Distribution and Potential Dispersal Corridors of Two Onychodactylus Species in the Republic of Korea
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Preparing Data for Species Distribution Modeling
2.2.1. Online Database
2.2.2. Environmental Variables
2.3. Species Distribution Modeling
2.3.1. Model Construction and Parameter Tuning
2.3.2. Model Selection and Evaluation
2.4. Field Survey and Genetic Species Identification
2.5. Identification of Potential Dispersal Corridors
2.5.1. Niche Overlap Analysis
2.5.2. Habitat Connectivity Evaluation
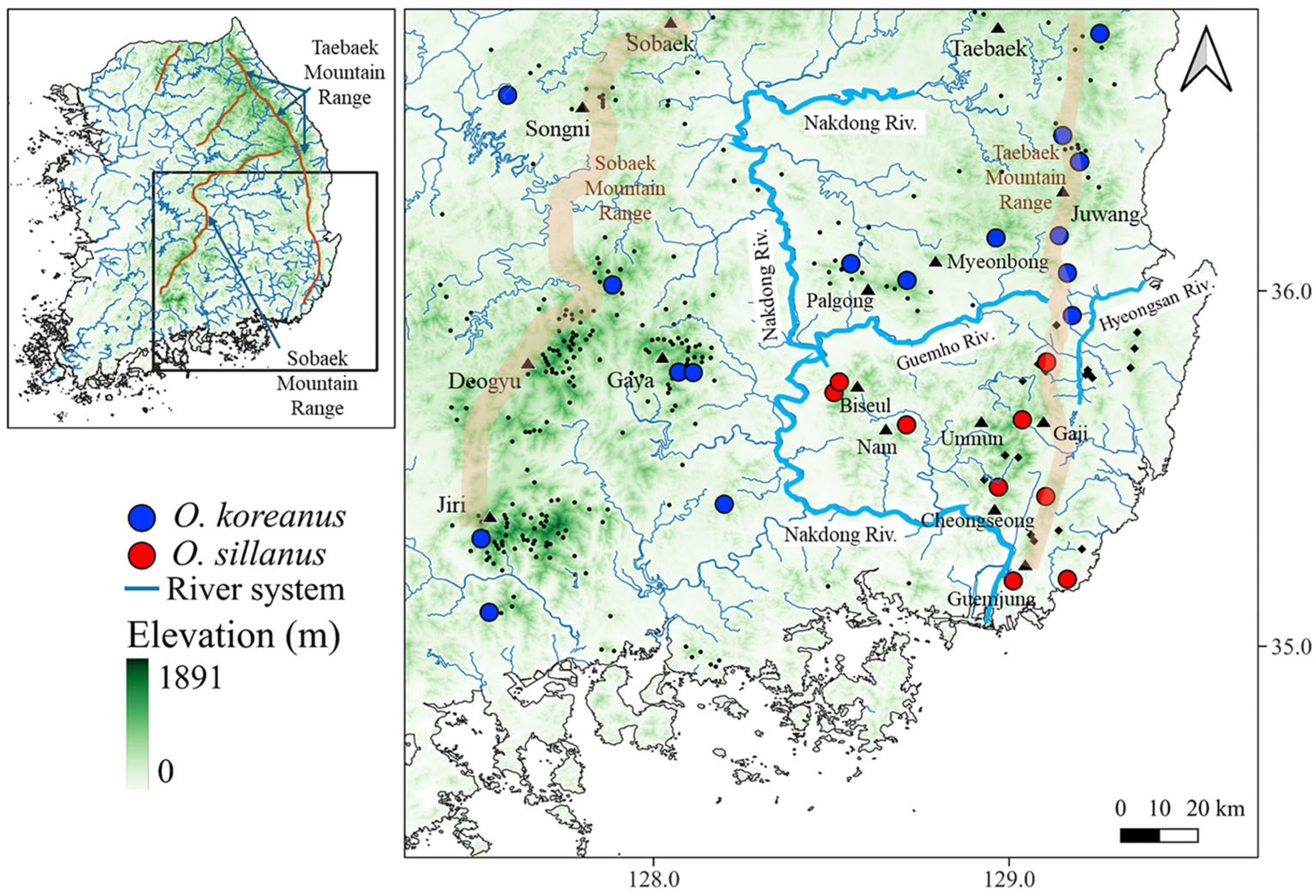
3. Results
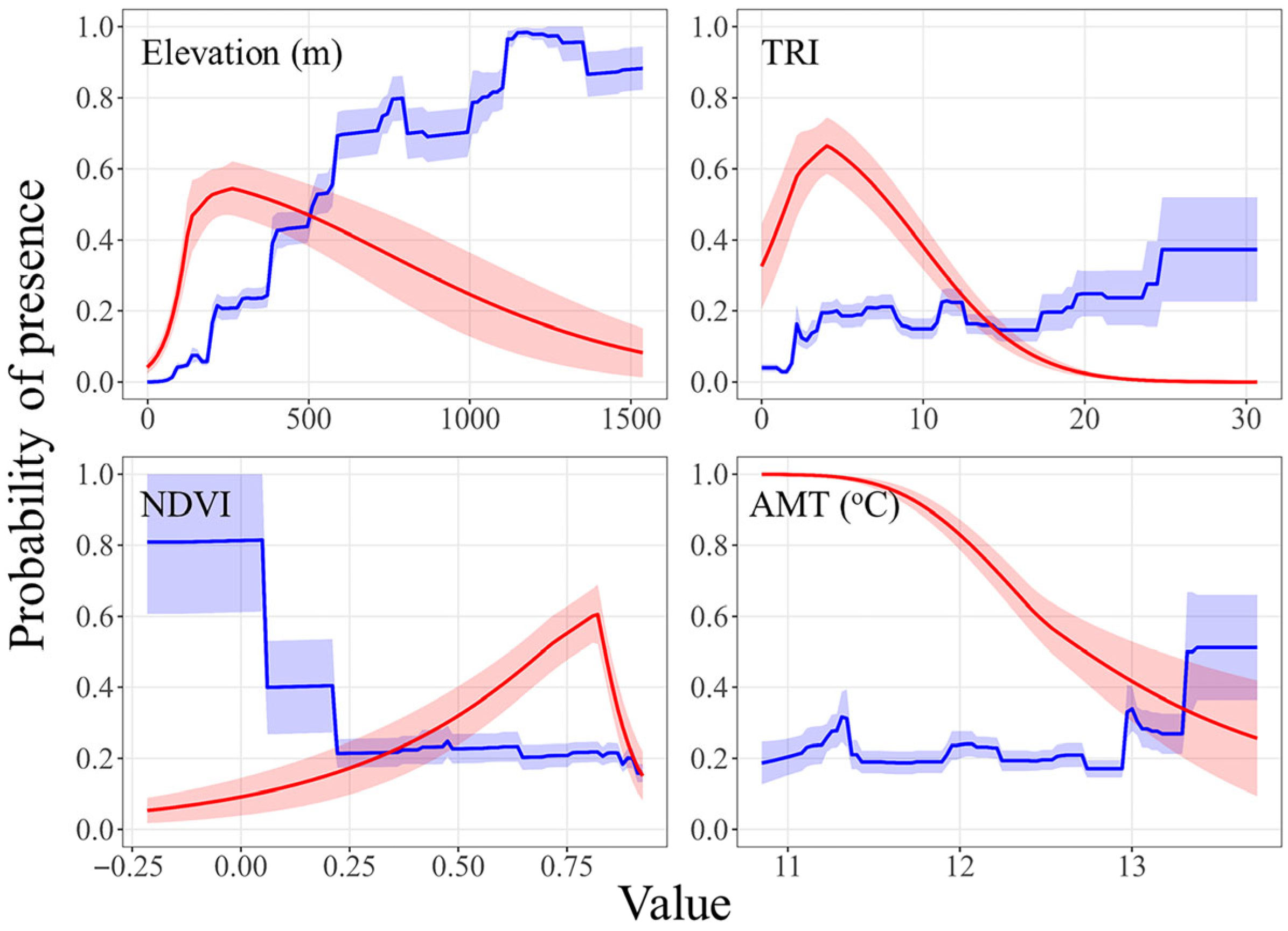
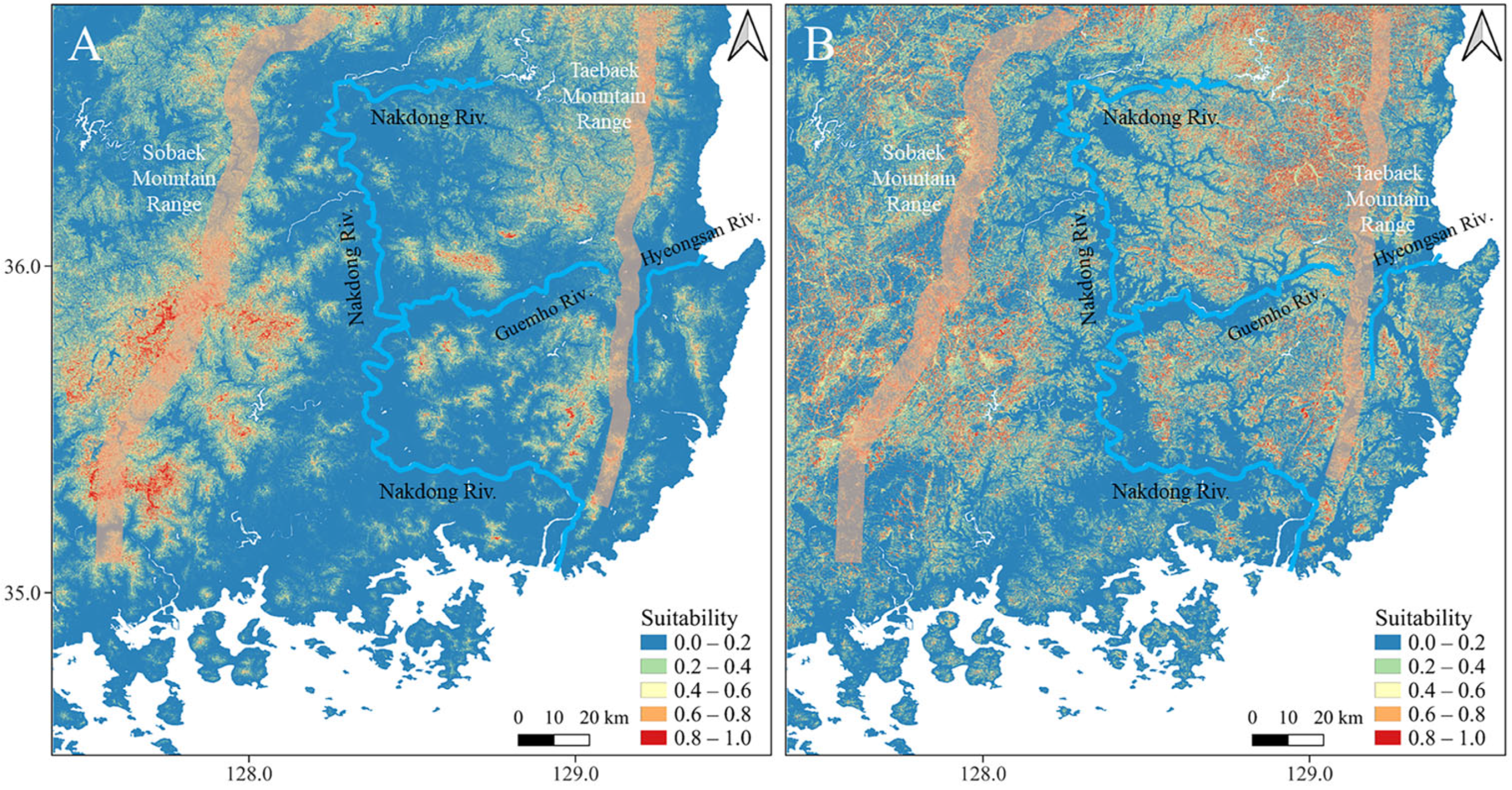
3.1. Species Distribution Modeling
3.2. Field Survey and Genetic Species Identification
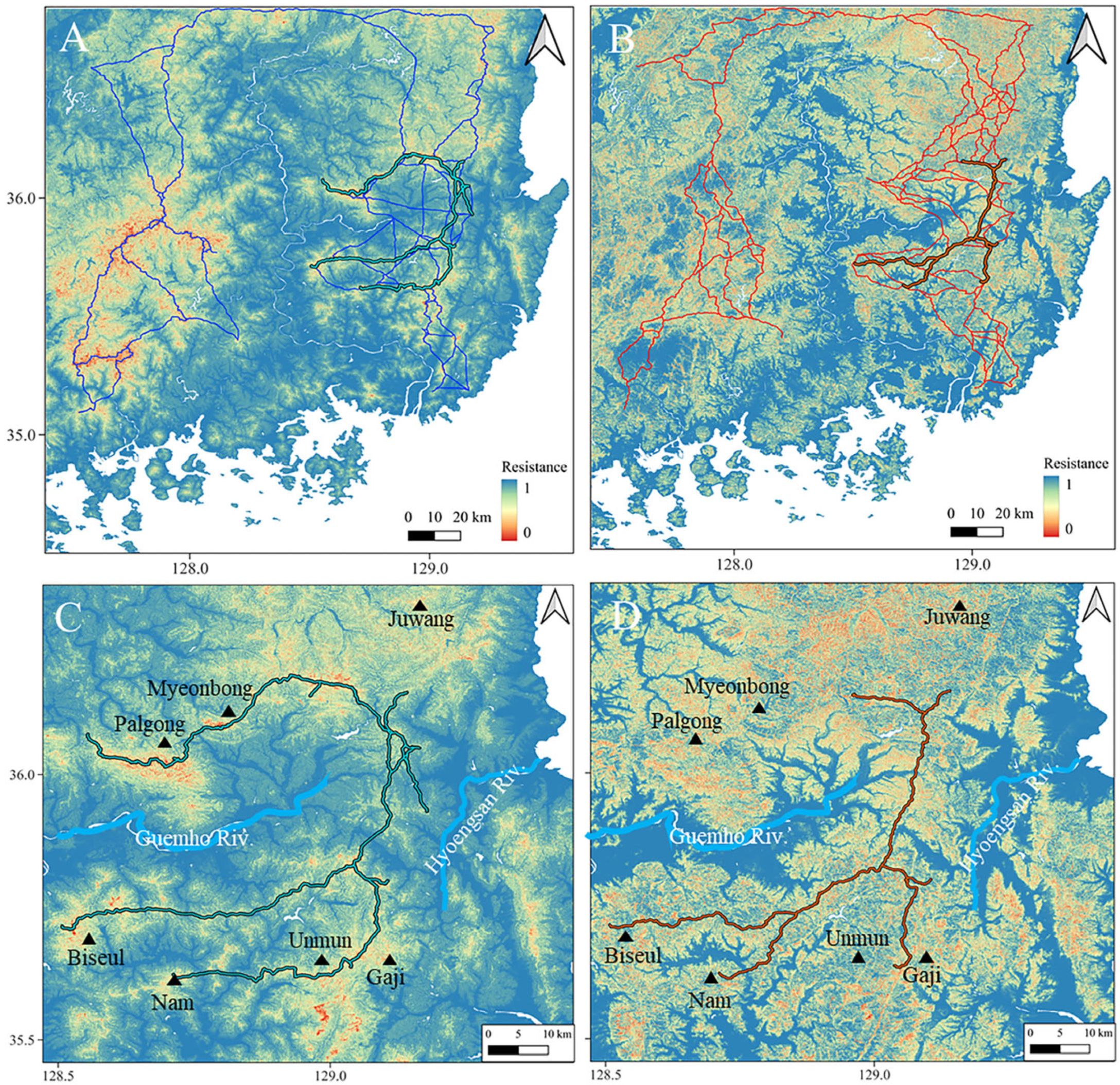
3.3. Identification of Potential Dispersal Corridors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMT | Annual mean temperature |
| AP | Annual precipitation |
| AUC | Area under the curve |
| CQ | Corridor quality |
| CWD | Cost-weighted distance |
| DEM | Digital elevation model |
| FC | Feature combination |
| H | Hinge |
| HIS | Habitat suitability index |
| KMA | Korea meteorological administration |
| KNPS | Korea national park service |
| L | Linear |
| LCP | Least cost path |
| MaxEnt | Maximum entropy |
| NDVI | Normalized difference vegetation index |
| NES | National ecosystem survey |
| OR10 | 10% omission rate |
| P | Product |
| PC | Principal component |
| PCA | Principal component analysis |
| Q | Quadratic |
| RF | Random forest |
| SDM | Species distribution model |
| T | Threshold |
| TPI | Topographic position index |
| TRI | Topographic roughness index |
| TSS | True skill statistic |
| USGS | United States geological survey |
References
- Brown, J.H. Macroecology; University of Chicago Press: Chicago, IL, USA, 1995; ISBN 0-226-07615-6. [Google Scholar]
- Lomolino, M.V. Biogeography: A Very Short Introduction; Oxford University Press: New York, NY, USA, 2020; ISBN 978-0-198-85006-9. [Google Scholar]
- Hanski, I. Metapopulation dynamics. Nature 1998, 396, 41–49. [Google Scholar] [CrossRef]
- Moilanen, A.; Hanski, I. On the use of connectivity measures in spatial ecology. Oikos 2001, 95, 147–151. [Google Scholar] [CrossRef]
- Crooks, K.R.; Sanjayan, M. Connectivity Conservation; Cambridge University Press: New York, NY, USA, 2006; ISBN 978-0-521-67381-5. [Google Scholar]
- Kusuma, W.E.; Ratmuangkhwang, S.; Kumazawa, Y. Molecular phylogeny and historical biogeography of the Indonesian freshwater fish Rasbora lateristriata species complex (Actinopterygii: Cyprinidae): Cryptic species and west-to-east divergences. Mol. Phylogenet. Evol. 2016, 105, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Hoang, C.V.; Nguyen, T.T.; Ninh, H.T.; Luong, A.M.; Pham, C.T.; Nguyen, T.Q.; Orlov, N.L.; Chen, Y.; Wang, B.; Ziegler, T.; et al. Two new cryptic species of Microhyla Tschudi, 1838 (Amphibia, Anura, Microhylidae) related to the M. heymonsi group from central Vietnam. ZooKeys 2021, 1036, 47–74. [Google Scholar] [CrossRef]
- Guo, P.; Wang, P.; Lyu, B.; Liu, Q.; Zheng, J.; Fu, C.; Wu, Y.; Shu, G.; Hou, S. Molecular phylogeny reveals cryptic diversity in Sibynophis from China (Serpentes: Sibynophiidae). Front. Ecol. Evol. 2023, 13, e10367. [Google Scholar] [CrossRef]
- Borzée, A.; Min, M.S. Disentangling the impacts of speciation, sympatry and the island effect on the morphology of seven Hynobius sp. salamanders. Animals 2021, 11, 187. [Google Scholar] [CrossRef]
- Niwa, K.; Nishikawa, K.; Matsui, M.; Kanamori, S.; Kuro-O, M. Taxonomic reassessment of salamanders (genus Hynobius) from Tsushima Islands, Japan, with a resurrection of Hynobius tagoi Dunn, 1923 (Amphibia: Caudata). Zootaxa 2023, 5339, 201–236. [Google Scholar] [CrossRef]
- Bergsten, J.; Bilton, D.T.; Fujisawa, T.; Elliott, M.; Monaghan, M.T.; Balke, M.; Hendrich, L.; Geijer, J.; Herrmann, J.; Foster, G.N.; et al. The effect of geographical scale of sampling on DNA barcoding. Syst. Biol. 2012, 61, 851–869. [Google Scholar] [CrossRef]
- Funk, W.C.; Caminer, M.; Ron, S.R. High levels of cryptic species diversity uncovered in Amazonian frogs. Proc. R. Soc. B Biol. Sci. 2012, 279, 1806–1814. [Google Scholar] [CrossRef]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.L.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef]
- Bortolus, A. Error cascades in the biological sciences: The unwanted consequences of using bad taxonomy in ecology. Ambio 2008, 37, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Angulo, A.; Icochea, J. Cryptic species complexes, widespread species and conservation: Lessons from Amazonian frogs of the Leptodactylus marmoratus group (Anura: Leptodactylidae). Syst. Biodivers. 2010, 8, 357–370. [Google Scholar] [CrossRef]
- Gaston, K.J. The Structure and Dynamics of Geographic Ranges; Oxford University Press: New York, NY, USA, 2003; ISBN 019-8-52641-5. [Google Scholar]
- Guisan, A.; Thuiller, W. Predicting species distribution: Offering more than simple habitat models. Ecol. Lett. 2005, 8, 993–1009. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Elith, J.; Graham, C.H.; Anderson, R.P.; Dudík, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A.; et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef]
- Raxworthy, C.J.; Martinez-Meyer, E.; Horning, N.; Nussbaum, R.A.; Schneider, G.E.; Ortega-Huerta, M.A.; Peterson, A.T. Predicting distributions of known and unknown reptile species in Madagascar. Nature 2003, 426, 837–841. [Google Scholar] [CrossRef]
- Kozak, K.H.; Wiens, J.J. Does niche conservatism promote speciation? A case study in North American salamanders. Evolution 2007, 60, 2604–2621. [Google Scholar] [CrossRef]
- Rondinini, C.; Di Marco, M.; Chiozza, F.; Santulli, G.; Baisero, D.; Visconti, P.; Hoffmann, M.; Schipper, J.; Stuart, S.N.; Tognelli, M.F.; et al. Global habitat suitability models of terrestrial mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 2633–2641. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Trivedi, M.R.; Berry, P.M.; Morecroft, M.D.; Dawson, T.P. Spatial scale affects bioclimate model projections of climate change impacts on mountain plants. Glob. Change Biol. 2008, 14, 1089–1103. [Google Scholar] [CrossRef]
- Randin, C.F.; Engler, R.; Normand, S.; Zappa, M.; Zimmermann, N.E.; Pearman, P.B.; Vittoz, P.; Thuiller, W.; Guisan, A. Climate change and plant distribution: Local models predict high-elevation persistence. Glob. Change Biol. 2009, 15, 1557–1569. [Google Scholar] [CrossRef]
- Lembrechts, J.J.; Lenoir, J.; Roth, N.; Hattab, T.; Milbau, A.; Haider, S.; Pellissier, L.; Pauchard, A.; Backes, A.R.; Dimarco, R.D.; et al. Comparing temperature data sources for use in species distribution models: From in-situ logging to remote sensing. Glob. Ecol. Biogeogr. 2019, 28, 1578–1596. [Google Scholar] [CrossRef]
- Franklin, J.; Davis, F.W.; Ikegami, M.; Syphard, A.D.; Flint, L.E.; Flint, A.L.; Hannah, L. Modeling plant species distributions under future climates: How fine scale do climate projections need to be? Glob. Change Biol. 2013, 19, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Storlie, C.J.; Phillips, B.L.; VanDerWal, J.; Williams, S.E. Improved spatial estimates of climate predict patchier species distributions. Divers. Distrib. 2013, 19, 1106–1113. [Google Scholar] [CrossRef]
- Zeilhofer, P.; Cezar, A.; Tôrres, N.M.; de Almeida Jácomo, A.T.; Silveira, L. Jaguar Panthera onca habitat modeling in landscapes facing high land-use transformation pressure—Findings from Mato Grosso, Brazil. Biotropica 2014, 46, 98–105. [Google Scholar] [CrossRef]
- Connor, T.; Hull, V.; Viña, A.; Shortridge, A.; Tang, Y.; Zhang, J.; Wang, F.; Liu, J. Effects of grain size and niche breadth on species distribution modeling. Ecography 2018, 41, 1270–1282. [Google Scholar] [CrossRef]
- Karami, P.; Tavakoli, S.; Esmaeili, M. Fine-scale habitat suitability and connectivity analysis for the core populations of yellow-spotted mountain pond-breeding newt (Neurergus derjugini) in the west of Iran and east of Iraq. Glob. Ecol. Conserv. 2023, 43, e02429. [Google Scholar] [CrossRef]
- Shin, Y.; Min, M.S.; Borzée, A. Driven to the edge: Species distribution modeling of a clawed salamander (Hynobiidae: Onychodactylus koreanus) predicts range shifts and drastic decrease of suitable habitats in response to climate change. Ecol. Evol. 2021, 11, 14669–14688. [Google Scholar] [CrossRef]
- Borzée, A.; Shin, Y.; Poyarkov, N.A.; Jeon, J.Y.; Baek, H.J.; Lee, C.H.; An, J.; Hong, Y.J.; Min, M.S. Dwindling in the mountains: Description of a critically endangered and microendemic Onychodactylus species (Amphibia, Hynobiidae) from the Korean Peninsula. Zool. Res. 2022, 43, 750–755. [Google Scholar] [CrossRef]
- Kuzmin, S.L. The Clawed Salamanders of Asia: Genus Onychodactylus: Biology, Distribution, and Conservation; Westarp Wissenschaften: Magdeburg, Germany, 1995; ISBN 978-3-894-32425-4. [Google Scholar]
- Poyarkov, N.A.; Che, J.; Min, M.S.; Kuro-o, M.; Yan, F.; Li, C.; Iizuka, K.; Vieites, D.R. Review of the systematics, morphology and distribution of Asian clawed salamanders, genus Onychodactylus (Amphibia, Caudata: Hynobiidae), with the description of four new species. Zootaxa 2012, 3465, 1–106. [Google Scholar] [CrossRef]
- Lee, J.H.; Ra, N.Y.; Eom, J.; Park, D. Population dynamics of the long-tailed clawed salamander larva, Onychodactylus fischeri, and its age structure in Korea. J. Ecol. Environ. 2008, 31, 31–36. [Google Scholar] [CrossRef]
- Suk, H.Y.; Lee, M.Y.; Bae, H.G.; Lee, S.J.; Poyarkov, N.A.; Lee, H.; Min, M.S. Phylogenetic structure and ancestry of Korean clawed salamander, Onychodactylus koreanus (Caudata: Hynobiidae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2018, 29, 650–658. [Google Scholar] [CrossRef]
- QGIS.org. QGIS, version 3.40.14; QGIS Association: Bern, Switzerland, 2025. [Google Scholar]
- Kim, H.W.; Yoon, S.; Kim, M.; Shin, M.; Yoon, H.; Kim, K. EcoBank: A flexible database platform for sharing ecological data. Biodivers. Data J. 2021, 9, e61866. [Google Scholar] [CrossRef] [PubMed]
- Hijmans, R.J. Package ‘Terra’. 2024. Available online: https://cran.r-project.org/web/packages/terra/index.html (accessed on 9 October 2025).
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2025. [Google Scholar]
- Boria, R.A.; Olson, L.E.; Goodman, S.M.; Anderson, R.P. Spatial filtering to reduce sampling bias can improve the performance of ecological niche models. Ecol. Model. 2014, 275, 73–77. [Google Scholar] [CrossRef]
- Aiello-Lammens, M.E.; Boria, R.A.; Radosavljevic, A.; Vilela, B.; Anderson, R.P. spThin: An R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 2015, 38, 541–545. [Google Scholar] [CrossRef]
- Park, D.; Jeong, H.; Park, J.; Park, I.K. Distribution and habitat assessments of the slender racer, Orientocoluber spinalis, for the registration of nationally endangered species in the Republic of Korea. Sci. Rep. 2023, 13, 12025. [Google Scholar] [CrossRef]
- Park, I.K.; Shin, Y.; Baek, H.J.; Kim, J.; Kim, D.I.; Seok, M.; Oh, Y.; Park, D. Establishment potential across South Korea for two gecko species, Gekko japonicus and G. swinhonis, adapted to different climates. NeoBiota 2024, 93, 39–62. [Google Scholar] [CrossRef]
- U.S. Geological Survey. USGS 3D Elevation Program Digital Elevation Model. Available online: https://elevation.nationalmap.gov/arcgis/rest/services/3DEPElevation/ImageServer (accessed on 9 October 2025).
- Amatulli, G.; Domisch, S.; Tuanmu, M.N.; Parmentier, B.; Ranipeta, A.; Malczyk, J.; Jetz, W. A suite of global, cross-scale topographic variables for environmental and biodiversity modeling. Sci. Data 2018, 5, 180040. [Google Scholar] [CrossRef]
- Pettorelli, N.; Vik, J.O.; Mysterud, A.; Gaillard, J.M.; Tucker, C.J.; Stenseth, N.C. Using the satellite-derived NDVI to assess ecological responses to environmental change. Trends Ecol. Evol. 2005, 20, 503–510. [Google Scholar] [CrossRef]
- Hengl, T.; Walsh, M.G.; Sanderman, J.; Wheeler, I.; Harrison, S.P.; Prentice, I.C. Global mapping of potential natural vegetation: An assessment of machine learning algorithms for estimating land potential. PeerJ 2018, 6, e5457. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Phillips, S.; Leathwick, J.; Elith, J. Dismo: Species Distribution Modeling. R Package Version 1.3-15. 2024. Available online: https://cran.r-project.org/web/packages/dismo/index.html (accessed on 9 October 2025).
- Phillips, S.J.; Dudík, M. Modeling of species distributions with Maxent: New extensions and a comprehensive evaluation. Ecography 2008, 31, 161–175. [Google Scholar] [CrossRef]
- Swets, J.A. Measuring the accuracy of diagnostic systems. Science 1988, 240, 1285–1293. [Google Scholar] [CrossRef] [PubMed]
- Radosavljevic, A.; Anderson, R.P. Making better Maxent models of species distributions: Complexity, overfitting and evaluation. J. Biogeogr. 2014, 41, 629–643. [Google Scholar] [CrossRef]
- Yost, A.C.; Petersen, S.L.; Gregg, M.; Miller, R. Predictive modeling and mapping sage grouse (Centrocercus urophasianus) nesting habitat using Maximum Entropy and a long-term dataset from Southern Oregon. Ecol. Inform. 2008, 3, 375–386. [Google Scholar] [CrossRef]
- Yang, X.Q.; Kushwaha, S.P.S.; Saran, S.; Xu, J.; Roy, P.S. Maxent modeling for predicting the potential distribution of medicinal plant, Justicia adhatoda L. in Lesser Himalayan foothills. Ecol. Eng. 2013, 51, 83–87. [Google Scholar] [CrossRef]
- Lee, S.Y.; Choi, S.-Y.; Bae, Y.-S.; Suh, J.-H.; Jang, H.J.; Do, M.S. Distribution prediction of Korean clawed salamander (Onychodactylus koreanus) according to the climate change. Korean J. Environ. Ecol. 2021, 35, 480–489. (In Korean) [Google Scholar] [CrossRef]
- Yoshikawa, N.; Matsui, M.; Nishikawa, K.; Kim, J.B.; Kryukov, A. Phylogenetic relationships and biogeography of the Japanese clawed salamander, Onychodactylus japonicus (Amphibia: Caudata: Hynobiidae), and its congener inferred from the mitochondrial cytochrome b gene. Mol. Phylogenet. Evol. 2008, 49, 249–259. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Broennimann, O.; Fitzpatrick, M.C.; Pearman, P.B.; Petitpierre, B.; Pellissier, L.; Yoccoz, N.G.; Thuiller, W.; Fortin, M.J.; Randin, C.; Zimmermann, N.E.; et al. Measuring ecological niche overlap from occurrence and spatial environmental data. Glob. Ecol. Biogeogr. 2012, 21, 481–497. [Google Scholar] [CrossRef]
- Warren, D.L.; Matzke, N.J.; Cardillo, M.; Baumgartner, J.B.; Beaumont, L.J.; Turelli, M.; Glor, R.E.; Huron, N.A.; Simões, M.; Iglesias, T.L.; et al. ENMTools 1.0: An R package for comparative ecological biogeography. Ecography 2021, 44, 504–511. [Google Scholar] [CrossRef]
- Schoener, T.W. The anolis lizards of Bimini: Resource partitioning in a complex fauna. Ecology 1968, 49, 704–726. [Google Scholar] [CrossRef]
- Warren, D.L.; Glor, R.E.; Turelli, M. Environmental niche equivalency versus conservatism: Quantitative approaches to niche evolution. Evolution 2008, 62, 2868–2883. [Google Scholar] [CrossRef] [PubMed]
- van Etten, J.R. Package gdistance: Distances and routes on geographical grids. J. Stat. Softw. 2017, 76, 1–21. [Google Scholar] [CrossRef]
- Li, J.; Liang, L.; Chao, Y.; Wang, X.; Qiu, M.; Luo, P.; Zhu, Y. Construction of the ecological security pattern of Mu Us sandy land on the basis of the “Source − Resistance − Corridor” theory. Ecol. Indic. 2025, 171, 113162. [Google Scholar] [CrossRef]
- Adriaensen, F.; Chardon, J.P.; De Blust, G.; Swinnen, E.; Villalba, S.; Gulinck, H.; Matthysen, E. The application of ‘least-cost’ modelling as a functional landscape model. Landsc. Urban Plan. 2003, 64, 233–247. [Google Scholar] [CrossRef]
- Dutta, T.; Sharma, S.; McRae, B.H.; Roy, P.S.; DeFries, R. Connecting the dots: Mapping habitat connectivity for tigers in central India. Reg. Environ. Change 2016, 16, 53–67. [Google Scholar] [CrossRef]
- Wang, I.J.; Savage, W.K.; Shaffer, H.B. Landscape genetics and least-cost path analysis reveal unexpected dispersal routes in the California tiger salamander (Ambystoma californiense). Mol. Ecol. 2009, 18, 1365–1374. [Google Scholar] [CrossRef]
- Peterman, W.E.; Connette, G.M.; Semlitsch, R.D.; Eggert, L.S. Ecological resistance surfaces predict fine-scale genetic differentiation in a terrestrial woodland salamander. Mol. Ecol. 2014, 23, 2402–2413. [Google Scholar] [CrossRef]
- Peng, Y.; Li, Y.; Cao, G.; Li, H.; Shin, Y.; Piao, Z.; Perez, F.; Zhu, W.; Borzée, A. Estimation of habitat suitability and landscape connectivity for Liaoning and Jilin clawed salamanders (Hynobiidae: Onychodactylus) in the transboundary region between the People’s Republic of China and the Democratic People’s Republic of Korea. Glob. Ecol. Conserv. 2023, 48, e02694. [Google Scholar] [CrossRef]
- Shin, Y.; Borzée, A.; Park, D. Climatic data sources and limitations of ecological niche models impact the estimations of historical ranges and niche overlaps in distantly related Korean salamanders. BMC Ecol. Evol. 2025, 25, 105. [Google Scholar] [CrossRef]
- Soberón, J.; Peterson, A.T. Interpretation of models of fundamental ecological niches and species’ distributional areas. Biodivers Inform. 2005, 2, 1–10. [Google Scholar] [CrossRef]
- Colwell, R.K.; Rangel, T.F. Hutchinson’s duality: The once and future niche. Proc. Natl. Acad. Sci. USA 2009, 106, 19651–19658. [Google Scholar] [CrossRef] [PubMed]
- Harcourt, A.H.; Wood, M.A. Rivers as barriers to primate distributions in Africa. Int. J. Primatol. 2012, 33, 168–183. [Google Scholar] [CrossRef]
- Jackson, N.D.; Austin, C.C. The combined effects of rivers and refugia generate extreme cryptic fragmentation within the common ground skink (Scincella lateralis). Evolution 2010, 64, 409–428. [Google Scholar] [CrossRef] [PubMed]
- Stevens, V.M.; Verkenne, C.; Vandewoestijne, S.; Wesselingh, R.A.; Baguette, M. Gene flow and functional connectivity in the natterjack toad. Mol. Ecol. 2006, 15, 2333–2344. [Google Scholar] [CrossRef]
- Yan, F.; Zhou, W.; Zhao, H.; Yuan, Z.; Wang, Y.; Jiang, K.; Jin, J.; Murphy, R.W.; Che, J.; Zhang, Y. Geological events play a larger role than Pleistocene climatic fluctuations in driving the genetic structure of Quasipaa boulengeri (Anura: Dicroglossidae). Mol. Ecol. 2012, 22, 1120–1133. [Google Scholar] [CrossRef]
- Chough, S.K.; Kwon, S.T.; Ree, J.H.; Choi, D.K. Tectonic and sedimentary evolution of the Korean peninsula: A review and new view. Earth Sci. Rev. 2000, 52, 175–235. [Google Scholar] [CrossRef]
- Son, M.; Song, C.W.; Kim, M.C.; Cheon, Y.; Jung, S.; Cho, H.; Kim, H.G.; Kim, J.S.; Sohn, Y.K. Miocene crustal deformation, basin development, and tectonic implication in the southeastern Korean Peninsula. J. Geol. Soc. Korea 2013, 49, 93–118. (In Korean) [Google Scholar] [CrossRef]
- Cushman, S.A.; McKelvey, K.S.; Hayden, J.; Schwartz, M.K. Gene flow in complex landscapes: Testing multiple hypotheses with causal modeling. Am. Nat. 2006, 168, 486–499. [Google Scholar] [CrossRef]
- McRae, B.H.; Dickson, B.G.; Keitt, T.H.; Shah, V.B. Using circuit theory to model connectivity in ecology, evolution, and conservation. Ecology 2008, 89, 2712–2724. [Google Scholar] [CrossRef]
- Spear, S.F.; Peterson, C.R.; Matocq, M.D.; Storfer, A. Landscape genetics of the blotched tiger salamander (Ambystoma tigrinum melanostictum). Mol. Ecol. 2005, 14, 2553–2564. [Google Scholar] [CrossRef]
- Beerli, P.; Mashayekhi, S.; Sadeghi, M.; Khodaei, M.; Shaw, K. Population genetic inference with Migrate. Curr. Protoc. Bioinform. 2019, 68, e87. [Google Scholar] [CrossRef]
- Araújo, M.B.; Thuiller, W.; Pearson, R.G. Climate warming and the decline of amphibians and reptiles in Europe. J. Biogeogr. 2006, 33, 1712–1728. [Google Scholar] [CrossRef]
- Das, B.; Das, A.; Baishya, K.K. Predicting impressions of climate change on the distribution of Crocodile newt (Tylototriton verrucosus), a rare amphibian in the Darjeeling Himalayan sub-region, India. Int. J. Ecol. Environ. Sci. 2023, 49, 17–29. [Google Scholar] [CrossRef]
| Variable | Study Area | O. koreanus | O. sillanus |
|---|---|---|---|
| Elevation (m) | 163.42 ± 212.27 (0.000–1891.946) | 595.26 ± 293.22 (80.30–1519.15) | 315.62 ± 151.82 (128.86–744.44) |
| Slope (°) | 9.53 ± 10.52 (0.000–68.793) | 15.40 ± 8.07 (2.09–45.45) | 14.34 ± 6.92 (1.35–34.93) |
| Aspect | 178.95 ± 102.54 (0.00–360.00) | 180.50 ± 106.28 (0.34–359.28) | 176.50 ± 89.97 (26.38–337.36) |
| TPI | −3.17 × 10−5 ± 2.57 (−54.43–56.61) | −2.06 ± 3.73 (−17.60–20.44) | −1.66 ± 2.38 (−6.43–3.59) |
| TRI | 4.38 ± 4.50 (0.00–63.50) | 7.70 ± 3.87 (0.97–24.63) | 6.36 ± 2.72 (2.09–13.29) |
| NDVI | 0.73 ± 0.19 (−1.00–1.00) | 0.83 ± 0.10 (−0.05–0.92) | 0.77 ± 0.09 (0.49–0.88) |
| AMT | 12.61 ± 0.45 (10.01–14.32) | 12.21 ± 0.43 (11.05–13.36) | 12.29 ± 0.41 (11.12–13.14) |
| AP | 1285.33 ± 56.35 (1091.77–3070.31) | 1338.48 ± 138.81 (1188.87–2146.79) | 1269.04 ± 55.37 (1210.60–1520.31) |
| Variable | PC1 | PC2 | PC3 |
|---|---|---|---|
| Elevation | −0.456 | 0.437 | 0.156 |
| Slope | −0.497 | −0.348 | −0.141 |
| Aspect | −0.003 | 0.024 | 0.262 |
| Topographic position index | −0.046 | 0.255 | −0.339 |
| Topographic roughness index | −0.492 | −0.341 | −0.143 |
| Normalized difference vegetation index | −0.424 | −0.044 | −0.265 |
| Annual mean temperature | 0.346 | −0.478 | −0.509 |
| Annual precipitation | 0.032 | 0.525 | −0.65 |
| Eigenvalue | 3.095 | 1.151 | 1.069 |
| Variance explained (%) | 38.68 | 14.38 | 13.36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Kim, Y.-G.; Nam, H.; Park, J.; Park, J.; Park, D. Distribution and Potential Dispersal Corridors of Two Onychodactylus Species in the Republic of Korea. Diversity 2026, 18, 57. https://doi.org/10.3390/d18010057
Kim Y-G, Nam H, Park J, Park J, Park D. Distribution and Potential Dispersal Corridors of Two Onychodactylus Species in the Republic of Korea. Diversity. 2026; 18(1):57. https://doi.org/10.3390/d18010057
Chicago/Turabian StyleKim, Young-Guk, Hahyun Nam, Jaejin Park, Jiho Park, and Daesik Park. 2026. "Distribution and Potential Dispersal Corridors of Two Onychodactylus Species in the Republic of Korea" Diversity 18, no. 1: 57. https://doi.org/10.3390/d18010057
APA StyleKim, Y.-G., Nam, H., Park, J., Park, J., & Park, D. (2026). Distribution and Potential Dispersal Corridors of Two Onychodactylus Species in the Republic of Korea. Diversity, 18(1), 57. https://doi.org/10.3390/d18010057






