Abstract
We describe a new stick insect species, Pseudoparamenexenus beiliuensis sp. nov., by an integrated approach using morphological and molecular data. The mitochondrial genomes of this new species and Pseudoparamenexenus yangi collected from Jianfengling, Hainan, China, were fully sequenced and annotated. Both mitogenomes contained the standard metazoan gene set arranged in the ancestral phasmid order, with ATP8 showing the highest evolutionary rate, and COX1 the strongest purifying selection. Phylogenetic analyses were conducted based on 13 protein-coding genes, revealing the two species form a well-supported sister-group relationship. The systematic position of the genus Pseudoparamenexenus was resolved as follows: ((Pseudoparamenexenus beiliuensis sp. nov. + Pseudoparamenexenus yangi) + (Neohirasea stephanus + (Neohirasea japonica + Neohirasea hongkongensis))) + ((Pachyscia longicauda + Acanthophasma brevicercum) + ((Sinophasma brevipenne + Micadina phluctainoides) + (Micadina brevioperculina + Micadina brachyptera))). The discovery of this species not only advances our understanding of the genus Pseudoparamenexenus but also addresses knowledge gaps concerning the diversity of stick insects.
1. Introduction
Phasmatodea is a moderately species-diverse order comprising stick and leaf insects, with more than 3500 species worldwide, including 14 families and 531 genera [1,2]. Stick and leaf insects primarily occur in tropical, subtropical, and temperate regions [2]. The insects are known for their mimetic camouflage, variously appearing as twigs, leaves, and mosses [3,4,5]. The morphological convergence and extreme sexual dimorphism in Phasmatodea present significant taxonomic challenges, yet precise classification remains fundamental to elucidating the evolutionary relationships within this ecologically important insect order [6,7,8,9].
Paramenexenus yangi described by Chen and He in 2002 [10] exhibits distinct morphological differences compared to other members of the genus: P. yangi males are characterized by a smooth thorax and a medioventral carina on the femora bearing a small subapical spine, whereas other males of the genus Paramenexenus possess a distinctly spinose thorax. Moreover, P. yangi females display a strongly elongated, bifurcated posterior apex of the anal abdominal segment and a truncated posterior apex of the flattened subgenital plate, in contrast to other females of the genus Paramenexenus that have a more robust body shape and a strongly elongate subgenital plate. Eggs further differentiate this species from its congeners: those of P. yangi have a pear-shaped capsule and an oblong micropylar plate, while the eggs of other Paramenexenus species are distinguished by an oval capsule and an elongate micropylar plate. Based on these morphological distinctions, Ho re-examined a female specimen collected from the type locality (viz. Wuzhishan Tropical Rainforest National Park, Hainan, China) and transferred P. yangi to a newly established genus, Pseudoparamenexenus [11].
With the development of next-generation sequencing technology, researchers have begun to utilize molecular data to study the phylogenetic relationships of the order Phasmatodea [12,13,14,15,16,17,18,19]. Insect mitochondrial genomes typically contain 37 genes, 14–20 kb in length, including 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and a control region (CR) [20,21,22,23,24].
The mitochondrial genome has emerged as an important focus of research due to its characteristics of rapid evolution, low recombination rates, and matrilineal inheritance [25,26,27,28,29,30]. Mitochondrial DNA has been extensively used in studies of insect molecular evolution, population genetics, phylogenetics, and species identification [31,32,33,34].
To date, Pseudoparamenexenus has been considered a monotypic genus of the family Lonchodidae. Although numerous mitochondrial genomes of stick insects have been sequenced and analyzed, the genus Pseudoparamenexenus remains uncharacterized in terms of its mitogenomic features and phylogenetic relationships. By analyzing mitochondrial genomes, this study aimed to resolve the phylogenetic position of the genus Pseudoparamenexenus within the family Lonchodidae and its related taxa. The results contribute to the taxonomic refinement of Lonchodidae while supporting further investigation into phasmid evolution, phylogenetic reconstruction, and conservation genetics. In this study, we describe Pseudoparamenexenus beiliuensis sp. nov. as a new species. We present the first two sequences of the mitogenome of the genus Pseudoparamenexenus, along with a preliminary comparative analysis of its genetic composition and structural features. Finally, to clarify the taxonomic status of the genus Pseudoparamenexenus, viz. the new species and P. yangi, we conducted a phylogenetic analysis of 36 mitochondrial genomes of Necrosciinae (including two newly sequenced and 34 previously published) and 2 outgroups. These integrated morphological and molecular approaches not only advance our systematic understanding of Phasmatodea diversity but also establish a solid framework for future evolutionary studies of these ecologically important insects.
2. Materials and Methods
2.1. Sample Collection and Photography
Stick insects were collected by netting at night and preserved in absolute alcohol at Guangxi Normal University, Guilin, China. All the specimens were examined using a Nikon (Nikon Corporation, Tokyo, Japan) SMZ745T stereomicroscope and photographed with a Nikon (Nikon Corporation, Tokyo, Japan) D7200 with a 105-mm f/2.8 G IF–ED macro lens of Nikon (Nikon Corporation, Tokyo, Japan). The photographs were processed in Photoshop CS 2018 (Adobe Inc., San Jose, CA, USA).
2.2. Sequencing and Mitogenome Assembly
Total genomic DNA was extracted from the hind legs of adult specimens using a TIANamp Genomic DNA Kit (TIANGEN, Beijing, China), and then high-throughput sequenced using 150-bp PE on the Illumina NovaSeq platform (Berry Genomics, Beijing, China) and the DNBSEQ platform (Shenzhen BGI Genomics Co., Ltd., Shenzhen, China). The high-quality clean reads were compared with whole mitochondrial genome sequences of closely related species in the NCBI database using the CLC Genomics Workbench 12 [35] to identify the most closely related species (Neohirasea stephanus OL405132 [36]), which was used as a reference. The mitochondrial genome sequences were assembled using NOVOPlasty v.4.2.1 [37] and annotated by the MITOS2 web server based on the Galaxy platform [38]. A circular map of the mitochondrial genome was visualized using Chloroplot [39].
2.3. Mitogenome Annotation and Characteristics Analysis
The nucleotide composition, composition skew, codon usage of PCGs, relative synonymous codon usage (RSCU), and mitogenomic organization tables were obtained using PhyloSuite v.1.2.3 [40]. Synonymous (Ks) and non-synonymous (Ka) substitution rates were calculated for the PCGs of P. beiliusis using DnaSP 6.0 [41]. The sequence heterogeneity within datasets was analyzed by using AliGROOVE [42], with the default sliding window size. A saturation test was performed using DAMBE 7 [43]. Saturation was based on the values of Iss (simple index of substitution saturation) and Iss.c (critical Iss value), with Iss < Iss.c indicating that the genetic marker was not saturated.
2.4. Construction of Phylogenetic Trees
For the phylogenetic analysis, we utilized 36 stick insect mitochondrial genomes as the ingroup, including 34 publicly available mitogenomes of Lonchodidae from the GenBank database and two newly sequenced genomes obtained in this study (Table S1). Two mitochondrial genomes from closely related taxa (family Bacillidae) were selected as outgroups. We aligned 13 PCGs and two ribosomal RNA genes (12S and 16S rRNA) to construct four distinct datasets: PCG123 (all codon positions, 11,059 bp), PCG123 + 2R (all codon positions + 2 rRNA, 12, 625 bp), PCG12 (first and second codon positions, 7372 bp), and PCG12 + 2R (PCG12 + 2 rRNA, 8, 938 bp). The most suitable dataset for phylogenetic reconstruction was subsequently selected through comparative analyses (Table S2). We aligned the mitochondrial genome sequences using MEGA 11 [44] and concatenated the resulting alignments with SEQUENCEMATRIX v.1.7.8 [45]. PhyloSuite v.1.2.3 was used to construct phylogenetic trees using the Bayesian inference (BI) and maximum likelihood (ML) methods [40]. For the BI tree, GTR + F + I + G4 [46] was used as the best substitution model, and the tree was reconstructed by the Markov chain Monte Carlo method in MrBayes 3.2.7 [40], with 2 million generations, sampling 1000 times, and discarding the first 25% of generations as a burn-in. For the ML analysis, IQ-TREE v2.2.0 was used with the best substitution model of GTR + F + I + G4 and 1000 bootstrap replicates [40]. The resulting evolutionary trees were visualized using the Interactive Tree Of Life (iTOL) (https://itol.embl.de/) [47].
3. Taxonomy
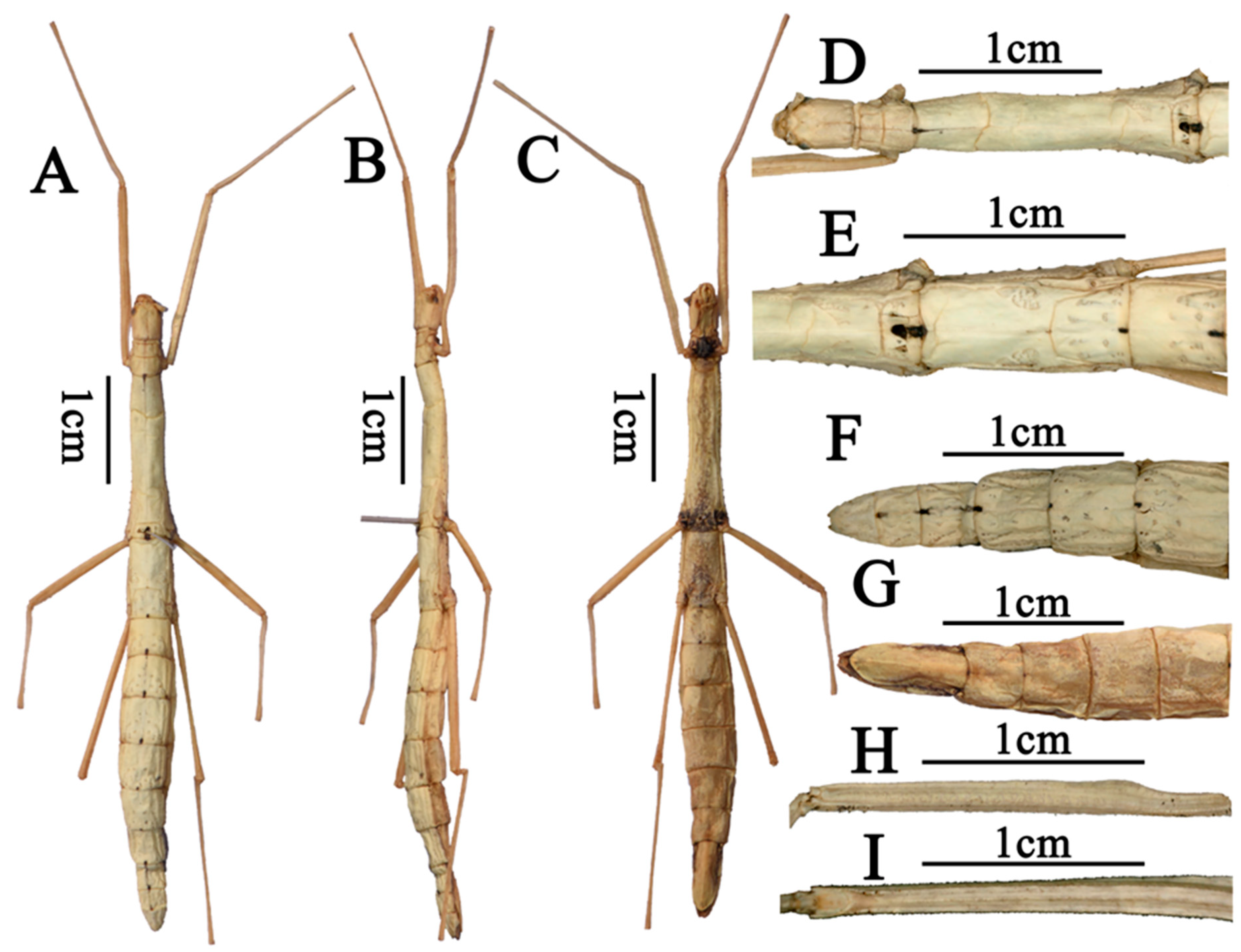
- Order: Phasmatodea Leach, 1815Family: Lonchodidae Brunner von Wattenwyl, 1893Subfamily: Necrosciinae Brunner von Wattenwyl, 1893Tribe: Necrosciini Brunner von Wattenwyl, 1893Genus: Pseudoparamenexenus Ho, 2016 [11]Species: Pseudoparamenexenus beiliuensis sp. nov.Diagnosis. Medium-sized Necrosciinae. Female apterous with scale-like wing rudiments. Spineless, female with few short tubercles on the thorax laterally and male unarmed. Head oval, with four to six swellings on the posterior margin. Vertex flat. Pronotum rectangular. Mesonotum constricted at the anterior region, moderately swollen pre-medially and slightly narrowing in posterior half in female; Abdomen smooth, medially and laterally carinate. Subgenital plate scoop-shaped, posterior margin truncate, surpassing the midlength of anal segment. Legs lacking distinct armature. Pseudoparamenexenus beiliuensis sp. nov. is closely related to Pseudoparamenexenus yangi (Chen and He, 2002) [10] (Figure 1), but can be distinguished by the following characters: mid-ventral carina of the femora lacking minute spines at the apex; female anal segment longer than tergum IX, sparsely covered with short bristles, posterior margin nearly truncate, slightly concave medially. In contrast, the female of Pseudoparamenexenus yangi has a strongly elongated and bifurcated posterior apex of the anal abdominal segment, along with a flattened subgenital plate that has a truncate posterior apex (Figure 1E–G).Tpye material. Holotype: ♀, China, Guangxi, Darongshan, Beiliu, alt. 930 m, 5 August 2022, coll. Shan Li and Qianwen Zhang.Etymology. The new species is named after the type locality (Beiliu, Guangxi Zhuang Autonomous Region).Description. Female. Mid-sized. Body rod-shaped, ash-white (Figure 2A–C).
Head. Ash-white, oblong, longer than wide, nearly as long as the pronotum. Occiput flat. Compound eyes prominent, strongly hemispherical projecting. Ocelli lacking. Antennal sockets distinct and well-defined (Figure 2D); scape prominent, subtriangular in cross-section; pedicel shorter than scape, rice-shaped; antennae filiform, extending to tergum V, with a distinct median longitudinal furrow extending to the apex of the abdomen (Figure 2D).
Thorax. Pronotum with a distinct transverse sulcus anteriorly, flanked by longitudinal sulci extending to the midlength of the pronotum; a distinct cruciform sulcus centrally located (Figure 2D); transverse sulcus complete, reaching the lateral margins of the pronotum. Defensive gland openings visible at the lateral front margins of the pronotum. Mesonotum subtrapezoidal, with a distinct arcuate transverse line at the anterior quarter (Figure 2D); posteriorly expanded and widened; lateral margins with irregularly arranged fine denticles. Median longitudinal carina at the junction of the pronotum and mesonotum bears black marking, and the median longitudinal furrow with irregular rugae. No wing remnants of the mesonotum without wings. Metanotum nearly rectangular and bears scale-like wing rudiments (Figure 2E). Median segment prominent, approximately one-third the length of the metanotum. Metanotum lateral margins are lined with irregularly arranged fine teeth. Surface of median segment scattered with small pits.
Abdomen. Each abdominal segment bears a black marking at its posterior margin. Tergum II–VI with scattered depressions at posterior ends. Abdominal tergum III longer than II, tergum IV longer than III (Figure 2F,G), and tergum V shorter than IV. The anal segment longer than tergum IX, covered with setae; its posterior margin nearly truncate, slightly concave centrally. Subgenital plate scoop-shaped, posterior margin truncate, not extending beyond the apex of the anal segment. Cerci cylindrical, tapering to a pointed apex, densely covered with fine setae, slightly surpassing the posterior margin of the anal segment.
Legs. Slender. Femora slightly longer than the corresponding tibiae. Tibia have no area apicalis. Profemora distinctly curved basally, inner and outer ridges of all femora tipped with two or three small spines, mid-ventral carina of the femora lacking minute spines at the apex (Figure 2H,I).
- Male. Unknown.Eggs. Unknown.Pseudoparamenexenus yangi (Chen and He, 2002) [10]
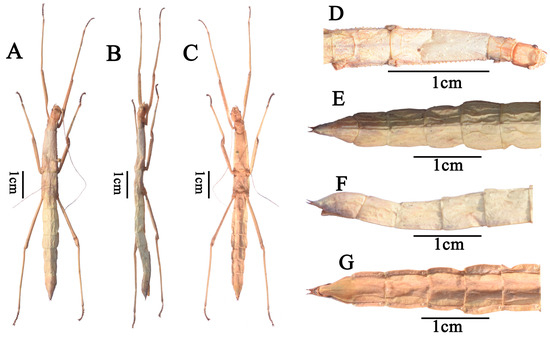
Figure 1.
Pseudoparamenexenus yangi, female. (A). habitus, dorsal view; (B). habitus, lateral view; (C). habitus, ventral view; (D). head and thorax, dorsal view; (E). apex of abdomen, cerci, dorsal view; (F). apex of abdomen, cerci, lateral view; (G). apex of abdomen, cerci, ventral view. Scale bars = 1 cm.
Figure 1.
Pseudoparamenexenus yangi, female. (A). habitus, dorsal view; (B). habitus, lateral view; (C). habitus, ventral view; (D). head and thorax, dorsal view; (E). apex of abdomen, cerci, dorsal view; (F). apex of abdomen, cerci, lateral view; (G). apex of abdomen, cerci, ventral view. Scale bars = 1 cm.
- Material examined. 1♀, China, Hainan, Wuzhishan, alt. 784 m, 18 May 2024, coll. Yanting Qin and Yizhen Yao. 2♀, China, Hainan, Ledong, Jianfengling, alt. 1268 m, 28 May 2024, coll. Yanting Qin and Yizhen Yao. 2♀ China, Hainan, Changjiang, Bawangling, alt. 508 m, 28 May 2024, coll. Yanting Qin and Yizhen Yao.
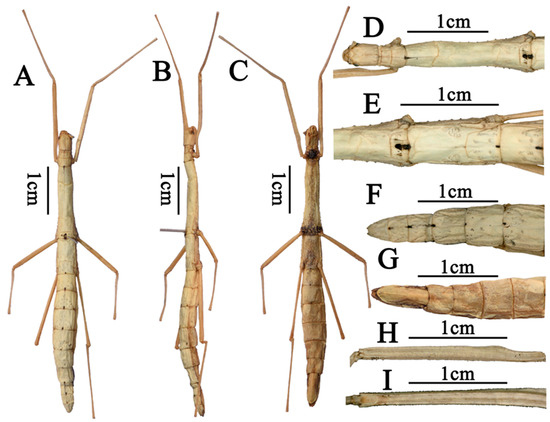
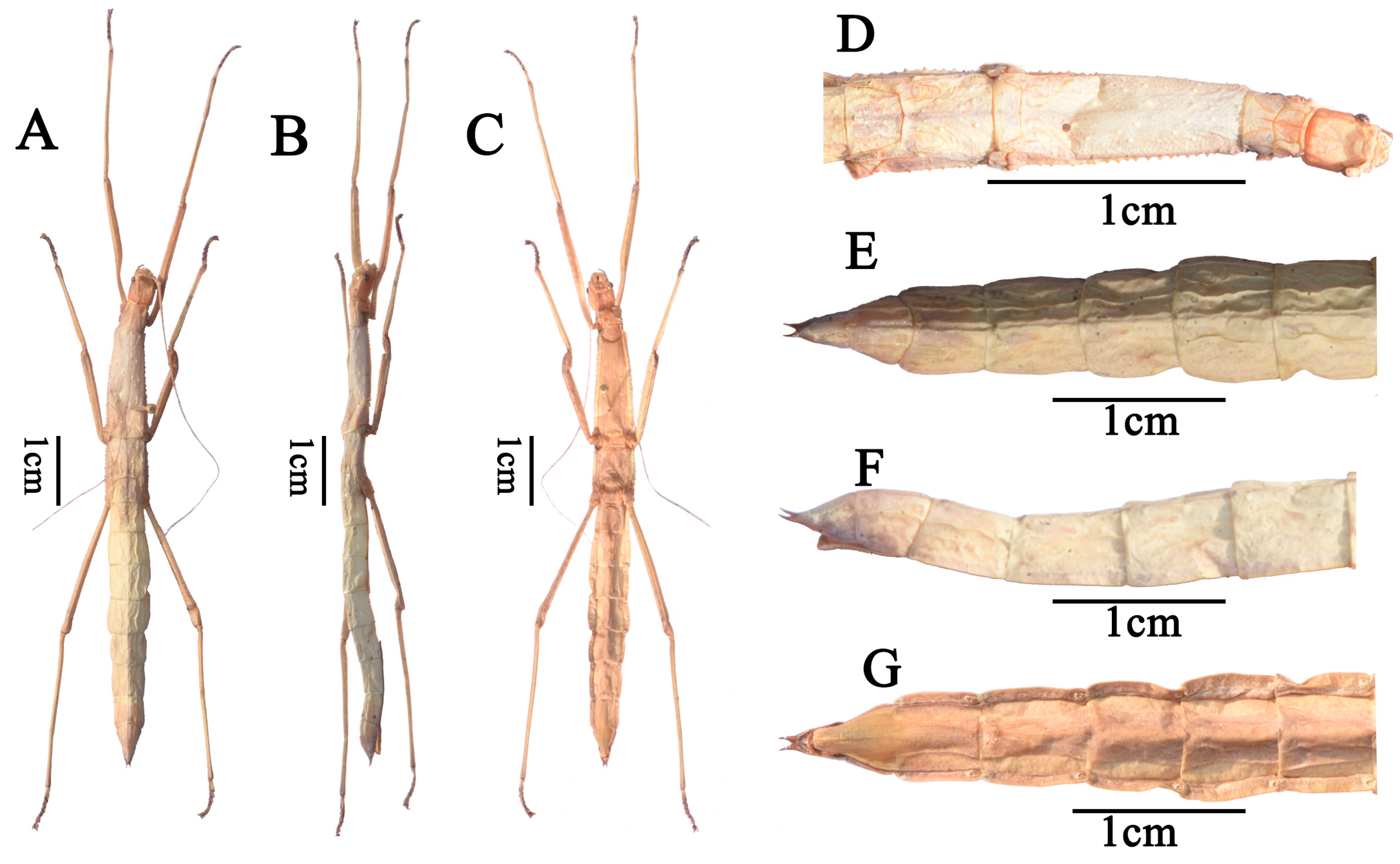
Figure 2.
Pseudoparamenexenus beiliuensis sp. nov., holotype, female. (A). habitus, dorsal view, holotype, female; (B). habitus, lateral view, holotype, female; (C). habitus, ventral view, holotype, female; (D). Head, pro– and mesothorax, dorsal view, holotype, female; (E). meso– and metathorax, dorsal view, holotype, female; (F). apex of abdomen, cerci, dorsal view, holotype, female; (G). apex of abdomen, cerci, ventral view, holotype, female; (H). profemora, dorsal view, holotype, female; (I). profemora, lateral view holotype, female. Scale bars = 1 cm.
Figure 2.
Pseudoparamenexenus beiliuensis sp. nov., holotype, female. (A). habitus, dorsal view, holotype, female; (B). habitus, lateral view, holotype, female; (C). habitus, ventral view, holotype, female; (D). Head, pro– and mesothorax, dorsal view, holotype, female; (E). meso– and metathorax, dorsal view, holotype, female; (F). apex of abdomen, cerci, dorsal view, holotype, female; (G). apex of abdomen, cerci, ventral view, holotype, female; (H). profemora, dorsal view, holotype, female; (I). profemora, lateral view holotype, female. Scale bars = 1 cm.
- Measurements. Body 71–78 mm; head 3.5–4 mm; antennae 73–80 mm; pronotum 3.5–4 mm; mesonotum 17–20 mm; metanotum 6.5–7 mm; median segment 3 mm; profemora 22–25 mm; mesofemora 15 mm; metafemora 22–25 mm; protibiae 23–24 mm; mesotibiae 14–15 mm; metatibiae 24–29 mm.
4. Results
4.1. Characteristics of Pseudoparamenexenus beiliuensis sp. nov. and Pseudoparamenexenus yangi Mitochondrial Genomes
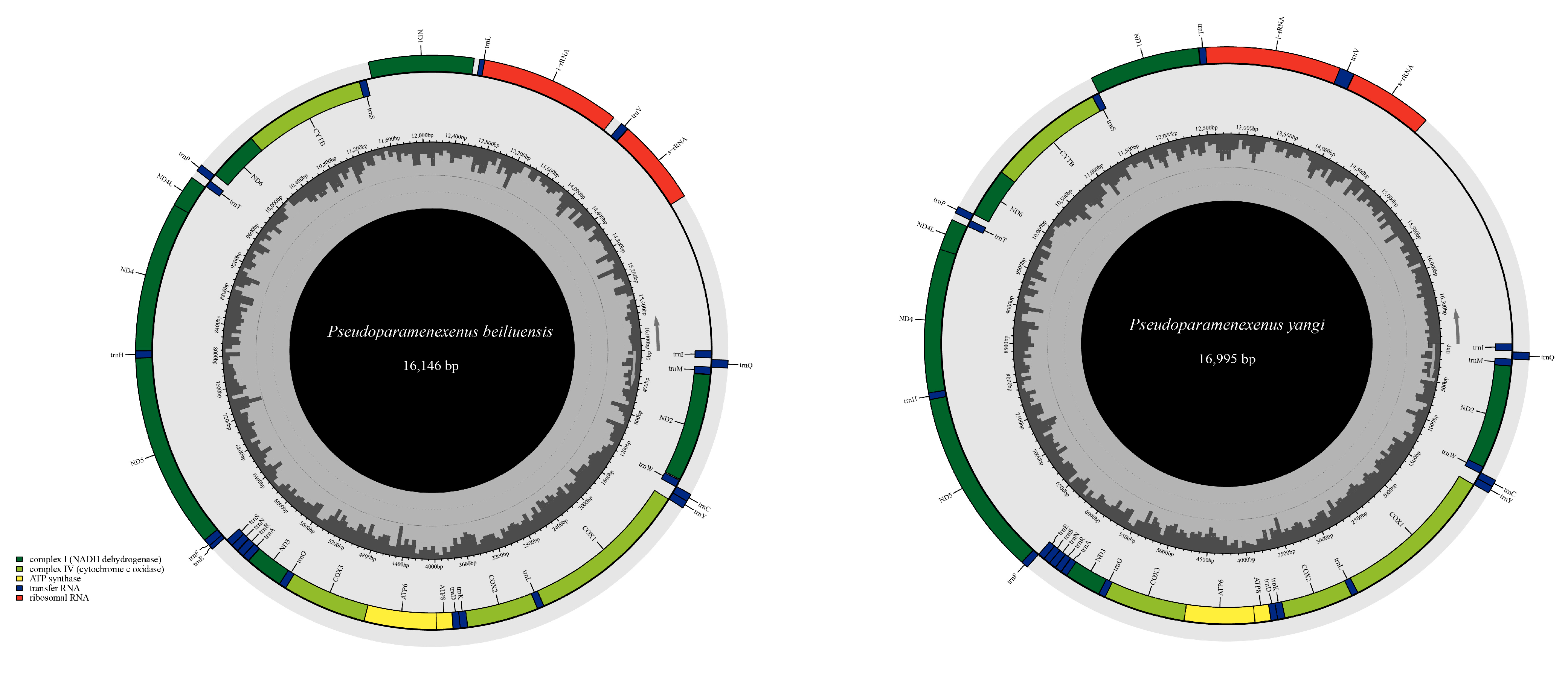
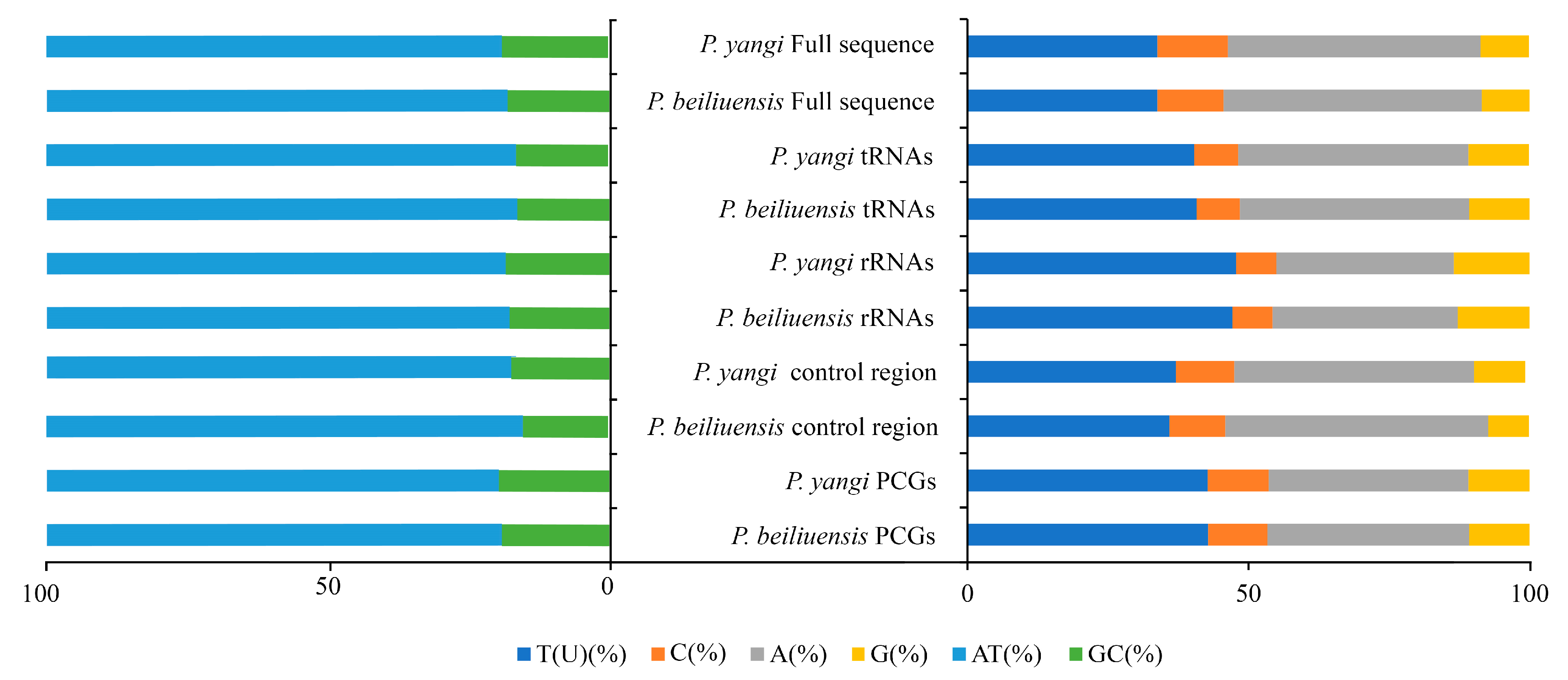
Due to incomplete annotation of the control region in the mitochondrial genome attributed to the partial deletion of the control region, the obtained total lengths of the mitochondrial genomes of P. beiliuensis sp. nov. (Table S3) and P. yangi (Table S4) are 16,146 bp and 16,995 bp, respectively (Figure 3). The mitochondrial genomes of both species exhibit a notably high AT content (Figure 4). The total length of the 13 PCGs of P. beiliuensis sp. nov. is 11,058 bp, with 78.7% AT content, while that of P. yangi spans 11,166 bp, with marginally lower AT richness (78.2%). The typical 22 tRNA genes maintain comparable AT content (81.5% and 81.3%) but differ in total length (1499 bp and 1531 bp) for P. beiliuensis sp. nov. and P. yangi, respectively. The two rRNA components demonstrates species-specific variation, with P. beiliuensis sp. nov. possessing a 1237 bp 16S rRNA (80.0% AT content) and 764-bp 12S rRNA (80.1% AT content), while P. yangi has a longer 16S rRNA (1289 bp and 80.6% AT content) and shorter 12S rRNA (753 bp and 78.3% AT content). Both species share typical ATN initiation codons for all PCGs: P. beiliuensis sp. nov.: ATG, ATT, ATA, and ATC; P. yangi: ATG, ATT, and ATA. Except for the NAD3 and NAD5 genes of P. yangi, which use TAG as stop codons, the other PCGs are terminated with TAA.
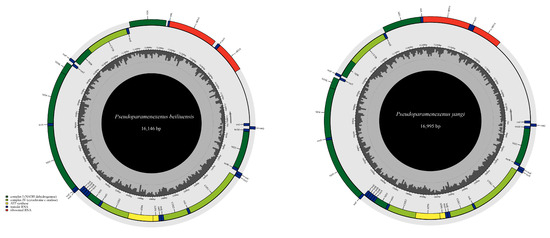
Figure 3.
Complete mitochondrial genome map of P. beiliuensis sp. nov. and P. yangi, with 13 PCGs, 22 transfer RNAs (tRNAs), and two ribosomal RNAs (rRNAs).
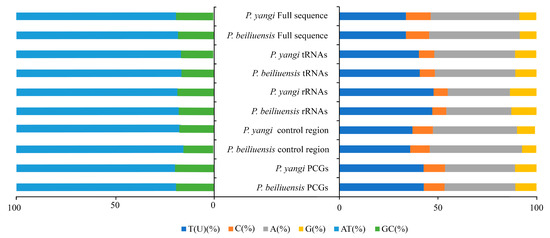
Figure 4.
Comparative analysis of nucleotide composition in mitochondrial functional regions between P. yangi and P. beiliuensis sp. nov. Horizontal axis: mitogenomic components; vertical axis: relative proportion (%).
4.2. Comparative Analysis
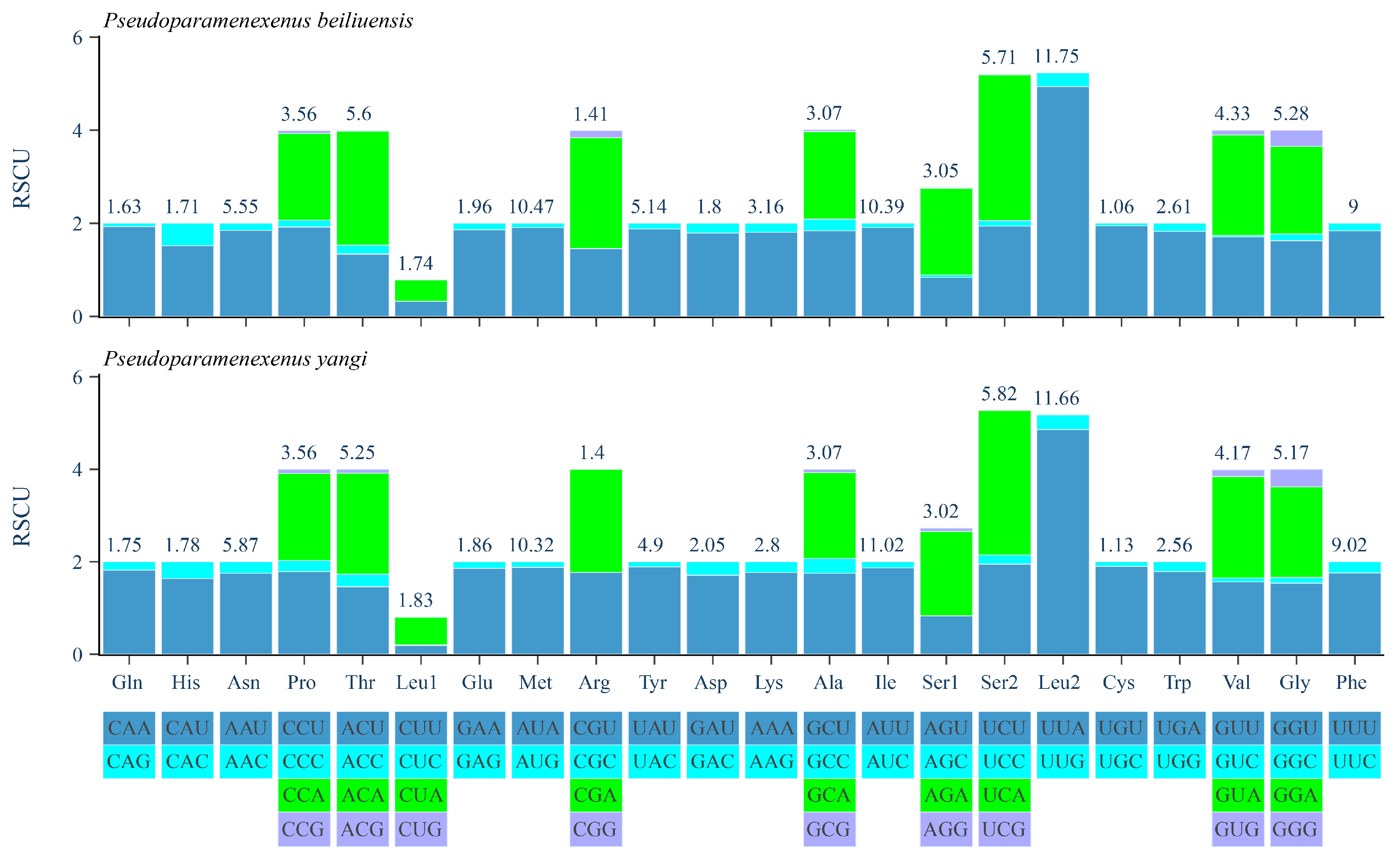
The relative synonymous codon usage (RSCU) analysis of the PCGs of P. beiliuensis sp. nov. and P. yangi revealed a pronounced AT bias, with the five most frequent codons being AAU, AUA, AUU, UUA, and UUU (Figure 5). Among these, leucine (Leu), isoleucine (Ile), and methionine (Met) are the most highly represented amino acids, while cysteine (Cys) and arginine (Arg) are the least common (Figure 5).
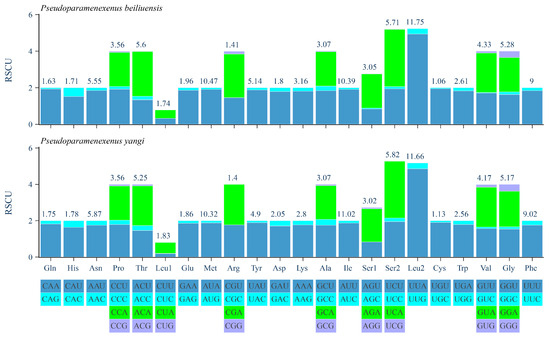
Figure 5.
Relative synonymous codon usage (RSCU) of the PCGs in the P. beiliuensis sp. nov. and P. yangi mitogenomes.
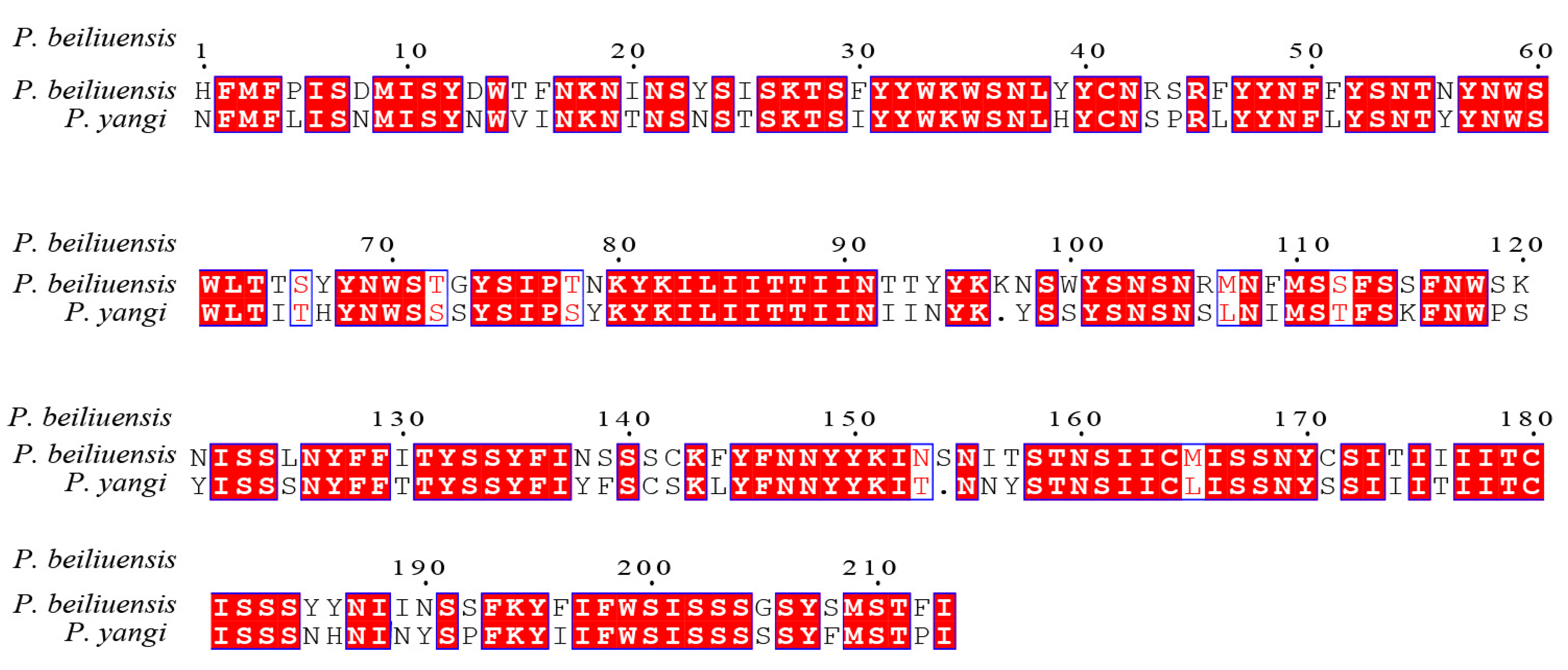
In previous studies [48,49,50,51], the standard 658-base fragment of the 5′ end of mitochondrial gene cytochrome c oxidase subunit I (COI) has been identified as one of the most evolutionarily conserved mitochondrial genes; thus, it is widely employed as a molecular barcode for species identification and phylogenetic inference. To assess the sequence divergence between P. beiliuensis sp. nov. and P. yangi, their COI barcodes were aligned and compared with homologous sequences retrieved from the NCBI database [52]. Following length optimization to ensure comparability, the pairwise sequence similarity was calculated, revealing 88.8% identity between the two species (Figure 6). Genetic distance analysis further supported this divergence, with a Kimura 2-parameter (K2P) distance of 0.115, consistent with the interspecific differentiation observed in related taxa [52,53,54].
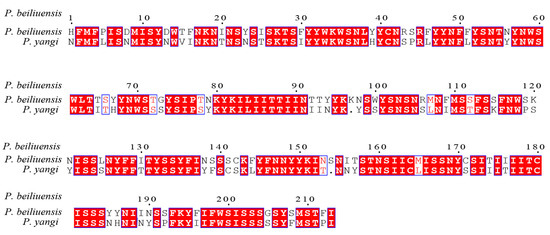
Figure 6.
Sequence similarity of the amino acid of P. beiliuensis sp. nov. and P. yangi COX1 sequences.
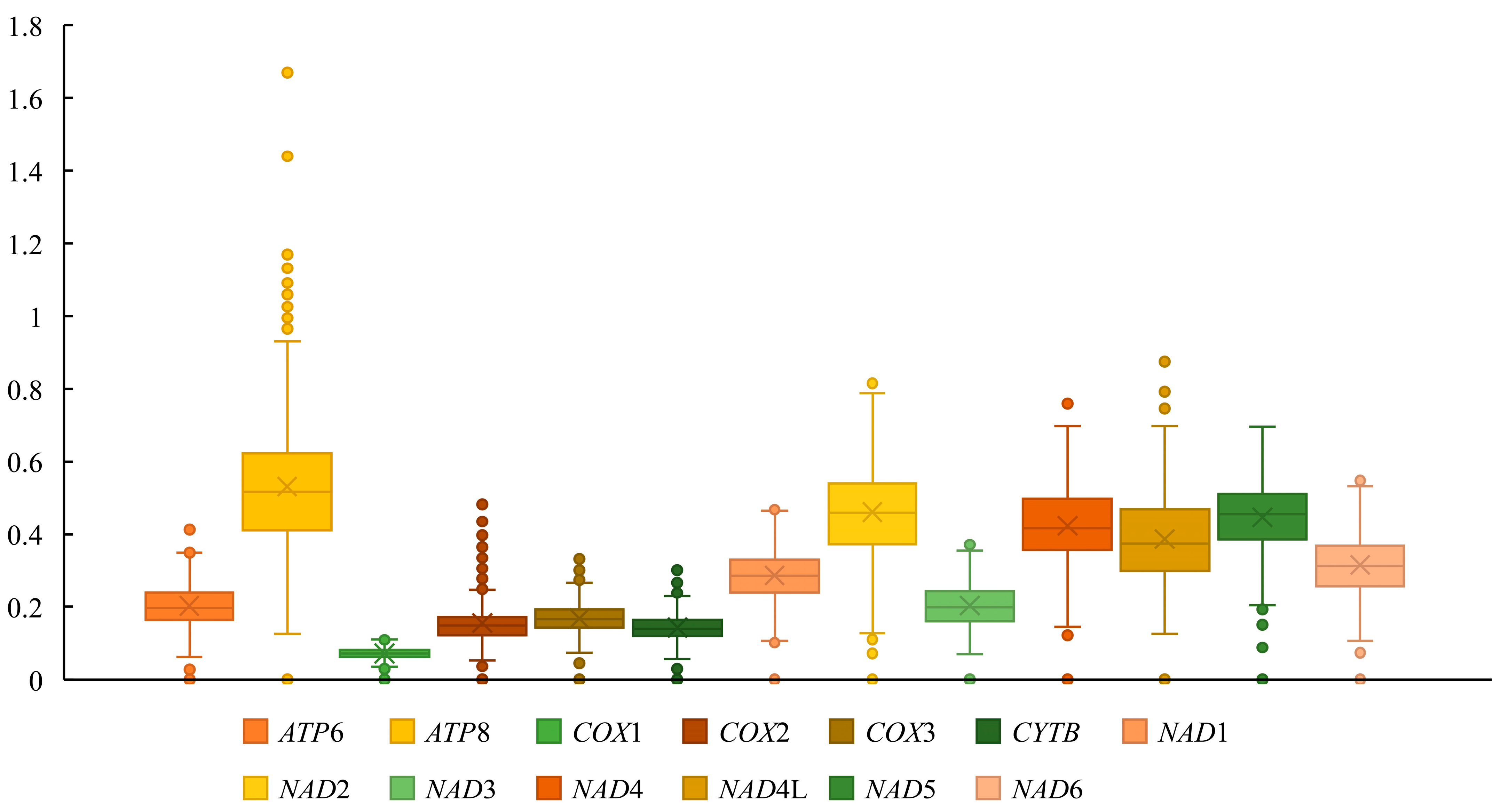
We conducted a Ka/Ks analysis on the 13 PCGs to investigate potential differences in selection pressures acting on these genes during evolution (Figure 7). All 13 PCGs have Ka/Ks values less than 1 and the ATP8 gene displays the highest Ka/Ks ratio (0.489) while COX1 has the lowest ratio (0.071).
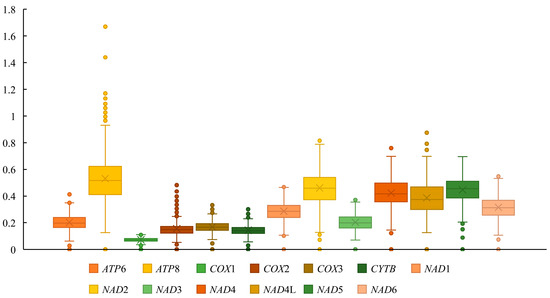
Figure 7.
The ratio of non-synonymous to synonymous substitutions (Ka/Ks) of the 13 PCGs of 36 Lonchodidae mitogenomes.
4.3. Phylogenetic Analysis
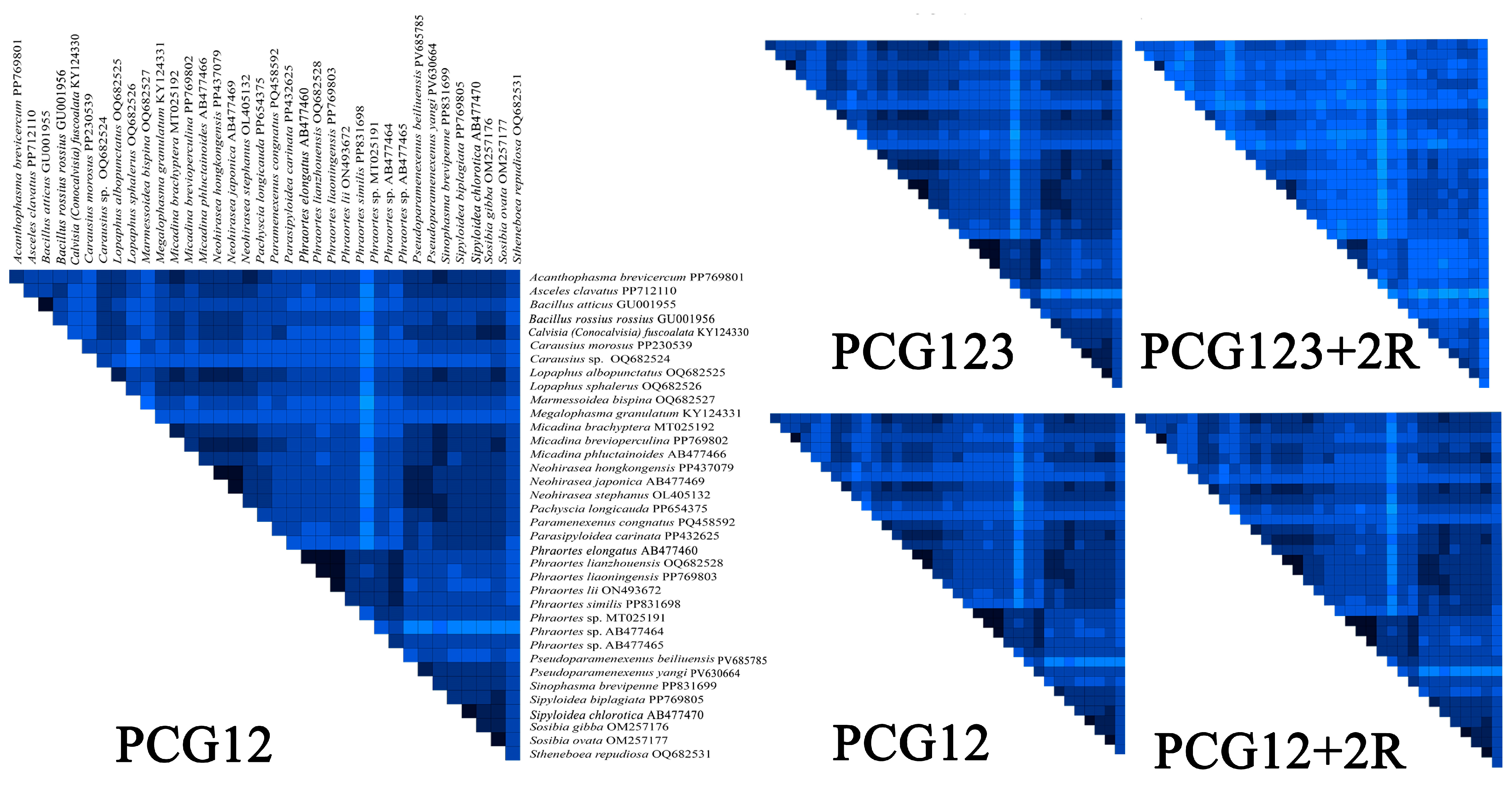
The comparative analysis identified distinct heterogeneity patterns across the datasets. The PCG12 and PCG12 + 2R (PCG12 with 2 rRNA genes) datasets exhibited low heterogeneity in most pairwise comparisons, while the PCG12 dataset (first and second codon positions) showed significantly greater heterogeneity (Figure 8). Therefore, we ultimately chose the PCG12 dataset to construct a phylogenetic tree. In addition, by examining the nucleotide substitution saturation of all 13 mitochondrial PCGs, it was shown that the PCGs substitution saturation test Iss < Iss.c indicated that the sequence substitution was not saturated and that the datasets were suitable for phylogenetic analysis (Figure S1).
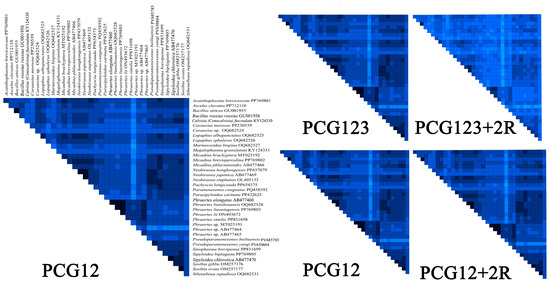
Figure 8.
AliGROOVE analysis of various datasets. Heatmap showing mean pairwise similarity scores for four datasets: PCG12, PCG12 + 2R, PCG123, and PCG123 + 2R. The PCG12 dataset was selected based on its highest mean similarity score.
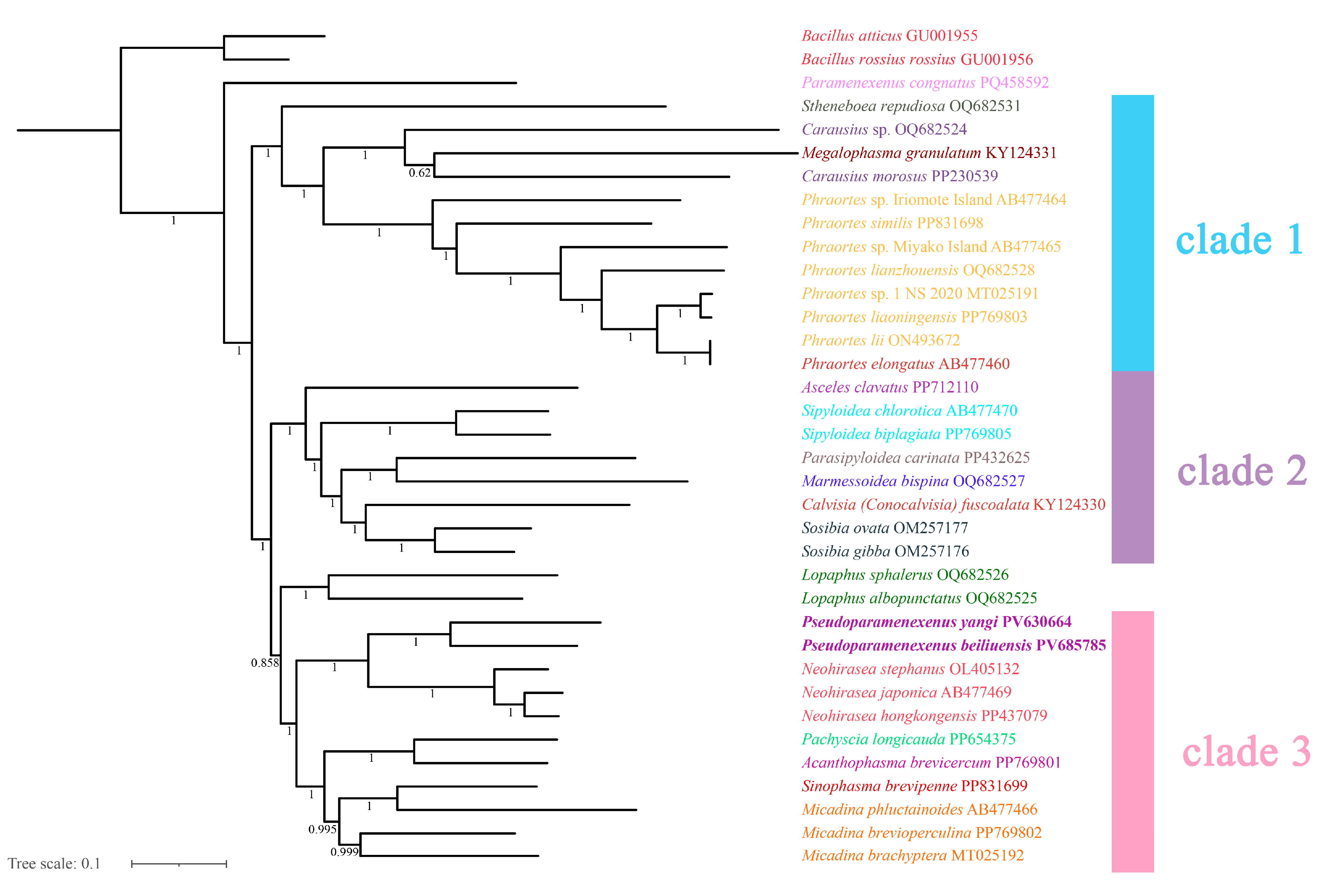
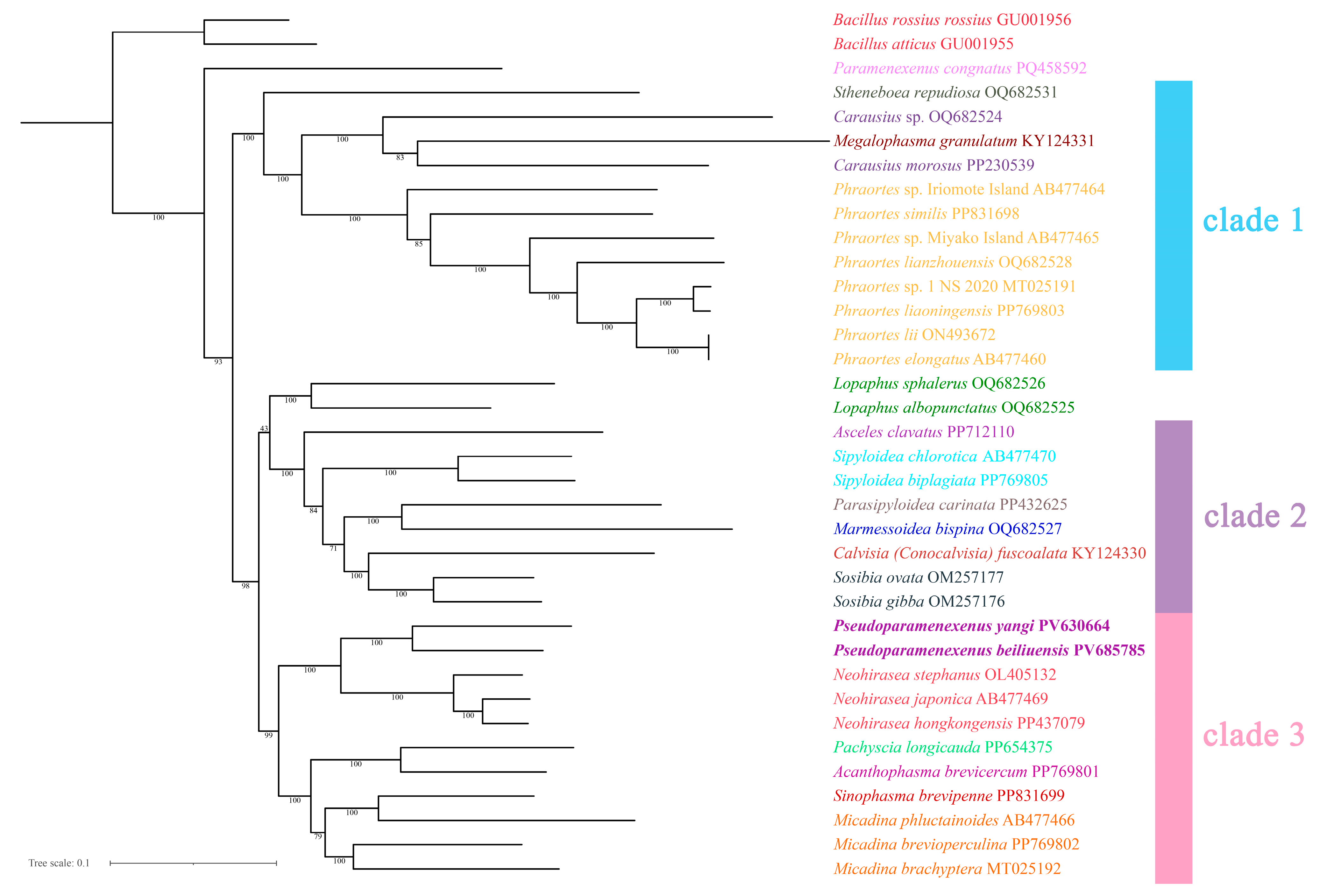
The topologies inferred using the two tree-building methods are largely congruent (Figure 9 and Figure 10), with the monophyly of Lonchodinae consistently recovered. Necrosciinae was not supported as monophyletic, and its members were resolved into two distinct clades. The primary topological discrepancy between methods involved the placement of the genus Lopaphus: in the BI tree (Figure 9), two species of the genus were recovered as the sister group to clade 3, which was further resolved with high support. In addition, the BI clade 3 structure resolved the genera Pseudoparamenexenus and Neohirasea as sister genera that together formed a sister group to a well-defined subclade containing Pachyscia longicauda, Acanthophasma brevicercum, Sinophasma brevipenne, and three species of the genus Micadina (namely M. phluctainoides, M. brevioperculina, and M. brachyptera), with the following internal structure: (Pachyscia longicauda + Acanthophasma brevicercum + (Sinophasma brevipenne + Micadina phluctainoides + (Micadina brevioperculina + Micadina brachyptera))).
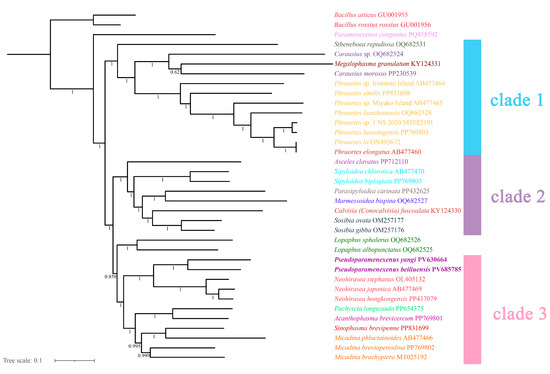
Figure 9.
The phylogenetic tree obtained by Bayesian inference based on the PCG12 dataset (including the first and second codon positions; 7372-bp sequences), with the numbers on the branches indicating bootstrap percentages.
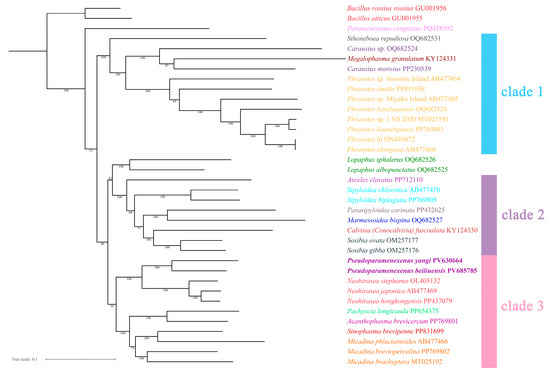
Figure 10.
The phylogenetic tree obtained by maximum likelihood ML based on the PCG12 dataset (including the first and second codon positions; 7372-bp sequences), with numbers on the branches indicating bootstrap percentages.
In the ML tree (Figure 10), the genus Pseudoparamenexenus remained consistent with clade 3 as described above. However, the placement of the genus Lopaphus differed, as follows: (Lopaphus sphalerus + Lopaphus albopunctatus) + (Asceles clavatus + ((Sipyloidea chlorotica + Sipyloidea biplagiata) + ((Parasipyloidea carinata + Marmessoidea bispina) + (Calvisia (Conocalvisia) fuscoalata + (Sosibia ovata + Sosibia gibba))))). This arrangement highlights the divergent topologies between the ML and BI trees, underscoring the complexity of the evolutionary relationships within these taxa.
5. Discussion
In this study, we report the sequence and analyze the mitochondrial genome of the stick insect genus Pseudoparamenexenus for the first time. In examining the start and stop codons of the 13 PCGs, we found that most PCGs in the genus Pseudoparamenexenus begin with standard ATN codons (ATG, ATT, ATA, and ATC). ATN is the recognized canonical start codon for insect mitochondrial genomes. Among stick insects having ATN as the start codon, ATA, ATG, and ATT predominate, with only a minority employing ATC [13,55,56,57]. All PCGs are terminated by TAN stop codons (TAA or TAG).
During the RSCU analysis, we observed significantly high encoding of methionine (Met), isoleucine (Ile), and leucine (Leu) in the mitochondrial genomes of the genus Pseudoparamenexenus. The high-frequency occurrence of codons for these amino acids aligns with the known coding patterns observed in other mitochondrial genomes of the order Phasmatodea [57,58,59,60]. Furthermore, our study revealed that the mitochondrial genomes of the genus Pseudoparamenexenus exhibit an AT basis, a characteristic commonly found in insect mitogenomes [61,62]. These findings underscore the conserved nature of codon usage biases and genomic composition in the evolution of mitogenomes within insects [63,64,65].
The Lonchodidae, the most species-rich group referred to as a family within Phasmatodea, exhibits persistent taxonomic uncertainties that demand systematic revision, particularly following recent species discoveries [2,66,67]. The genus Pseudoparamenexenus exhibits closer morphological similarities to the genus Paramenexenus species in overall body structure [11]. Morphological similarities include: a tubercle on the occiput, small spines on the body sides, longitudinal rows of sparse granular protrusions; fine teeth in longitudinal rows on the lateral plates of the meso- and metathorax; a slender abdomen with multiple longitudinal ridges, scale-like wing remnants, small pits scattered on the surface of the median segment, and short, pointed cerci.
Phylogenetic analyses provided some support for the monophyly of the genus Pseudoparamenexenus and suggested a possible sister relationship with the genus Neohirase, with both genera sharing derived traits: medium-sized Necrosciinae, Body cylindrical, elongate. Head oval, posterior margin with swellings. Pronotum rectangular; mesonotum medially elevated. Subgenital plate scoop-shaped, posterior margin truncate, reaching the middle of anal segment; supra-anal plate indistinct; cerci short and flattened. femora thicker than tibiae, lacking a distinct armature. Apterous, with scale-like rudiments. capsule oval, micropylar plate oval. The genus Neohirasea is diagnostically characterized by a mesonotal bi-spined hump, subapical femoral serrations, and a female praeopercular organ [67], whereas the genus Pseudoparamenexenus exhibits male-specific unarmed nota with a single femoral spine and has females with bifurcate anal segments and truncate subgenital plates. During our examination of the egg morphology in the genus Pseudoparamenexenus and the genus Neohirasea, we observed notable similarities, specifically the brown-shaded egg capsules, rounded or oval micropylar plate, indistinct median line, and operculum with a distinctly elevated central area [11,67]. Growing evidence [68,69,70] suggests that egg morphology in stick insects (Phasmatodea) is closely associated with their oviposition strategies, providing valuable taxonomic characters for species identification and classification. These distinct morphological adaptations not only reflect ecological specialization but also serve as reliable diagnostic traits in stick insect taxonomy. Traditional classification heavily relies on adult morphology, yet cryptic speciation and sexual dimorphism often complicate identification. Egg characteristics, however, remain consistent within species and can help resolve taxonomic ambiguities, particularly in poorly studied or morphologically similar groups [5,69,70,71,72,73]. Previous studies have reported incongruences between morphological characteristics and molecular phylogenies across various phasmid groups. Our phylogenetic analyses revealed a significant repositioning of the genus Pseudoparamenexenus, the genus Paramenexenus, and the genus Neohirasea compared to previous studies [8,68]. While earlier research suggested a closer relationship between the genus Paramenexenus and the genus Neohirasea, our data robustly support a sister-group relationship between the genus Pseudoparamenexenus and the genus Neohirasea (BP = 100/PP = 1.0). This incongruence may be attributed to the limited taxon sampling currently available for the genus Pseudoparamenexenus and the genus Paramenexenus, which can constrain the analytical resolution and lead to potential biases in the phylogenetic reconstruction. To address these limitations and provide a more comprehensive understanding of the intergeneric relationships within this clade, future studies should incorporate: Expanded taxonomic sampling across their biogeographic ranges, multi-locus data integration using both mitochondrial (COI, 16S rRNA) and nuclear (28S rRNA, H3) markers. Such an approach will not only enhance the robustness of the phylogenetic hypotheses but also facilitate a more nuanced understanding of the evolutionary history and diversification patterns of these genera. Additionally, incorporating morphological and ecological data could further elucidate the factors driving the observed phylogenetic relationships and provide insights into the adaptive radiation of these taxa.
In our phylogenetic tree, the three species of Micadina (Micadina phluctainoides, Micadina brevioperculina, and Micadina brachyptera) formed a monophyletic group with Sinophasma brevipenne, in contrast to previous studies [5,13,66,70,74]. Specifically, Megalophasma granulatum is nested between two species of this genus, which differs from the results of Chen et al. [74], who supported the relationship (Megalophasma granulatum + (Carausius morosus + Carausius sp.)). However, all species in the genus Phraortes are clustered in our phylogenetic tree, consistent with the findings of Chen et al. [74]. indicating a well-supported monophyly for this genus. Phylogenetic ambiguity surrounding the genus Lopaphus persisted in our ML reconstructions (bootstrap support <70%), whereas the BI frameworks resolved its placement with greater reliability, likely due to improved modeling of site heterogeneity and branch-length parameters [65,66,67,68,69,70,71,72,73,74,75,76,77]. In summary, our phylogenetic tree has discrepancies with prior studies in the classification and relationships of several genera [13,19,36,66,70,77,78], particularly Carausius and Micadina. Additionally, the genus Micadina remains a contentious topic. Given the low support values for the relevant branches, we recommend incorporating additional molecular markers, increasing the sample size, or employing alternative phylogenetic methods to corroborate these findings. For example, subsequent research could incorporate nuclear gene marker sequencing and comparative transcriptomic analyses.
6. Conclusions
Based on integrated morphological and molecular evidence, we herein describe Pseudoparamenexenus beiliuensis sp. nov. as a new species that expands the known diversity of Phasmatodea and provides novel insights into the evolutionary relationships within this order. The mitogenome of P. beiliuensis sp. nov. exhibited highly conserved composition, arrangement, and structure, maintaining ancestral genomic features consistent with those of basal phasmid lineages. Phylogenetic analyses provided some support for the monophyly of the genus Pseudoparamenexenus and suggested a possible sister relationship with the genus Neohirasea. However, due to the limited taxon sampling, these relationships should require further validation with additional data. suggesting shared evolutionary trajectories that warrant further investigation. This study highlights the importance of integrating morphological and molecular data in taxonomic research. Such integrative methodologies not only enhance the delimitation accuracy of species delimitation but also contribute to a more comprehensive understanding of stick insect biodiversity and evolutionary patterns.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d17090637/s1, Table S1. List of samples included in phylogenetic analysis; Table S2. The composition and lengths of the various datasets; Table S3. Mitochondrial genome content, organization, and codon information of P. beiliuensis sp. nov.; Table S4. Mitochondrial genome content, organization, and codon information of P. yangi.; Figure S1. Nucleotide substitution saturation plots of all 13 mitochondrial protein-coding genes. Saturation plot for transitions (green) and transversions (blue).
Author Contributions
Methodology, Y.Q.; Formal analysis, Y.Q.; Resources, X.B.; Writing—original draft, Y.Q.; Writing—review and editing, Y.Q. and X.B.; Supervision, Z.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (No. 32360126), Innovation Project of Guangxi Graduate Education (XYCS2025109, XYCS2025110 and XYCSR2023019).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
All data supporting this study are included in the article and its supplementary materials. The complete mitochondrial genomes of two species have been deposited in NCBI GenBank under accession numbers PV685785 (Pseudoparamenexenus beiliuensis sp. nov.) and PV630664 (Pseudoparamenexenus yangi).
Acknowledgments
We sincerely thank the reviewers for their constructive comments and gratefully acknowledge all contributors who facilitated the publication of this work.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- Bradler, S.; Buckley, T.R. Biodiversity of phasmatodea. Insect Biodivers. Sci. Soc. 2018, 2, 281–313. [Google Scholar] [CrossRef]
- Brock, P.D.; Büscher, T.H.; Baker, E. Phasmida Species File Online. Available online: https://phasmida.speciesfile.org/ (accessed on 25 January 2025).
- Bragg, P.E. The phasmid database version 1.5. Phasmid Stud. 1995, 3, 41–42. [Google Scholar]
- Brock, P.D.; Hasenpusch, J.W. The Complete Field Guide to Stick and Leaf Insects of Australia; CSIRO Publishing: Clayton, Australia, 2009; pp. 162–181. [Google Scholar] [CrossRef]
- Robertson, J.A.; Bradler, S.; Whiting, M.F. Evolution of oviposition techniques in stick and leaf insects (Phasmatodea). Front. Ecol. Evol. 2018, 6, 216. [Google Scholar] [CrossRef]
- Buckley, T.R.; Attanayake, D.; Bradler, S. Extreme convergence in stick insect evolution: Phylogenetic placement of the Lord Howe Island tree lobster. Proc. R. Soc. B Biol. Sci. 2009, 276, 1055–1062. [Google Scholar] [CrossRef]
- Komoto, N.; Yukuhiro, K.; Ueda, K.; Tomita, S. Exploring the molecular phylogeny of phasmids with whole mitochondrial genome sequences. Mol. Phylogenet. Evol. 2011, 58, 43–52. [Google Scholar] [CrossRef]
- Wang, B.C.; Jeng, M.L.; Tsai, J.F.; Wu, L.W. Genome skimming for improved phylogenetics of Taiwanese phasmids (Insecta: Phasmatodea). Mol. Phylogenet. Evol. 2025, 205, 108292. [Google Scholar] [CrossRef]
- Plazzi, F.; Ricci, A.; Passamonti, M. The mitochondrial genome of Bacillus stick insects (Phasmatodea) and the phylogeny of orthopteroid insects. Mol. Phylogenet. Evol. 2011, 58, 304–316. [Google Scholar] [CrossRef]
- Chen, S.C.; He, Y.H.; Li, Y. Phasmatodea. In Forest Insects of Hainan; Huang, F.S., Ed.; Science Press: Beijing, China, 2002; pp. 100–116. [Google Scholar]
- Ho, W.C.G. Contribution to the knowledge of Chinese Phasmatodea III: Catalogue of the phasmids of Hainan Island, China, with descriptions of one new genus, one new species and two new subspecies and proposals of three new combinations. Zootaxa 2016, 4150, 314–340. [Google Scholar] [CrossRef]
- Bradler, S.; Cliquennois, N.; Buckley, T.R. Single origin of the Mascarene stick insects: Ancient radiation on sunken islands? BMC Evol. Biol. 2015, 15, 196. [Google Scholar] [CrossRef]
- Xu, K.K.; Chen, Q.P.; Ayivi, S.P.G.; Guan, J.Y.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Three complete mitochondrial genomes of Orestes guangxiensis, Peruphasma schultei, and Phryganistria guangxiensis (Insecta: Phasmatodea) and their phylogeny. Insects 2021, 12, 779. [Google Scholar] [CrossRef]
- Simon, S.; Letsch, H.; Bank, S.; Buckley, T.R.; Donath, A.; Liu, S.; Machida, R.; Meusemann, K.; Miso, B.; Podsiadlowsk, L.; et al. Old World and New World Phasmatodea: Phylogenomics resolve the evolutionary history of stick and leaf insects. Front. Ecol. Evol. 2019, 7, 345. [Google Scholar] [CrossRef]
- Bank, S.; Cumming, R.T.; Li, Y.; Henze, K.; Le Tirant, S.; Bradler, S. A tree of leaves: Phylogeny and historical biogeography of the leaf insects (Phasmatodea: Phylliidae). Commun. Biol. 2021, 4, 932. [Google Scholar] [CrossRef]
- Cameron, S.; Beckenbach, A.; Dowton, M.; Whiting, M. Evidence from mitochondrial genomics on interordinal relationships in insects. Arthropod Syst. Phylogeny 2006, 64, 27–34. [Google Scholar] [CrossRef]
- Friedemann, K.; Wipfler, B.; Bradler, S.; Beutel, R.G. On the head morphology of Phyllium and the phylogenetic relationships of Phasmatodea (Insecta). Acta Zool. 2012, 93, 184–199. [Google Scholar] [CrossRef]
- Forni, G.; Plazzi, F.; Cussigh, A.; Conle, O.; Hennemann, F.; Luchetti, A.; Mantovani, B. Phylomitogenomics provides new perspectives on the Euphasmatodea radiation (Insecta: Phasmatodea). Mol. Phylogenet. Evol. 2021, 155, 106983. [Google Scholar] [CrossRef]
- Bank, S.; Bradler, S. A second view on the evolution of flight in stick and leaf insects (Phasmatodea). BMC Ecol. Evol. 2022, 22, 62. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Cameron, S. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst. Entomol. 2014, 39, 400–411. [Google Scholar] [CrossRef]
- Tihelka, E.; Cai, C.; Giacomelli, M.; Lozano-Fernandez, J.; Rota-Stabelli, O.; Huang, D.; Pisani, D. The evolution of insect biodiversity. Curr. Biol. 2021, 31, R1299–R1311. [Google Scholar] [CrossRef]
- Salvato, P.; Simonato, M.; Battisti, A.; Negrisolo, E. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC Genom. 2008, 9, 331. [Google Scholar] [CrossRef]
- Cameron, S.L.; Whiting, M.F. The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene 2008, 408, 112–123. [Google Scholar] [CrossRef]
- Masta, S.E.; Boore, J.L. Parallel evolution of truncated transfer RNA genes in arachnid mitochondrial genomes. Mol. Biol. Evol. 2008, 25, 949–959. [Google Scholar] [CrossRef]
- Caterino, M.S.; Cho, S.; Sperling, F.A. The current state of insect molecular systematics: A thriving Tower of Babel. Annu. Rev. Entomol. 2000, 45, 1–54. [Google Scholar] [CrossRef]
- Shao, R.; Barker, S.C. Chimeric mitochondrial minichromosomes of the human body louse, Pediculus humanus: Evidence for homologous and non-homologous recombination. Gene 2011, 473, 36–43. [Google Scholar] [CrossRef]
- Dong, Z.; Wang, Y.; Li, C.; Li, L.; Men, X. Mitochondrial DNA as a molecular marker in insect ecology: Current status and future prospects. Ann. Entomol. Soc. Am. 2021, 114, 470–476. [Google Scholar] [CrossRef]
- Johnson, K.P.; Dietrich, C.H.; Friedrich, F.; Beutel, R.G.; Wipfler, B.; Peters, R.S.; Yoshizawa, K. Phylogenomics and the evolution of hemipteroid insects. Proc. Natl. Acad. Sci. USA 2018, 115, 12775–12780. [Google Scholar] [CrossRef]
- Brown, W.M.; George, M., Jr.; Wilson, A.C. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 1967–1971. [Google Scholar] [CrossRef]
- Saccone, C.; De Giorgi, C.; Gissi, C.; Pesole, G.; Reyes, A. Evolutionary genomics in Metazoa: The mitochondrial DNA as a model system. Gene 1999, 238, 195–209. [Google Scholar] [CrossRef]
- Gissi, C.; Iannelli, F.; Pesole, G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 2008, 101, 301–320. [Google Scholar] [CrossRef]
- Macino, G.; Scazzocchio, C.; Waring, R.B.; Berks, M.M.; Wayne Davies, R. Conservation and rearrangement of mitochondrial structural gene sequences. Nature 1980, 288, 404–406. [Google Scholar] [CrossRef]
- Bernt, M.; Braband, A.; Schierwater, B.; Stadler, P.F. Genetic aspects of mitochondrial genome evolution. Mol. Phylogenet. Evol. 2013, 69, 328–338. [Google Scholar] [CrossRef]
- Matvienko, M. CLC Genomics Workbench. Plant Anim. Genome Sr. Field Appl. Sci. CLC Bio 2015, 1, 1–42. [Google Scholar]
- Luo, T.; Zhang, Q.; Pang, S.; Qin, Y.; Zhang, B.; Bian, X. Comparative Mitochondrial Genomic and Phylogenetic Study of Eight Species of the Family Lonchodidae (Phasmatodea: Euphasmatodea). Genes 2025, 16, 565. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Zheng, S.; Poczai, P.; Hyvönen, J.; Tang, J.; Amiryousefi, A. Chloroplot: An online program for the versatile plotting of organelle genomes. Front. Genet. 2020, 11, 576124. [Google Scholar] [CrossRef]
- Xiang, C.Y.; Gao, F.; Jakovlić, I.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. Imeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez.; Del-Barrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Kück, P.; Meid, S.A.; Groß, C. AliGROOVE—Visualization of heterogeneous sequence divergence within multiple sequence alignments and detection of inflated branch support. BMC Bioinform. 2014, 15, 294. [Google Scholar] [CrossRef]
- Xu, H.X. DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Norman, J.E.; Gray, M.W. The cytochrome oxidase subunit 1 gene (cox1) from the dinoflagellate, Crypthecodinium cohnii. FEBS Lett. 1997, 413, 333–338. [Google Scholar] [CrossRef]
- Hendrich, L.; Pons, J.; Ribera, I.; Balke, M. Mitochondrial cox1 sequence data reliably uncover patterns of insect diversity but suffer from high lineage-idiosyncratic error rates. PLoS ONE 2010, 5, e14448. [Google Scholar] [CrossRef]
- Rodrigues, M.S.; Morelli, K.A.; Jansen, A.M. Cytochrome c oxidase subunit 1 gene as a DNA barcode for discriminating Trypanosoma cruzi DTUs and closely related species. Parasites Vectors 2017, 10, 488. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (NCBI). National Library of Medicine (US). 1988. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 23 December 2024).
- Chantangsi, C.; Leander, B.S. An SSU rDNA barcoding approach to the diversity of marine interstitial cercozoans, including descriptions of four novel genera and nine novel species. Int. J. Syst. Evol. Microbiol. 2010, 60, 1962–1977. [Google Scholar] [CrossRef]
- Mata-Somarribas, C.; Cardoso das Graças, G.; de Oliveira, R.; Pereira, L.; Côrtes Boité, M.; Motta Cantanhêde, L.; Braga Filgueira; Cupolillo, E. Applying a cytochrome c oxidase I barcode for Leishmania species typing. PLoS ONE 2024, 19, e0309277. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, H.; Sanchez-Puerta, M.V.; Li, J. Horizontal gene transfer has impacted cox1 gene evolution in Cassytha filiformis. J. Mol. Evol. 2020, 88, 361–371. [Google Scholar] [CrossRef]
- Bank, S.; Buckley, T.R.; Büscher, T.H.; Bresseel, J.; Constant, J.; De Haan; Bradler, S. Reconstructing the nonadaptive radiation of an ancient lineage of ground-dwelling stick insects (Phasmatodea: Heteropterygidae). Syst. Entomol. 2021, 46, 487–507. [Google Scholar] [CrossRef]
- Zhou, Z.; Guan, B.; Chai, J.; Che, X. Next-generation sequencing data used to determine the mitochondrial genomes and a preliminary phylogeny of Verophasmatodea insects. J. Asia-Pac. Entomol. 2017, 20, 713–719. [Google Scholar] [CrossRef]
- Song, N.; Li, X.H.; Na, R.S. Mitochondrial genomes of stick insects (Phasmatodea) and phylogenetic considerations. PLoS ONE 2020, 15, e0240186. [Google Scholar] [CrossRef]
- Dong, Z.W.; Li, J.; He, J.W.; Liu, G.C.; Mao, C.Y.; Zhao, R.P.; Li, X.Y. The mitochondrial genome of a leaf insect Phyllium westwoodii (Phasmatodea: Phylliidae) in Southeast Asia. Mitochondrial DNA Part B Resour. 2021, 6, 888–890. [Google Scholar] [CrossRef]
- Mikheyev, A.S.; Zwick, A.; Magrath, M.J.L.; Grau, M.L.; Qiu, L.; Su, Y.N.; Yeates, D. Museum genomics confirms that the Lord Howe Island stick insect survived extinction. Curr. Biol. 2017, 27, 3157–3161. [Google Scholar] [CrossRef]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [CrossRef]
- Parvathy, S.T.; Udayasuriyan, V.; Bhadana, V. Codon usage bias. Mol. Biol. Rep. 2022, 49, 539–565. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.R.; Moriyama, E.N. Evolution of codon usage bias in Drosophila. Proc. Natl. Acad. Sci. USA 1997, 94, 7784–7790. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dai, X.; Xu, X.; Zhang, Z.; Yu, D.; Storey, K.B.; Zhang, J. The complete mitochondrial genomes of five longicorn beetles (Coleoptera: Cerambycidae) and phylogenetic relationships within Cerambycidae. PeerJ 2019, 7, e7633. [Google Scholar] [CrossRef] [PubMed]
- Tomita, S.; Yukuhiro, K.; Kômoto, N. The mitochondrial genome of a stick insect Extatosoma tiaratum (Phasmatodea) and the phylogeny of polyneopteran insects. J. Insect Biotechnol. Sericology 2011, 80, 3079–3088. [Google Scholar] [CrossRef]
- Linde, J.B. A Comprehensive Phylogeny of Stick and Leaf Insects (Insecta: Phasmatodea) Reveals Widespread Taxonomic Incongruence. Master’s Thesis, Brigham Young University, Provo, Utah, 2022. Available online: https://lib.byu.edu/about/copyright (accessed on 25 January 2025).
- Bradler, S.; Robertson, J.A.; Whiting, M.F. A molecular phylogeny of Phasmatodea with emphasis on Necrosciinae, the most species-rich subfamily of stick insects. Syst. Entomol. 2014, 39, 205–222. [Google Scholar] [CrossRef]
- Ho, W.C.G. The genus Neohirasea Rehn (Phasmatodea: Diapheromeridae: Necrosciinae) from Vietnam. Zool. Syst. 2018, 43, 37–51. [Google Scholar] [CrossRef]
- Boisseau, R.P.; Woods, H.A. Resource allocation strategies and mechanical constraints drive the diversification of stick and leaf insect eggs. Curr. Biol. 2024, 34, 2880–2892. [Google Scholar] [CrossRef]
- Büscher, T.H.; Bank, S.; Cumming, R.T.; Gorb, S.N.; Bradler, S. Leaves that walk and eggs that stick: Comparative functional morphology and evolution of the adhesive system of leaf insect eggs (Phasmatodea: Phylliidae). BMC Ecol. Evol. 2023, 23, 17. [Google Scholar] [CrossRef]
- Cumming, R.T.; Bank, S.; Le Tirant, S.; Bradler, S. Notes on the leaf insects of the genus Phyllium of Sumatra and Java, Indonesia, including the description of two new species with purple coxae (Phasmatodea, Phylliidae). ZooKeys 2020, 913, 89. [Google Scholar] [CrossRef]
- Keating, J.N.; Garwood, R.J.; Sansom, R.S. Phylogenetic congruence, conflict and consilience between molecular and morphological data. BMC Ecol. Evol. 2023, 23, 30. [Google Scholar] [CrossRef]
- Buckley, T.R.; Attanayake, D.; Nylander, J.A.A.; Bradler, S. The phylogenetic placement and biogeographical origins of the New Zealand stick insects (Phasmatodea). Syst. Entomol. 2010, 35, 207–225. [Google Scholar] [CrossRef]
- Bradler, S. The vomer of Timema Scudder, 1895 (Insecta: Phasmatodea) and its significance for phasmatodean phylogeny. Cour.-Forschungsinstitut Senckenberg 1999, 215, 43–48. [Google Scholar]
- Chen, Y.; Yuan, Y.; Yang, W.; Storey, K.B.; Zhang, J.; Yu, D. Complete mitochondrial genome of the introduced Indian walking stick Carausius morosus (Lonchodidae, Insecta) from California. Insects 2024, 15, 858. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, L.; Tian, S.; Liu, Y.; Jiang, Y.; Zhou; Wang, Y. Inconsistent performance of multi-type genomic data in phylogenomics of neuropteridan insects, with solutions toward conflicting results. Syst. Entomol. [CrossRef]
- Yuan, Y.; Zhang, L.; Li, K.; Hong, Y.; Storey, K.B.; Zhang, J.; Yu, D. Nine mitochondrial genomes of Phasmatodea with two novel mitochondrial gene rearrangements and phylogeny. Insects 2023, 14, 485. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Creedy, T.J.; Ding, Y.; Gregory, K.M.; Swaby, L.; Zhang, F.; Vogler, A.P. Bioinformatics of combined nuclear and mitochondrial phylogenomics to define key nodes for the classification of Coleoptera. BioRxiv 2024. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).