Abstract
Molecular-assisted alpha taxonomy has recently become an effective practice in reassessing biodiversity and floristics for a variety of different organisms. This paper presents a series of examples that have been drawn from biodiversity work being carried out on the marine red algae of Bermuda. Molecular sequencing of DNA from Bermuda samples has already begun to greatly alter the makeup of the flora as it was known just decades ago, and will help set a new database for future comparison as climate change affects species composition in the islands.
1. Introduction
Molecular-assisted alpha taxonomy (MAAT) has recently emerged as an effective technique due to its ability to conquer the challenges of classifying many organisms prone to simple or convergent morphologies. Here we review its use in marine macroalgae, particularly the red algae (Rhodophyta), a group whose individuals are infamously difficult to identify due to characteristic obstacles such as heteromorphic life cycles, evolutionary convergence, and the influence of environmental factors on phenotype expression. The incorporation of molecular data to the understanding of red algal classification has fundamentally altered the way in which we understand this group. Prior to the advent of gene sequencing, taxonomic placement among red algae often could not be definitive without reproductive structures. Individuals discovered with only vegetative characteristics were often classified by their relatedness to species only at the generic level, an inexact science at best. Likewise, convergent evolution and recent speciation often produce cryptic species—impossible to differentiate at the gross morphological level, but in fact genetically distinct. Molecular analyses can now be used to reveal discrete algal ancestries obscured by seemingly identical appearances as a result of converging on a similar morphology. At times, however, the reverse is true. Many species exhibit great morphological diversity based upon the environmental conditions under which a population of marine algae grows (protected vs. exposed environments, shallow subtidal vs. deep-water habitats) and this diversity is often misinterpreted as multiple species. Thus, two fundamental questions guide molecular-assisted alpha taxonomy: Will the alleged conspecifics hold up under molecular scrutiny (Figure 1a), or are distinct entities present (Figure 1b)? Or, conversely, will the data confirm or reject apparent differences between uniquely classified individuals (Figure 1c)?
Although alpha taxonomy is generally performed in similar ways across algal groups, the molecular element of MAAT studies can vary depending on the taxa in question. In some cases, little is known about the species being examined. This is particularly true in instances where reproductive features are unknown, and in such cases, more ‘traditional’ molecular phylogenetic methods are often used. These include the use of full, or nearly full, gene or ribosomal RNA (rRNA) sequences and in-depth likelihood and Bayesian analysis. In cases where the goal is examining potentially closely-related species, the approach might be to use an established “DNA barcode”, such as the 5’ portion of the cytochrome oxidase I gene (COI-5’) encoded in the mitochondrion, the internal transcribed spacer (ITS) of the nuclear rRNA or a portion of the plastid 23S ribosomal subunit (UPA). These DNA regions may be used individually, or in combination, to determine whether there is an obvious distinction between intra- and interspecies genetic variation, but this method can be subject to sampling biases [e.g., 1]. Accordingly, the sampling strategy for barcoding studies is geared towards obtaining multiple samples of each possible species, in order to better assess species boundaries at the morphological and molecular level. Whereas none of the above methods will give an absolute identity to an alga in every instance, the molecular results can form a framework in which it is possible to re-assess recognized “taxonomically informative” characters or establish novel ones. In combination, each technique can inform the other to establish a more robust classification scheme.
Over the past decade, two of the authors (CWS, CEL) have been applying MAAT to the marine flora of Bermuda in order to better understand the algal biodiversity of this isolated and small island grouping. The Bermudas are among the northernmost islands in the world to support a tropical marine biota [2,3], including the highest latitude coral reefs in the western Atlantic [4]. Eighty-seven percent of the species living in Bermuda are known from the Caribbean Sea, and despite their small size and distance from the tropics proper, the islands of Bermuda support approximately 30% of the 1,442 species known in the tropical and subtropical western Atlantic [5], the bulk of them residing in the Caribbean proper. The “lifeline” of the biota of Bermuda is the swift-flowing Gulf Stream, which brings tropical waters from the Gulf of Mexico and Caribbean via the Florida and Antilles Currents, these originating with the Northern and Southern Equatorial Currents off northern South America [2,4,6]. Because of seasonal water temperature oscillations, the macroalgal assemblage of Bermuda is made up of warm-water tolerant species from the western mid-Atlantic that have persisted since re-colonization during the last ice age [7], cool-water tolerant Caribbean species carried by the current northward and an additional 3% that are endemic to this “postage stamp” in the Atlantic Ocean.
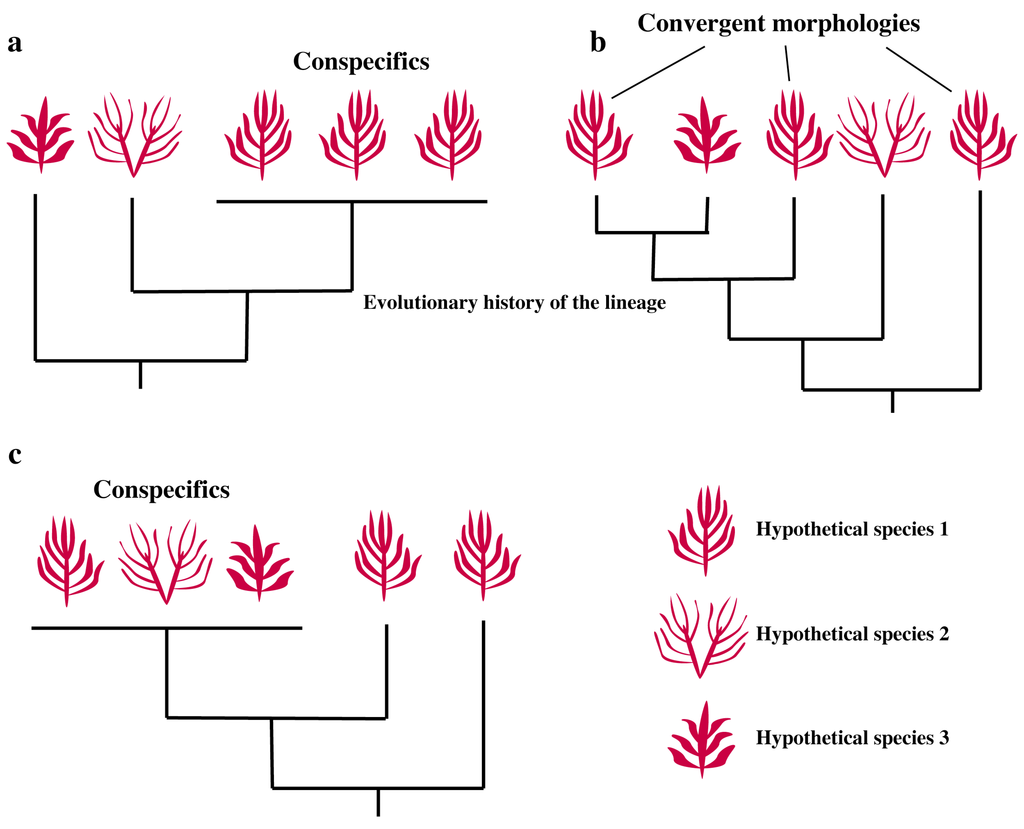
Figure 1.
Possible scenarios resulting from the application of molecular-assisted alpha taxonomy. Species of algae, classified based on morphology, are represented diagrammatically. In many cases, the traditional morphological classification reflects the molecular data (a). However, convergent evolution (b) can produce distantly related species that are morphologically similar. Molecular data has been instrumental in clarifying this type of situation, as well as instances where morphologically dissimilar specimens represent the same species (c). This occurs most commonly either when the complete life history of an organism with heteromorphic alternation of generations is unknown, or in some species that exhibit a large degree of environmentally-influenced morphological plasticity.
Bermuda is a particularly good example of how the use of MAAT can redefine a marine flora because of its location, the history of taxonomic work on the islands and the manageable number of taxa. As a small and isolated archipelago, dispersal to and from Bermuda is relatively rare, leading to an environment amenable to the evolution of new species through genetic isolation. Additionally, despite the heavy influence of the Caribbean on the marine flora of Bermuda, a major obstacle to understanding seaweed biodiversity surrounding the islands has been the North Atlantic biases of early visiting botanists. All of the early descriptions of algal species from Bermuda were reported by individuals from either New England or Europe [8] and many species were misidentified based on a superficial likeness to North Atlantic algae. Many of Bermuda’s algal species, therefore, have had to be painstakingly decoupled from the names of their northern relatives. Investigations based on morphology alone, for reasons outlined above, are often inadequate to convincingly reclassify species that cannot be found reproductive, thus making use of MAAT in this process a critical step towards understanding Bermuda’s marine biodiversity.
The first compiled list of species in the marine flora of Bermuda was “The Algae of Bermuda” in 1917 by F.S. Collins and A.B. Hervey [9]. This report included 342 species of marine algae, many with names of recognized eastern North Atlantic species. This study [9] was the last complete flora strictly dedicated to the algae of Bermuda. A year later, M.A. Howe [10] contributed the algal section to N.L. Britton’s 1918 Flora of Bermuda, but he only included the more common and more conspicuous algae occurring in the islands and again exhibited a northern bias. In the 1940s, W.R. Taylor first visited Bermuda with his student, A.J. Bernatowicz [11], and he included their data along with previous collectors in his 1960 comprehensive Marine Algae of the Eastern Tropical and Subtropical Coasts of the Americas [12]. Nearly a decade later, Taylor and Bernatowicz [13] produced an annotated list of the most common shallow water macroscopic seaweeds of the Bermudas. At that time, 50 years after the Collins and Hervey report, 40 new species of algae were added to the Bermuda flora and, within the context of the Caribbean, many Bermudian seaweed names were changed. These works brought the total marine red, green and brown species in the islands to 380.
From the time of Taylor’s work until late in the 20th century, additions to the Bermuda marine flora have been published only sporadically. However, beginning in 1997, more than 30 new species and new records to the islands were added to the flora culminating in a checklist of taxa [8]. Since then, nearly 50 more have been added [14,15,16,17,18,19,20,21], bringing the total flora to 449 species. Of this number, 258 are red algae. Several additional questionable records of European taxa reported in the first half of the 20th century have yet to be confirmed and are not included in the total. It is important to note that 66 species in the flora have their type localities (origin of the specimen used to define a species) in Bermuda, several of which are presently synonymized with other species described from distant geographic locations. Below we review the significant impacts MAAT has recently had on the understanding of Bermudian and western Atlantic seaweed diversity.
2. Convergence, Hidden Diversity and Sorting It Out
Classification and taxonomy in red algae has traditionally been based on reproductive structures. In particular, the development of female gametangia, or “carpogonia”, both before and after fertilization, and the development of tetrasporangia, when found, are extremely valuable taxonomically. The sporophytic stage in the well-known triphasic red algal life history develop sporangia that meiotically produce four tetraspores. These sporangia divide in one of three distinct division patterns typical for genera, and at times, higher taxonomic units. Characters derived from these stages of the life history are critical in differentiating the majority of red algal species, but many collections have either only male reproductive features or none at all, making morphological convergence of two species at the gross anatomical level a vexing problem.
2.1. Convergence
A red algal species recently discovered in Bermuda, Chondracanthus saundersii C.W. Schneid. et C.E. Lane (Gigartinales, Gigartinaceae), was discovered devoid of female gametangia or tetrasporangia [15]. Although male spermatangia were present in some collections, these were not helpful for generic assignment. As a genus, the taxonomy of Chondracanthus Kütz. was first revised morphologically [22], then recently using the large subunit of the chloroplast-encoded RuBisCo gene (rbcL) [23]. After rbcL sequencing of Bermuda specimens from Walsingham Pond and Harrington Sound collected at various times of the year, these collections grouped within, but were not identical to, other Chondracanthus sequences [15]. The collections of C. saundersii were found to be morphologically identical to specimens historically collected in Bermuda, and throughout much of the Caribbean, known as C. acicularis (Roth) Fredericq. However, at the molecular level the western Atlantic specimens are quite distinct from sequences produced from isolates collected in Europe, the area from which C. acicularis was originally described. Based on the morphological re-examination of material from both sides of the Atlantic, character differences were discovered. The primary morphological distinction between C. acicularis and C. saundersii is the narrower, flattened lubricous axes and less dense medulla of C. saundersii, but these characters alone would not have convinced all taxonomists of the distinction between the two species using strictly traditional practices.
Chondracanthus saundersii is also smaller and less copiously branched than another European Chondracanthus once also thought to occur in Bermuda, C. teedei (Mert. ex Roth) Kütz. [15]. Chondracanthus teedei was likewise removed from the flora of Bermuda after morphological observations of historical specimens from the islands also showed them to belong to the newly described C. saundersii. Thus, molecular work in Bermuda became the platform upon which European C. acicularis and C. teedei were questioned or removed as members of a western Atlantic flora. Collections of C. acicularis from Haiti, Cuba, and Brazil were also annotated as C. saundersii, thus affecting their floras. The Bermuda-focused study [15] did report two western Atlantic records (North Carolina, Florida) that for the present remain as C. acicularis, but even these locations should be sampled and their genetics compared against European sequences of this species in order to confirm that this species remains as a true member of both the eastern and western Atlantic floras.
2.2. Hidden Diversity
In a similar manner, Botryocladia pyriformis (Børgesen) Kylin is an example of a Caribbean red algal species name being applied to a wide range of morphologies in the Atlantic Ocean. Botryocladia bermudana C.W. Schneid. et C.E. Lane was one of the species previously known as B. pyriformis in Bermuda [17] prior to the advent of MAAT. Collections of B. pyriformis from the Canary Islands, Gulf of Mexico and the Caribbean, but not from its type locality in the West Indies, nor Bermuda, had previously been examined using MAAT. Molecular analyses had already carved out four species from specimens previously attributed to B. pyriformis: B. bahamense D.L. Ballant. et Aponte, B. canariensis Afonso-Carr. et Sobrino, B. ballantinei Gavio et Fredericq and B. caraibica Gavio et Fredericq [24,25,26,27]. Botryocladia bermudana, presently an endemic to Bermuda, recently became the sixth member of the “B. pyriformis complex,” again using MAAT [17] .This species is by far the most common of the three Botryocladia species in the islands [17], and is found at depths ranging from the intertidal to 73 m. But, since its first report in Bermuda by Collins and Hervey [9] and Howe [10], this taxon had been a cryptic species under the name of B. pyriformis. In reality, it clusters closely with newly described and sequenced B. caraibica from Atlantic Panama and Florida in a molecular phylogeny, not B. pyriformis. Morphologically, these sister species are differentiated by the greater thickness of B. bermudana vesicle medullary cells and walls, and occasional club-shaped secretory cells, although no single defining, non-overlapping feature stands alone to separate B. bermudana from B. caraibica [17]. Thus, molecular sequencing allowed the discovery of this new Botryocladia with minimal anatomical differences from the species it had previously been assigned to in Bermuda [17].
To make matters worse for traditional taxonomists, herbivores in the natural environment of Botryocladia bermudana beget mostly populations of algal individuals under 1 cm tall, creating the illusion of a second species when compared with 6.5 cm, highly branched individuals residing in a protected environments, such as a reef tank in the Bermuda National Aquarium. The Bermuda Aquarium has only ever housed anything in its tanks that has been collected locally, making it an occasional sanctuary for native marine algae that end up in tanks lacking herbivores. It appears that the parrot fish which graze B. bermudana in “wild” habitats have been an important recent force shaping the scattered B. bermudana populations, as Collins and Hervey [9] never collected its smaller “cropped” form, instead collecting luxurious large specimens in inshore environments where today they cannot be found [17]. Stunted or cropped growth has also been observed among offshore Botryocladia exquisita C.W. Schneid. et C.E. Lane. Despite over a decade of collecting Bermudian algae in all habitats, large, developed plants have only ever been collected in the reef tanks of the Bermuda Aquarium [17]. The small number of cell layers in the B. exquisita vesicle walls, which are less than 36 µm thick, and the production of both specialized and non-specialized secretory cell bearing cells in B. exquisita, distinguish it from a similar Caribbean and Gulf of Mexico Botryocladia species, B. monoica Schnetter [17]. The cropped small offshore specimens of B. exquisita could easily have been misidentified as B. monoica had molecular analysis not been performed.
Despite its relative rarity in Bermuda, a third species of Botryocladia, B. flookii C.W. Schneid. et C.E. Lane, has been described from the intertidal to 30 m. Although the gross morphology of B. flookii is virtually identical to that of B. macaronesica Afonso-Carrillo, Sobrino, Tittley et Neto of the Canary Islands, it differs from the eastern Atlantic species in the lateral attachment of its vesicles, its large secretory bearing cells and the development of the medulla into the vesicle cavity below carposporophytes [17]. Unfortunately, no molecular data exist for B. macaronesica nor was any available for DNA extraction at the time of the study [17]. While there is no single outstanding morphological feature which separates B. flookii from all other Botryocladia species, the genus Irvinea was split from Botryocladia on the basis of molecular genetics alone [28]. Since this split, confidence in the use of morphology to separate Botryocladia species has seriously diminished.
Another red alga that contained cryptic species within its binomial in a variety of locations worldwide, including Bermuda, was Asteromenia peltata (W.R. Taylor) Huisman et A. Millar [28]. This species was first described from Venezuela based solely on vegetative material [30, as Fauchea peltata W.R. Taylor]. Since that original report from the Caribbean, A. peltata has been reported as a cosmopolitan tropical species [29]. Recent molecular work [21], however, has carved up ‘A. peltata’ into five regional Asteromenia species aligned with the Hymenocladiaceae rather than the Rhodymeniaceae [31]. During this recent study [21], two species of Asteromenia were reported in Bermuda, A. peltata and A. bermudensis G.W. Saunders, C.E. Lane, C.W. Schneid. et Kraft where previously only the former was known. New samples of ‘Asteromenia peltata’ from Hawaii, Fiji, French Polynesia and other locations where it has been reported from and not sequenced in this study are now in need of molecular analysis to discover which species they actually represent, as A. peltata has only been verified using gene sequencing in the western Atlantic Ocean [21]. Once again, molecular work created a domino effect of species in need of additional clarification.
Centroceras clavulatum (C. Agardh) Mont. (Ceramiales, Ceramiaceae) has been a reported cosmopolitan alga in the biogeographical literature [3,32]. This species has been recorded from the Pacific, Atlantic and Indian Oceans and places such as Pacific South America, California, North Carolina and Bermuda to Brazil, southeast Asia, Australia, the South Seas and all of the coasts of Africa, to name a few [33]. Thus, it ranges from cool temperate to tropical waters [3]. This was the case until, once again, MAAT greatly restricted the biogeography of C. clavulatum, in this case to the Pacific Ocean [32]. Molecular phylogenetic trees illustrating the relationships between various Centroceras specimens were produced using small subunit (SSU) and large subunit (LSU) nuclear encoded genes, and ribulose bisphosphate carboxylase (rbcL) chloroplast DNA. Three significant changes were made in the taxonomy of this species described in the 19th century [34, as Ceramium clavulatum C. Agardh]. First, true Centroceras clavulatum was found to be restricted to the Pacific coasts of the Americas from southern California to Chile, southern Australia and New Zealand. Because C. clavulatum had previously been reported from Bermuda, these collections now required analysis both morphologically and molecularly in order to determine their true identity. Next, this study [32] resurrected three western Atlantic species from the ‘C. clavulatum complex’ which had previously been retired as synonyms of C. clavulatum: C. gasparrinii (Menegh.) Kütz., C. hyalacanthum Kütz. and C. micracanthum Kütz. [35]. Lastly, Won et al. [32] also added two new species to the genus, one from South Africa and the other from southern Chile.
2.3. Morphological Plasticity
Halymenia pseudofloresii Collins et M. Howe is a noteworthy Bermudian red algal species because of its distinctive morphological plasticity. Prior to molecular analysis, a similar species, H. floresii (Clemente) C. Agardh, was also a member of the Bermuda flora [8]. Nuclear LSU ribosomal DNA and a protein-coding elongation factor were sequenced and compared with sequences from other Halymeniales using Bayesian and likelihood molecular phylogenetic methods. The cytochrome oxidase subunit I (COI-5P) from the mitochondria and universal plastid amplicon (UPA) were likewise sequenced from five Bermuda isolates including H. pseudofloresii and H. floresii morphs in order to investigate the possibility of intraspecific divergence [19]. The type locality of H. pseudofloresii is not precise, being described as from “a grotto near Walsingham” [36], clearly within the national park of that name in Bermuda. There are a number of ponds and pools (or grottos) in the Walsingham area, all of which are connected by a system of underground saltwater caves. Several specimens from this tidally fed system were collected for molecular and morphological analysis. Molecular analysis showed no more than a single base pair difference between the COI-5P of any two of the isolates, which included distinct morphologies such as narrowly pinnate (H. floresii morph), intermediate, and typical broad fronds (H. pseudofloresii morph). Likewise, no differences were observed in the UPA of any of the Bermuda specimens tested [19]. Thus despite the range of appearance, molecular data from several common markers clearly indicates that they all represent a single, phenotypically plastic species in the islands [19]. Additionally, all of the Bermuda plants were distinct from a sequence obtained from freshly collected material of H. floresii from near its type locality in Spain, thus bringing into question other reports of this European species from the western Atlantic. Further comparisons with sequences from other members of the Halymeniales in this study [19] suggest that Halymenia is a polyphyletic genus as it currently stands, opening up another avenue for molecular work. Clearly, other species found in Bermuda and elsewhere which are observed as morphological continua between distinct morphologies could benefit from the refined techniques of MAAT [19]. Some may, in fact, represent distinct species with overlapping morphologies, while others such as H. pseudofloresii outlined here represent species with highly variable mophologies.
2.4. Remaining Barriers to Sorting It All Out
Clarifying taxonomic issues often requires more than just obtaining samples of the species in question. It is often necessary to examine type specimens and then critical to collect and process DNA samples from the type locality and from potentially related species. For that reason, many species remain in limbo while comparative collections are being made. This process can take years, because some species have only been collected once or exist in remote locations. Additionally, habitats have changed since the time early 20th century collections were made and locating comparative material can be impossible. This is especially a problem because early vegetative algal collections, in particular, had their broader taxonomic placements misjudged as often as they were erroneously classified at the genus or species level. For example, when Taylor [37] described the genus and species Rhododictyon bermudensis W.R. Taylor from Bermuda, he tentatively placed it in the family Dasyaceae (Ceramiales). This new genus and species was later moved to the Ceramiaceae prompted by the discovery of tetrasporangia in offshore collections from North Carolina [38]. The tetrasporangia of R. bermudensis form at the ends of filaments obtruding from the lower, older cells of the blade. Such an arrangement is similar to that of Compsothamnion Nägeli and was used to propose an alliance with the tribe Compsothamnieae [38]. Because taxonomically critical female gametangia have yet to be discovered for this genus, molecular analysis is required to firmly establish the taxonomic position of this monotypic genus in its family and tribe [38]. Yet this species is only found at great depth and in small numbers when found, an example of a microscopic taxon difficult to find enough material for both DNA and morphological study.
Flahaultia tegetiformans W.R. Taylor (Gigartinales, Solieriaceae) provides an example of a red alga that may not have been placed in the correct genus. Despite numerous collections from a variety of locations in Jamaica none were ever collected with gametangia [39]. Taylor described this species as a prominent member of the Jamaican flora from three subtidal sites. Again, Taylor’s pressed herbarium collections were made prior to sample preservation for DNA extraction, and no recent workers have collected fresh material for genetic sequencing. The Jamaican vegetative specimens sat upon Taylor’s lab bench for years awaiting fertile collections that were never found; eventually, he assigned the new species to the genus Flahaultia Bornet on the basis of its resemblance to the eastern Atlantic F. appendiculata Bornet. Although a recent report of F. tegetiformans from the Greater and Lesser Antilles has been made [40], no specimens have been sequenced from either the type locality nor these recently collected sites. Such an analysis is necessary to ensure that the Antillean specimens are indeed the same as those collections from the type locality in Jamaica, and also whether the species is properly assigned to Flahaultia. Thus, the taxonomic placement of F. tegetiformans remains uncertain.
Although Crassitegula walsinghamii C.W. Schneid., C.E. Lane et G.W. Saunders appears in habit and vegetative morphology very similar to Flahaultia tegetiformans, sequences of its SSU nuclear DNA from Bermuda were used to firmly place it in the Sebdeniaceae (Sebdeniales), rather than the Solieriaceae. This classification was affirmed by the remarkable similarity in female sexual structures and post-fertilization stages observed between C. walsinghamii and Sebdenia (J. Agardh) Berthold when female material was found in a collection discovered after molecular sequencing was completed [18].
Another recent discovery in Bermuda was the endemic Griffithsia aestivana C.W. Schneid. et C.E. Lane [16]. It was identified without the benefit of gametophytes or gene sequencing, but was placed in Griffithsia C. Agardh owing to its obvious morphological resemblance to G. capitata Børgesen from the Canary Islands and other like members of the genus. The new species was based upon the formation of tetrasporangia located on whorled stalk cells at the distal ends of axial cells, among other things. Whorls occur around the two penultimate cells at the tips in the similar G. capitata; whorls occur on long cylindrical cells from mid-axis to a few cells below an apex in G. aestivana. Although G. aestivana seems to have unique characteristics, molecular analysis of G. aestivana is still necessary to ensure that it is a unique species, and the taxonomy will not change until that is completed [16].
Despite the fact that MAAT is becoming a standard procedure in floristics and phylogenetics, not all new species are being preserved for DNA extraction. Some are simply so small and of unknown identification in the field, that they remain to be determined in the lab long after the collections are made. Woelkerlingia sterreri C.W. Schneid. et M.J. Wynne was recently described from Bermuda [20]. This species was collected at 10 m, extremely well established on the unusual habitat created by a free-moving discarded linen tablecloth. The presence of distinctive 2-celled fertile female filaments and other characters of the procarp were critical in placing the specimens in the genus Woelkerlingia Alongi, Comaci et G. Furnari. However, no molecular work has been done within this genus to corroborate its taxonomic assignment; therefore, it is questionable whether or not all the genera of small fuzzy reds in the Wrangeliaceae are supported genetically, including Woelkerlingia [20].
3. Conclusions
The barriers to understanding algal biodiversity are problematic on a global scale for reasons covered here. Even relatively heavily studied genera in marine floras that were thought to be well understood using morphology and anatomy, have been revised based on recent MAAT studies [41,42,43], suggesting that substantial taxonomic revisions are likely over the next few decades. Although the MAAT method will almost certainly be superseded by novel techniques as methodologies advance, the combination of molecular data and microscopical observations has proven to be a robust approach to solving many long-standing taxonomic controversies among algal groups, particularly within red algae. Ultimately, the extent to which MAAT studies expand will likely depend on the stabilization of markers for various groups of the Tree of Life and continued training of students in alpha taxonomy—a skill that is rapidly declining in prevalence as descriptive morphological taxonomic work has fallen out of favor. The strengths of MAAT studies are, like any type of research, entirely dependent on the input data. DNA barcoding has been lauded as a major step forward in species identification, but several studies [1,44] have shown that sampling biases, either geographic or taxonomic, can result in incorrect species delimitations. What is acceptable in one lineage of organisms, however, may not work in another. Ironically, the very diversity of eukaryotes confounds our attempts to apply a universal marker to understand that diversity.
However, the short-term importance of understanding global biodiversity in an era when it is being depleted means that methods to do so must be identified, even if we have to adapt to each lineage independently. The examples we present here, as well as many others, have shown that the ability to definitively place vegetative algal samples in a taxonomic framework is an enormous asset to biodiversity studies in places where some species are only ever found as vegetative thalli. The accuracy of biotic inventories may be exceedingly important in the face of global climate change, as inaccurate baseline surveys will mask species loss. Numerous recent publications (including a special section of the August 11 issue of Science in 2006) suggest that rising sea temperature is already affecting the ranges of various marine animals, from turtles to fish. However, migrating animals may be responding directly to temperature or to any number of indirect factors, such as currents or prey items. In order to get a better understanding of the potential effect of rising sea temperature on the coastline, attached, rather than swimming, organisms would seem to be obvious environmental indicator species. Thus, an accurate and complete understanding of marine biodiversity in places like Bermuda, which sit at a climatic boundary, may be the first solid indicators of larger problems.
Acknowledgements
We gratefully acknowledge Roger Hollis, Chris Flook, LeeAnne Hinton and Patrick Talbot (BAMZ) provided logistical support while in Bermuda and travel support provided by a Charles A. Dana Professorship. Three anonymous reviewers are also acknowledged for their helpful comments.
References
- Meyer, C.P.; Paulay, G. DNA barcoding: Error rates based on comprehensive sampling. PLoS Biol. 2005, 3, 2229–2238. [Google Scholar]
- Briggs, J.C. Marine Zoogeography; McGraw-Hill: New York, NY, USA, 1974; pp. 1–475. [Google Scholar]
- Lüning, K. Seaweeds. Their Environment, Biogeography, and Ecophysiology; John Wiley & Sons: New York, NY, USA, 1990; pp. 1–527. [Google Scholar]
- Ekman, S. Zoogeography of the Sea; Sidgwick & Jackson Ltd.: London, UK, 1953; pp. 1–417. [Google Scholar]
- Wynne, M.J. A checklist of benthic marine algae of the tropical and subtropical western Atlantic: second revision. Nova Hedw. Beif. 2005, 129, 1–152. [Google Scholar]
- Searles, R.B.; Schneider, C.W. Observations on the deep water flora of Bermuda. Hydrobiologia 1987, 151-152, 261–266. [Google Scholar] [CrossRef]
- Schneider, C.W.; Searles, R.B. Notes on the marine algae of the Bermudas. 3. Avrainvillea sylvearleae, Discosporangium mesarthrocarpum and Peyssonnelia valentinii. J. Phycol. 1998, 34, 180–188. [Google Scholar]
- Schneider, C.W. An annotated checklist and bibliography of the marine macroalgae of the Bermuda islands. Nova Hedw. 2003, 76, 275–361. [Google Scholar] [CrossRef]
- Collins, F.S.; Hervey, A.B. The algae of Bermuda. Proc. Am. Acad. Arts Sci. 1917, 53, 1–195. [Google Scholar]
- Howe, M.A. Algae. In Flora of Bermuda; Britton, N.L., Ed.; Charles Scribner’s Sons: New York, NY, USA, 1918; pp. 489–540. [Google Scholar]
- Taylor, W.R. Survey of the marine algae of Bermuda. Yearbook Am. Phil. Soc. 1952, 1951, 167–171. [Google Scholar]
- Taylor, W.R. Marine Algae of the Eastern Tropical and Subtropical Coasts of the Americas; University of Michigan Press: Ann Arbor, MI, USA, 1960; pp. 1–879. [Google Scholar]
- Taylor, W.R.; Bernatowicz, A.J. Distribution of marine algae about Bermuda. Berm. Biol. Sta. Res. Spec. Publ. 1969, 1, 1–42. [Google Scholar]
- Schneider, C.W. Notes on the marine algae of the Bermudas. 6. Some rare or newly reported Ceramiales (Rhodophyta), including Crouania elisiae sp. nov. Phycologia 2004, 13, 563–578. [Google Scholar] [CrossRef]
- Schneider, C.W.; Lane, C.E. Notes on the marine algae of the Bermudas. 7. Additions to the flora including Chondracanthus saundersii sp. nov. (Rhodophyta, Gigartinaceae) based on rbcL sequence analysis. Phycologia 2005, 44, 72–83. [Google Scholar] [CrossRef]
- Schneider, C.W.; Lane, C.E. Notes on the marine algae of the Bermudas. 8. Further additions to the flora, including Griffithsia aestivana sp. nov. (Ceramiaceae, Rhodophyta) and an update on the alien Cystoseira compressa (Sargassaceae, Heterokontophyta). Bot. Mar. 2007, 50, 128–140. [Google Scholar]
- Schneider, C.W.; Lane, C.E. Notes on the marine algae of the Bermudas. 9. The genus Botryocladia (Rhodophyta, Rhodymeniaceae), including B. bermudana, B. exquisita and B. flookii spp. nov. Phycologia 2008, 47, 614–629. [Google Scholar] [CrossRef]
- Schneider, C.W.; Lane, C.E.; Saunders, G.W. Crassitegula walsinghamii (Sebdeniaceae, Halymeniales), a new red algal genus and species from Bermuda based upon morphology and SSU rDNA sequence analyses. Eur. J. Phycol. 2006, 41, 115–124. [Google Scholar] [CrossRef]
- Schneider, C.W.; Lane, C.E.; Saunders, G.W. Notes on the marine algae of the Bermudas. 11. More additions to the benthic flora and a phylogenetic assessment of Halymenia pseudofloresii Collins & M. Howe (Halymeniales, Rhodophyta) from its type locality. Phycologia 2010, 49, 154–168. [Google Scholar] [CrossRef]
- Schneider, C.W.; Wynne, M.J. Notes on the marine algae of the Bermudas. 10. Woelkerlingia sterreri sp. nov. (Rhodophyta, Ceramiaceae), a first record of the genus in the western Atlantic. Carib. J. Sci. 2009, 44, 301–310. [Google Scholar]
- Saunders, G.W.; Lane, C.E.; Schneider, C.W.; Kraft, G.T. Unraveling the Asteromenia peltata species complex with clarification of the genera Halichrysis and Drouetia (Rhodymeniaceae, Rhodophyta). Can. J. Bot. 2006, 84, 1581–1607. [Google Scholar] [CrossRef]
- Hommersand, M.H.; Guiry, M.D.; Fredericq, S.; Leister, G.L. New perspectives in the taxonomy of the Gigartinaceae (Gigartinales, Rhodophyta). Hydrobiologia 1993, 260-261, 105–120. [Google Scholar] [CrossRef]
- Hommersand, M.H.; Fredericq, S.; Freshwater, D.W.; Hughey, J. Recent developments in the systematics of the Gigartinaceae (Gigartinales, Rhodophyta) based on rbcL sequence analysis and morphological evidence. Phycol. Res. 1999, 47, 139–151. [Google Scholar] [CrossRef]
- Ballantine, D.L.; Aponte, N.E. Botryocladia bahamense sp. nov. (Rhodymeniaceae, Rhodophyta) from the Bahamas, western Atlantic. Crypt. Algol. 2002, 23, 123–130. [Google Scholar]
- Afonso-Carrillo, J.; Sobrino, C. Vegetative and reproductive morphology of Botryocladia botryoides, B. occidentalis and B. canariensis sp. nov. (Rhodymeniaceae, Rhodophyta) from the Canary Islands. Phycologia 2003, 42, 138–150. [Google Scholar] [CrossRef]
- Gavio, B.; Fredericq, S. Botryocladia caraibica (Rhodymeniales, Rhodophyta), a new species from the Caribbean. Cryptogam. Algol. 2003, 24, 93–106. [Google Scholar]
- Gavio, B.; Fredericq, S. New species and new records of offshore members of the Rhodymeniales (Rhodophyta) in the northern Gulf of Mexico. Gulf Mex. Sci. 2005, 23, 58–83. [Google Scholar]
- Saunders, G.W.; Strachan, I.M.; Kraft, G.T. The families of the order Rhodymeniales (Rhodophyta): a molecular-systematic investigation with a description of Faucheaceae fam. nov. Phycologia 1999, 38, 23–40. [Google Scholar] [CrossRef]
- Huisman, J.M.; Millar, A.J.K. Asteromenia (Rhodymeniaceae, Rhodymeniales), a new red algal genus based on Fauchea peltat. J. Phycol. 1996, 32, 138–145. [Google Scholar]
- Taylor, W.R. Caribbean marine algae of the Allan Hancock Expedition, 1939. Allan Hancock Atl. Exp. Rep. 1942, 2, 1–193. [Google Scholar]
- Le Gall, L.; Dalen, J.L.; Saunders, G.W. Phylogenetic analyses of the red algal order Rhodymeniales supports recognition of the Hymenocladiaceae fam. nov., Fryeellaceae fam. nov., and Neogastroclonium gen. nov. J. Phycol. 2008, 44, 1556–1571. [Google Scholar] [CrossRef]
- Won, B.Y.; Cho, T.O.; Fredericq, S. Morphological and molecular characterization of species of the genus Centroceras (Ceramiaceae, Ceramiales), including two new species. J. Phycol. 2009, 45, 227–250. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. AlgaeBase; www electronic publication, National University of Ireland: Galway, Ireland, 2010. Available online: http://www.algaebase.org (accessed on Mar 1, 2010).
- Kunth, C.S. Synopsis plantarum, quas, in itinere ad plagam aequinoctialem orbis novi, collegerunt Al. de Humboldt et Am. Bonpland; F.G. Levrault: Parisiis [Paris], France, 1822; Vol. 1, pp. 1–491. [Google Scholar]
- Kützing, F.T. Tabulae phycologicae. Vol. 13; Gedruckt auf kosten des Verfassers: Nordhausen, Germany, 1863; pp. 1-31,100. [Google Scholar]
- Collins, F.S.; Howe, M.A. Notes on species of Halymenia. Bull. Torrey Bot. Club 1916, 43, 169–182. [Google Scholar]
- Taylor, W.R. Notes on three Bermudian marine algae. Hydrobiologia 1961, 18, 277–283. [Google Scholar] [CrossRef]
- Schneider, C.W. North Carolina marine algae. V. Additions to the flora of Onslow Bay, including the reassignment of Fauchea peltata Taylor to Weberella Schmitz. Brit. Phycol. 1975, 10, 129–138. [Google Scholar] [CrossRef]
- Taylor, W.R. Notes on algae from the tropical Atlantic Ocean—VII. Rev. Algol. Nouv. Sér. 1974, 11, 58–71. [Google Scholar]
- Littler, D.S.; Littler, M.M. Caribbean Reef Plants. An Identification Guide to the Reef Plants of the Caribbean, Bahamas, Florida and Gulf of Mexico; OffShore Graphics, Inc.: Washington, DC, USA, 2000; pp. 1–542. [Google Scholar]
- Kucera, H.; Saunders, G.W. Assigning morphological variants of Fucus (Fucales, Phaeophyceae) in Canadian waters to recognized species using DNA barcoding. Botany 2008, 86, 1065–1079. [Google Scholar] [CrossRef]
- Saunders, G.W. Routine DNA barcoding of Canadian Gracilariales (Rhodophyta) reveals the invasive species Gracilaria vermiculophylla in British Columbia. Mol. Ecol. Resour. 2009, 9, 140–150. [Google Scholar] [CrossRef]
- Walker, R.H.; Brodie, J.; Russell, S.; Irvine, L.M.; Orfanidis, S. Biodiversity of coralline algae in the northeastern Atlantic including Corallina caespitosa sp. nov. (Corallinoideae, Rhodophyta). J. Phycol. 2009, 45, 287–297. [Google Scholar] [CrossRef]
- Wiemers, M.; Fiedler, K. Does the DNA barcoding gap exist?—A case study in blue butterflies (Lepidoptera: Lycaenidae). Front. Zool. 2007, 4, 8. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).