Loricariidae. Funct. Ecol. 2011, 28, 1327–1338. [Google Scholar]
- Lynd, L.R.; Weimer, P.J.; van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef]
- Mouchet, M.A.; Bouvier, C.; Bouvier, T.; Troussellier, M.; Escalas, A.; Mouillot, D. Genetic difference but functional similarity among fish gut bacterial communities through molecular and biochemical fingerprints. FEMS Microbiol. Ecol. 2012, 79, 568–580. [Google Scholar] [CrossRef]
- Leschine, S.B. Cellulose degradation in anaerobic environments. Ann. Rev. Microbiol. 1995, 8, 237–299. [Google Scholar]
- Wilson, D.B. Microbial diversity of cellulose hydrolysis. Curr. Opin. Microbiol. 2011, 14, 259–263. [Google Scholar]
- Breznak, J.A.; Brune, A. Role of microorganisms in the digestion of lignocellulose by termites. Annu. Rev. Entomol. 1994, 39, 453–487. [Google Scholar] [CrossRef]
- Cleveland, L.R. Symbiosis between termites and their intestinal protozoa. Proc. Natl. Acad. Sci. USA 1923, 9, 424–428. [Google Scholar] [CrossRef]
- Distel, D.L.; Beaudoin, D.J.; Morrill, W. Coexistence of multiple proteobacterial endosymbionts in the gills of the wood-boring bivalve Lyrodus pedicellatus (Bivalvia: Teredinidae). Appl. Environ. Microbiol. 2002, 68, 6292–6299. [Google Scholar] [CrossRef]
- Pellens, R.; Grandcolas, P.; da Silva Neto, I.D. A new and independently evolved case of xylophagy and the presence of intestinal flagellates in the cockroach Parasphaeria boleiriana (Dictyoptera, Blaberidae, Zetoborinae) from the remnants of the Brazilian Atlantic forest. Can. J. Zool. 2002, 80, 350–360. [Google Scholar] [CrossRef]
- Waterbury, J.B.; Turner, R.D.; Calloway, C.B. A cellulolytic nitrogen-fixing bacterium cultured from the gland of Deshayes in shipworms (Bivalvia: Teredinidae). Science 1983, 221, 1401–1403. [Google Scholar]
- Bayer, E.A.; Chanzy, H.; Lamed, R.; Shoham, Y. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 1998, 8, 548–557. [Google Scholar] [CrossRef]
- Talbot, J.M.; Treseder, K.K. Interactions among lignin, cellulose, and nitrogen drive litter chemistry–decay relationship. Ecology 2011, 93, 345–354. [Google Scholar] [CrossRef]
- Zaldivar, J.; Nielsen, J.; Olsson, L. Ethanol fuel production from lignocellulose: A challenge for metabolic engineering and process integration. Appl. Microbiol. Biotechnol. 2001, 56, 17–34. [Google Scholar] [CrossRef]
- Ragauskas, A.J.; Williams, C.K.; Davison, B.H.; Britovsek, G.; Cairney, J.; Eckert, C.A.; Frederick, W.J.; Hallett, J.P.; Leak, D.J.; Liotta, C.L.; et al. The path forward for biofuels and biomaterials. Science 2006, 311, 484–489. [Google Scholar] [CrossRef]
- McDonald, R.; Schreier, H.J.; Watts, J.E.M. Phylogenetic Analysis of Microbial Communities in different regions of the gastrointestinal tract in Panaque nigrolineatus, a wood-eating fish. PLoS One 2012, 7, e48018. [Google Scholar] [CrossRef]
- Sowers, K.R.; Noll, K.M. Techniques for Anaerobic Growth. In Archaea: A Laboratory Manual; Robb, F.T., Sowers, K.R., DasSharma, S., Place, A.R., Schreier, H.J., Fleischmann, E.M., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1995; pp. 15–48. [Google Scholar]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Ed.; John Wiley and Sons Ltd.: Cambridge, UK, 1991; pp. 115–175. [Google Scholar]
- Ferris, M.J.; Muyzer, G.; Ward, D.M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 1996, 62, 340–346. [Google Scholar]
- Warshaw, J.E.; Leschine, S.B.; Canale-Parola, E. Anaerobic cellulolytic bacteria from wetwood of living trees. Appl. Environ. Microbiol. 1985, 50, 807–811. [Google Scholar]
- Neufeld, J.D.; Vohra, J.; Dumont, M.G.; Lueders, T.; Manefield, M.; Friedrich, M.W.; Murrell, J.C. DNA stable-isotope probing. Nat. Protocols 2007, 2, 860–866. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar]
- Swidsinski, A.; Weber, J.; Loening-Baucke, V.; Hale, L.P.; Lochs, H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 2005, 43, 3380–3389. [Google Scholar] [CrossRef]
- Rogers, G.M.; Baecker, A.A.W. Clostridium xylanolyticum sp. nov., an anaerobic xylanolytic bacterium from decayed Pinus patula wood chips. Int. J. Syst. Bacteriol. 1991, 41, 140–143. [Google Scholar]
- Jeong, H.; Lim, Y.W.; Yi, H.; Sekiguchi, Y.; Kamagata, Y.; Chun, J. Anaerosporobacter mobilis gen. nov., sp. nov., isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2007, 57, 1784–1787. [Google Scholar]
- Manter, D.; Hunter, W.; Vivanco, J. Enterobacter soli sp. nov.: A lignin-degrading γ-Proteobacteria isolated from soil. Curr. Microbiol. 2011, 62, 1044–1049. [Google Scholar]
- Yoon, M.-H.; Ten, L.N.; Im, W.-T.; Lee, S.-T. Cellulomonas chitinilytica sp. nov., a chitinolytic bacterium isolated from cattle-farm compost. Int. J. Syst. Evol. Microbiol. 2008, 58, 1878–1884. [Google Scholar]
- Kim, C.H. Characterization and substrate specificity of an endo-beta-1,4-D-glucanase I (Avicelase I) from an extracellular multienzyme complex of Bacillus circulans. Appl. Environ. Microbiol. 1995, 61, 959–965. [Google Scholar]
- Singh, B.; Bhat, T.K.; Sharma, O.P.; Kanwar, S.S.; Rahi, P.; Gulati, A. Isolation of tannase-producing Enterobacter ludwigii GRT-1 from the rumen of migratory goats. Small Rumin. Res. 2012, 102, 172–176. [Google Scholar] [CrossRef]
- Buchman, A.L.; Pickett, M.J.; Mann, L.; Ament, M.E. Central venous catheter infection caused by Moraxella osloensis in a patient receiving home parenteral nutrition. Diagn. Microbiol. Infect. Dis. 1993, 17, 163–166. [Google Scholar] [CrossRef]
- Debelian, G.J.; Olsen, I.; Tronstad, L. Profiling of Propionibacterium acnes recovered from root canal and blood during and after endodontic treatment. Endod. Dent. Traumatol. 1992, 8, 248–254. [Google Scholar] [CrossRef]
- Figueira, V.; Vaz-Moreira, I.; Silva, M.; Manaia, C.M. Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water Res. 2011, 45, 5599–5611. [Google Scholar] [CrossRef]
- Wang, X.; Feng, Y.; Wang, H.; Qu, Y.; Yu, Y.; Ren, N.; Li, N.; Wang, E.; Lee, H.; Logan, B.E. Bioaugmentation for electricity generation from corn stover biomass using microbial fuel cells. Environ. Sci. Technol. 2009, 43, 6088–6093. [Google Scholar] [CrossRef]
- Lednická, D.; Mergaert, J.; Cnockaert, M.C.; Swings, J. Isolation and identification of cellulolytic bacteria involved in the degradation of natural cellulosic fibres. Syst. Appl. Microbiol. 2000, 23, 292–299. [Google Scholar] [CrossRef]
- Halsall, D.M.; Gibson, A.H. Comparison of two cellulomonas strains and their interaction with azospirillum brasilense in degradation of wheat straw and associated nitrogen fixation. Appl. Environ. Microbiol. 1986, 51, 855–861. [Google Scholar]
- Gößner, A.S.; Küsel, K.; Schulz, D.; Trenz, S.; Acker, G.; Lovell, C.R.; Drake, H.L. Trophic interaction of the aerotolerant anaerobe Clostridium intestinale and the acetogen Sporomusa rhizae sp. nov. isolated from roots of the black needlerush Juncus roemerianus. Microbiology 2006, 152, 1209–1219. [Google Scholar] [CrossRef]
- Chen, Y.; Vohra, J.; Murrell, J.C. Applications of DNA-Stable Isotope Probing in Bioremediation Studies. In Bioremediation, Methods in Molecular Biology; Cummings, S.P., Ed.; Humana Press: New York, NY, USA, 2010; pp. 129–138. [Google Scholar]
- Leadbetter, J.R. Cultivation of recalcitrant microbes: Cells are alive, well and revealing their secrets in the 21st century laboratory. Curr. Opin. Microbiol. 2003, 6, 274–281. [Google Scholar] [CrossRef]
- Doolittle, W.F. Phylogenetic classification and the universal tree. Science 1999, 284, 2124–2128. [Google Scholar] [CrossRef]
- Roeselers, G.; Mittge, E.K.; Stephens, W.Z.; Parichy, D.M.; Cavanaugh, C.M.; Guillemin, K.; Rawls, J.F. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011, 5, 1595–1608. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).