The Maghreb as a Hotspot of Diversity for the Freshwater Crab Genus Potamon (Decapoda, Potamidae)
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wallace, A.R. The Geographical Distribution of Animals; Harper: New York, NY, USA, 1876. [Google Scholar]
- Marabuto, E.; Pina-Martins, F.; Rebelo, M.T.; Paulo, O.S. Ancient Divergence, a Crisis of Salt and Another of Ice Shaped the Evolution of the West Mediterranean Butterfly Euchloe tagis. Biol. J. Linn. Soc. 2020, 131, 487–504. [Google Scholar] [CrossRef]
- Trájer, A.J.; Sebestyén, V.; Padisák, J. The Impacts of the Messinian Salinity Crisis on the Biogeography of Three Mediterranean Sandfly (Diptera: Psychodidae) Species. Geobios 2021, 65, 51–66. [Google Scholar] [CrossRef]
- Carranza, S.; Arnold, E.N.; Pleguezuelos, J.M. Phylogeny, Biogeography, and Evolution of Two Mediterranean Snakes, Malpolon monspessulanus and Hemorrhois hippocrepis (Squamata, Colubridae), Using MtDNA Sequences. Mol. Phylogenet. Evol. 2006, 40, 532–546. [Google Scholar] [CrossRef] [PubMed]
- Gaubert, P.; Machordom, A.; Morales, A.; López-Bao, J.V.; Veron, G.; Amin, M.; Barros, T.; Basuony, M.; Djagoun, C.A.M.S.; San, E.D.L.; et al. Comparative Phylogeography of Two African Carnivorans Presumably Introduced into Europe: Disentangling Natural versus Human-Mediated Dispersal across the Strait of Gibraltar. J. Biogeogr. 2011, 38, 341–358. [Google Scholar] [CrossRef]
- Dinis, M.; Merabet, K.; Martínez-Freiría, F.; Steinfartz, S.; Vences, M.; Burgon, J.D.; Elmer, K.R.; Donaire, D.; Hinckley, A.; Fahd, S.; et al. Allopatric Diversification and Evolutionary Melting Pot in a North African Palearctic Relict: The Biogeographic History of Salamandra algira. Mol. Phylogenet. Evol. 2019, 130, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Husemann, M.; Schmitt, T.; Zachos, F.E.; Ulrich, W.; Habel, J.C. Palaearctic Biogeography Revisited: Evidence for the Existence of a North African Refugium for Western Palaearctic Biota. J. Biogeogr. 2014, 41, 81–94. [Google Scholar] [CrossRef]
- Jaskuła, R. The Maghreb—One More Important Biodiversity Hot Spot for Tiger Beetle Fauna (Coleoptera, Carabidae, Cicindelinae) in the Mediterranean Region. Zookeys 2015, 482, 35–53. [Google Scholar] [CrossRef]
- Bespalaya, Y.V.; Vinarski, M.V.; Aksenova, O.V.; Babushkin, E.S.; Gofarov, M.Y.; Kondakov, A.V.; Konopleva, E.S.; Kropotin, A.V.; Mabrouki, Y.; Ovchankova, N.B.; et al. Phylogeny, Taxonomy, and Biogeography of the Sphaeriinae (Bivalvia: Sphaeriidae). Zool. J. Linn. Soc. 2024, 201, 305–338. [Google Scholar] [CrossRef]
- Mabrouki, Y.; Taybi, A.F.; El Alami, M.; Berrahou, A. Biotypology of Stream Macroinvertebrates from North African and Semi Arid Catchment: Oued Za (Morocco). Knowl. Manag. Aquat. Ecosyst. 2019, 420, 17. [Google Scholar] [CrossRef]
- Mabrouki, Y.; Taybi, A.F.; Skalli, A.; Sánchez-Vialas, A. Amphibians of the Oriental Region and the Moulouya River Basin of Morocco: Distribution and Conservation Notes. Basic Appl. Herpetol. 2019, 33, 19–32. [Google Scholar] [CrossRef]
- Taybi, A.F.; Mabrouki, Y.; Piscart, C. Distribution of Freshwater Alien Animal Species in Morocco: Current Knowledge and Management Issues. Diversity 2023, 15, 169. [Google Scholar] [CrossRef]
- Taybi, A.F.; Mabrouki, Y.; Berrahou, A.; Legssyer, B. Spatio-Temporal Typology of the Physico-Chemical Parameters of a Large North African River: The Moulouya and Its Main Tributaries (Morocco). Afr. J. Aquat. Sci. 2020, 45, 431–441. [Google Scholar] [CrossRef]
- Taybi, A.F.; Mabrouki, Y.; Dakki, M.; Berrahou, A.; Millán, A. Longitudinal Distribution of Macroinvertebrate in a Very Wet North African Basin: Oued Melloulou (Morocco). Ann. Limnol. 2020, 56, 17. [Google Scholar] [CrossRef]
- Cumberlidge, N. Freshwater Crabs of Africa: Biodiversity, Distribution, and Conservation. In The Diversity of Life in African Freshwaters: Under Water, Under Threat. An Analysis of the Status and Distribution of Freshwater Species Throughout Mainland Africa; Darwall, W.R.T., Smith, K.G., Allen, D.J., Holland, R.A., Harrison, I.J., Brooks, E.G.E., Eds.; IUCN: Cambridge, UK; Gland, Switzerland, 2011; pp. 178–199. [Google Scholar]
- Marrone, F.; Vecchioni, L.; Deidun, A.; Mabrouki, Y.; Arab, A.; Arculeo, M. DNA Taxonomy of the Potamid Freshwater Crabs from Northern Africa (Decapoda, Potamidae). Zool. Scr. 2020, 49, 473–487. [Google Scholar] [CrossRef]
- Ghanavi, H.R.; Rahimi, P.; Tavana, M.; Rezaei Tavabe, K.; Jouladeh-Roudbar, A.; Doadrio, I. The Evolutionary Journey of Freshwater Crabs of the Genus Potamon (Decapoda: Brachyura: Potamidae). Mol. Phylogenet. Evol. 2023, 180, 107690. [Google Scholar] [CrossRef]
- Gauthier, H. Recherches Sur la Faune des Eaux Continentales de L’algerie et de la Tunisie; Minerva: Alger, Algeria, 1928. [Google Scholar]
- Cumberlidge, N. Potamon algeriense. Available online: https://www.iucnredlist.org/species/134365/3945207 (accessed on 8 July 2025).
- Taybi, A.F.; Mabrouki, Y.; Berrahou, A.; El Abd, A.A. Bio–Ecology of Potamon algeriense (Herbst, 1785) (Crustacea, Decapoda) in Eastern Morocco. Anim. Biodivers Conserv. 2018, 41, 267–274. [Google Scholar] [CrossRef]
- Fadlaoui, S.; Melhaoui, M. Population Structure of the Freshwater Crab Potamon algeriense (Bott, 1967) Inhabiting Oued Zegzel, (Northeast of Morocco). Int. J. Ecol. 2019, 2019, 7281725. [Google Scholar] [CrossRef]
- Fadlaoui, S.; Melhaoui, M. Diet Composition of the North African Freshwater Crab, Potamon algeriense (Bott, 1967) in Oued Zegzel (Northeast of Morocco). Mar. Freshw. Behav. Physiol. 2022, 55, 73–86. [Google Scholar] [CrossRef]
- Fadlaoui, S.; El Asri, O.; Bouterfas, M.; Melhaoui, M. Effects of Physicochemical Variables of Superficial Waters on the Abundance of the North African Freshwater Crab Potamon algeriense (Bott, 1967). J. Toxicol. 2021, 2021, 6669919. [Google Scholar] [CrossRef] [PubMed]
- Mabrouki, Y.; Gourari, K.; Taybi, A.F. Atlantic Blue Crab Callinectes sapidus Rathbun, 1896 (Portunidae, Malacostraca) Could Lead to Local Extinction of Few Endemic Freshwater Species in the Eastern Morocco. Povolz. Ekol. Zh 2025, 2025, 91–99. [Google Scholar] [CrossRef]
- Fadlaoui, S.; Mahjoub, M.; El Asri, O.; Melhaoui, M. Allometric Growth of the Freshwater Crab Potamon algeriense (Bott, 1967) (Decapoda, Brachyura, Potamidae) in Oued Zegzel, a Mountain Stream, in the Northeast of Morocco. Int. J. Zool. 2019, 2019, 5168639. [Google Scholar] [CrossRef]
- QGIS Development Team QGIS Geographic Information System. Open-Source Geospatial Foundation Project. Available online: https://www.qgis.org/ (accessed on 2 July 2025).
- Beddek, M.; Zenboudji-Beddek, S.; Geniez, P.; Fathalla, R.; Sourouille, P.; Arnal, V.; Dellaoui, B.; Koudache, F.; Telailia, S.; Peyre, O.; et al. Comparative Phylogeography of Amphibians and Reptiles in Algeria Suggests Common Causes for the East-West Phylogeographic Breaks in the Maghreb. PLoS ONE 2018, 13, e0201218. [Google Scholar] [CrossRef]
- Brandis, D.; Storch, V.; Turkay, M. Türkay Michael Taxonomy and Zoogeography of the Freshwater Crabs of Europe, North Africa, and the Middle East (Crustacea, Decapoda, Potamidae). Senckenb. Biol. 2000, 80, 5–56. [Google Scholar]
- Vecchioni, L.; Marrone, F.; Deidun, A.; Adepo-Gourene, B.; Froglia, C.; Sciberras, A.; Bariche, M.; Burak, A.Ç.; Foka-Corsini, M.; Arculeo, M. DNA Taxonomy Confirms the Identity of the Widely-Disjunct Mediterranean and Atlantic Populations of the Tufted Ghost Crab Ocypode cursor (Crustacea: Decapoda: Ocypodidae). Zoolog. Sci. 2019, 36, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Schubart, C.D.; Huber, M.G.J. Genetic Comparisons of German Populations of the Stone Crayfish, Austropotamobius torrentium (Crustacea: Astacidae). BFPP—Bull. Fr. Peche Piscic. 2006, 380–381, 1018–1028. [Google Scholar] [CrossRef]
- Schubart, C.D. Mitochondrial DNA and Decapod Phylogenies; The Importance of Pseudogenes and Primer Optimization. In Decapod Crustacean Phylogenetics; Martin, J.W., Crandall, K.A., Felder, D.L., Eds.; Taylor & Francis/CRC Press: Boca Raton, FL, USA, 2009; pp. 47–65. [Google Scholar]
- Vecchioni, L.; Deidun, A.; Sciberras, J.; Sciberras, A.; Marrone, F.; Arculeo, M. The Late Pleistocene Origin of the Italian and Maltese Populations of Potamon fluviatile (Malacostraca: Decapoda): Insights from an Expanded Sampling of Molecular Data. Eur. Zool. J. 2017, 84, 575–582. [Google Scholar] [CrossRef]
- Keikhosravi, A.; Schubart, C.D. Description of a New Freshwater Crab Species of the Genus Potamon (Decapoda, Brachyura, Potamidae) from Iran, Based on Morphological and Genetic Characters. Crustac. Monogr. 2014, 19, 115–133. [Google Scholar] [CrossRef]
- Richterich, P. Estimation of Errors in “‘Raw’” DNA Sequences: A Validation Study. Genome. Res. 1998, 8, 251–259. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Jesse, R.; Schubart, C.D.; Klaus, S. Identification of a Cryptic Lineage within Potamon fluviatile (Herbst) (Crustacea:Brachyura:Potamidae). Invertebr. Syst. 2010, 24, 348–356. [Google Scholar] [CrossRef]
- Jesse, R.; Grudinski, M.; Klaus, S.; Streit, B.; Pfenninger, M. Evolution of Freshwater Crab Diversity in the Aegean Region (Crustacea: Brachyura: Potamidae). Mol. Phylogenet. Evol. 2011, 59, 23–33. [Google Scholar] [CrossRef]
- Jesse, R.; Pfenninger, M.; Fratini, S.; Scalici, M.; Streit, B.; Schubart, C.D. Disjunct Distribution of the Mediterranean Freshwater Crab Potamon fluviatile-Natural Expansion or Human Introduction? Biol. Invasions 2009, 11, 2209–2221. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.-J.; Forster, P.; Röhl, A. Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Veith, M.; Mayer, C.; Samraoui, B.; Barroso, D.D.; Bogaerts, S. From Europe to Africa and Vice Versa: Evidence for Multiple Intercontinental Dispersal in Ribbed Salamanders (Genus Pleurodeles). J. Biogeogr. 2004, 31, 159–171. [Google Scholar] [CrossRef]
- Stuckas, H.; Velo-Antón, G.; Fahd, S.; Kalboussi, M.; Rouag, R.; Arculeo, M.; Marrone, F.; Sacco, F.; Vamberger, M.; Fritz, U. Where Are You from, Stranger? The Enigmatic Biogeography of North African Pond Turtles (Emys orbicularis). Org. Divers. Evol. 2014, 14, 295–306. [Google Scholar] [CrossRef]
- Dufresnes, C.; Beddek, M.; Skorinov, D.V.; Fumagalli, L.; Perrin, N.; Crochet, P.A.; Litvinchuk, S.N. Diversification and Speciation in Tree Frogs from the Maghreb (Hyla meridionalis Sensu Lato), with Description of a New African Endemic. Mol. Phylogenet. Evol. 2019, 134, 291–299. [Google Scholar] [CrossRef] [PubMed]
- deMenocal, P.B. African Climate Change and Faunal Evolution during the Pliocene-Pleistocene. Earth Planet. Sci. Lett. 2004, 220, 3–24. [Google Scholar] [CrossRef]
- Joleaud, L. Étude Géologique de La Région de Bône et de La Calle. Bull. Serv. Cart. Géolog. L’algér. 1936, II, 199. [Google Scholar]
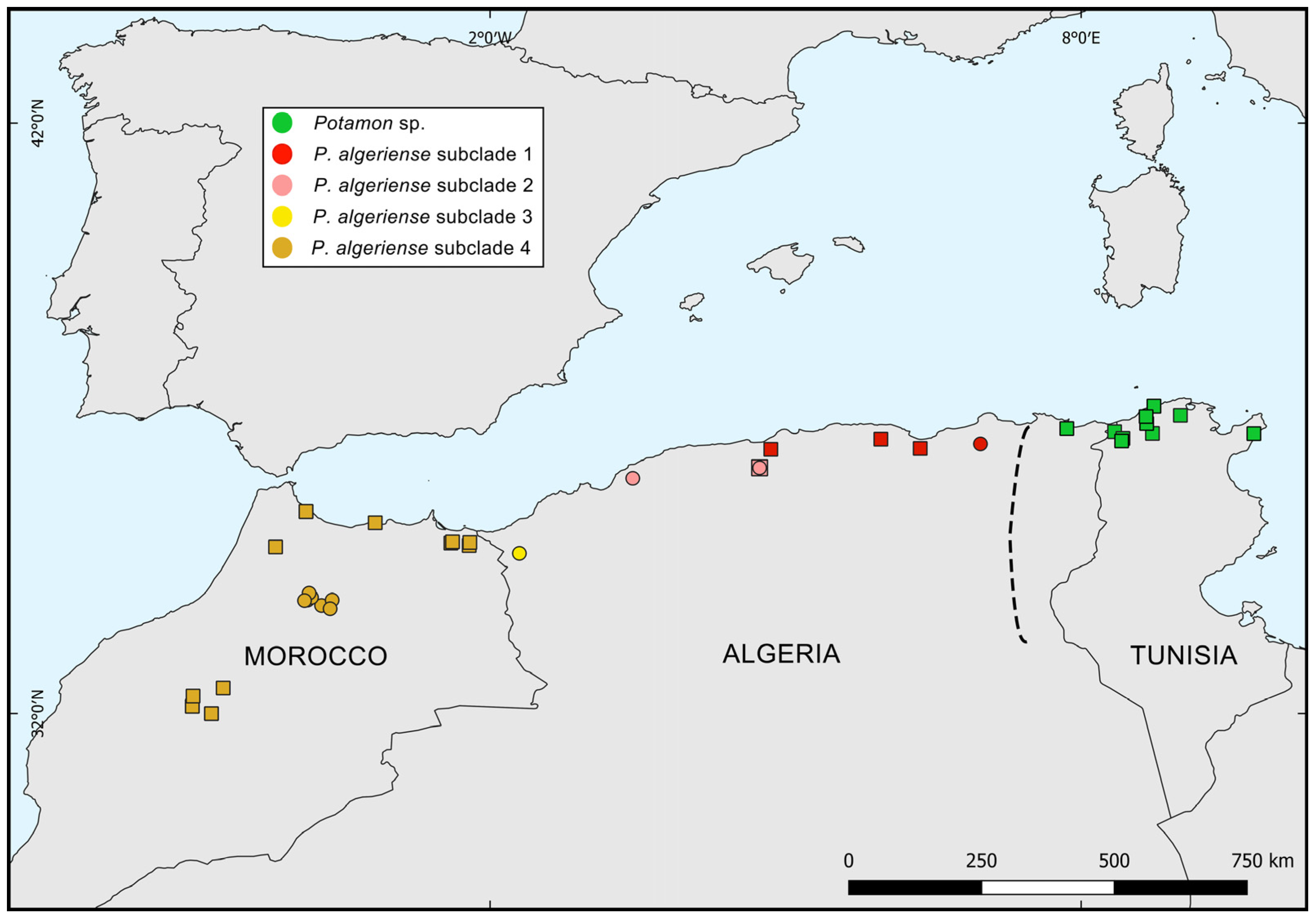
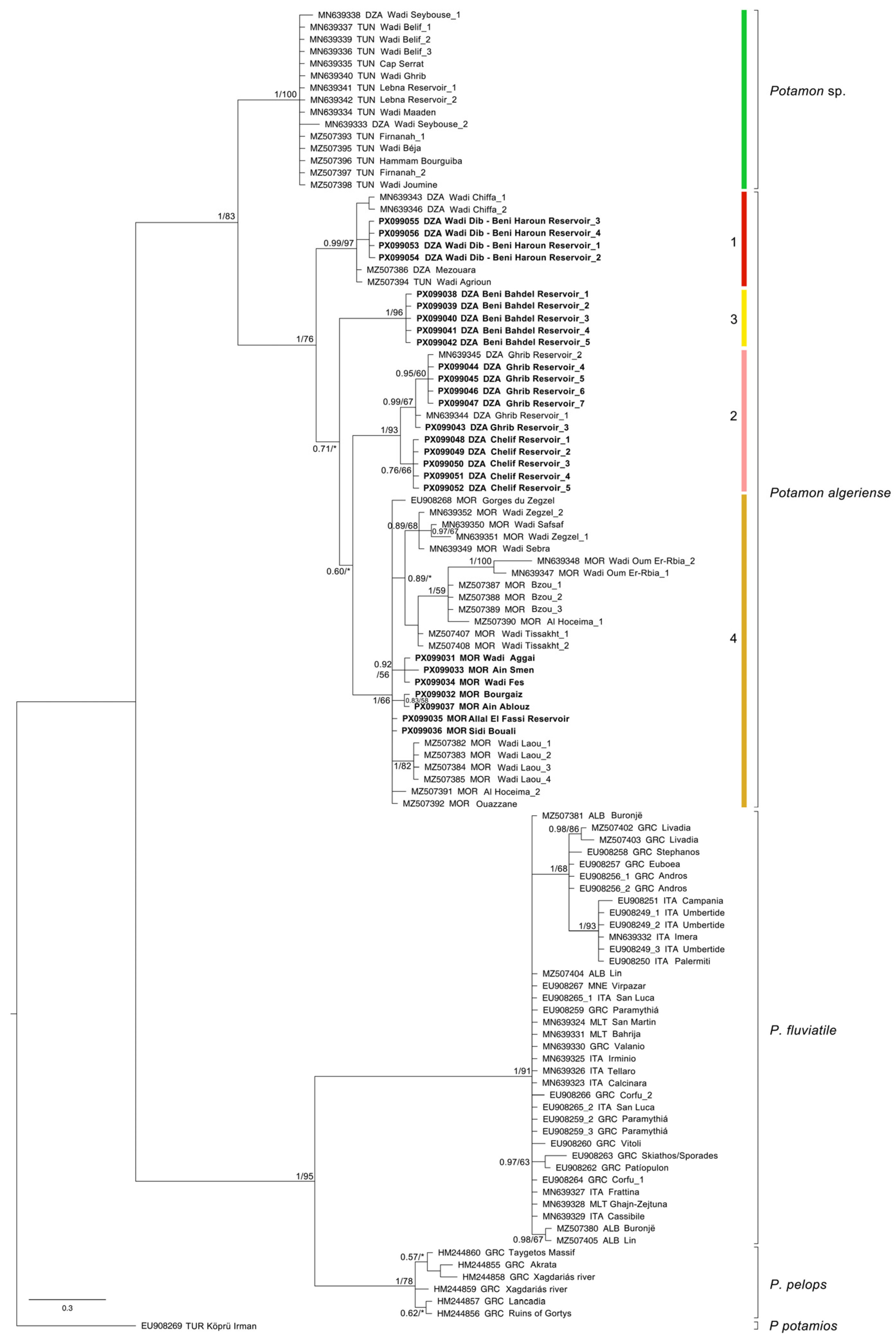
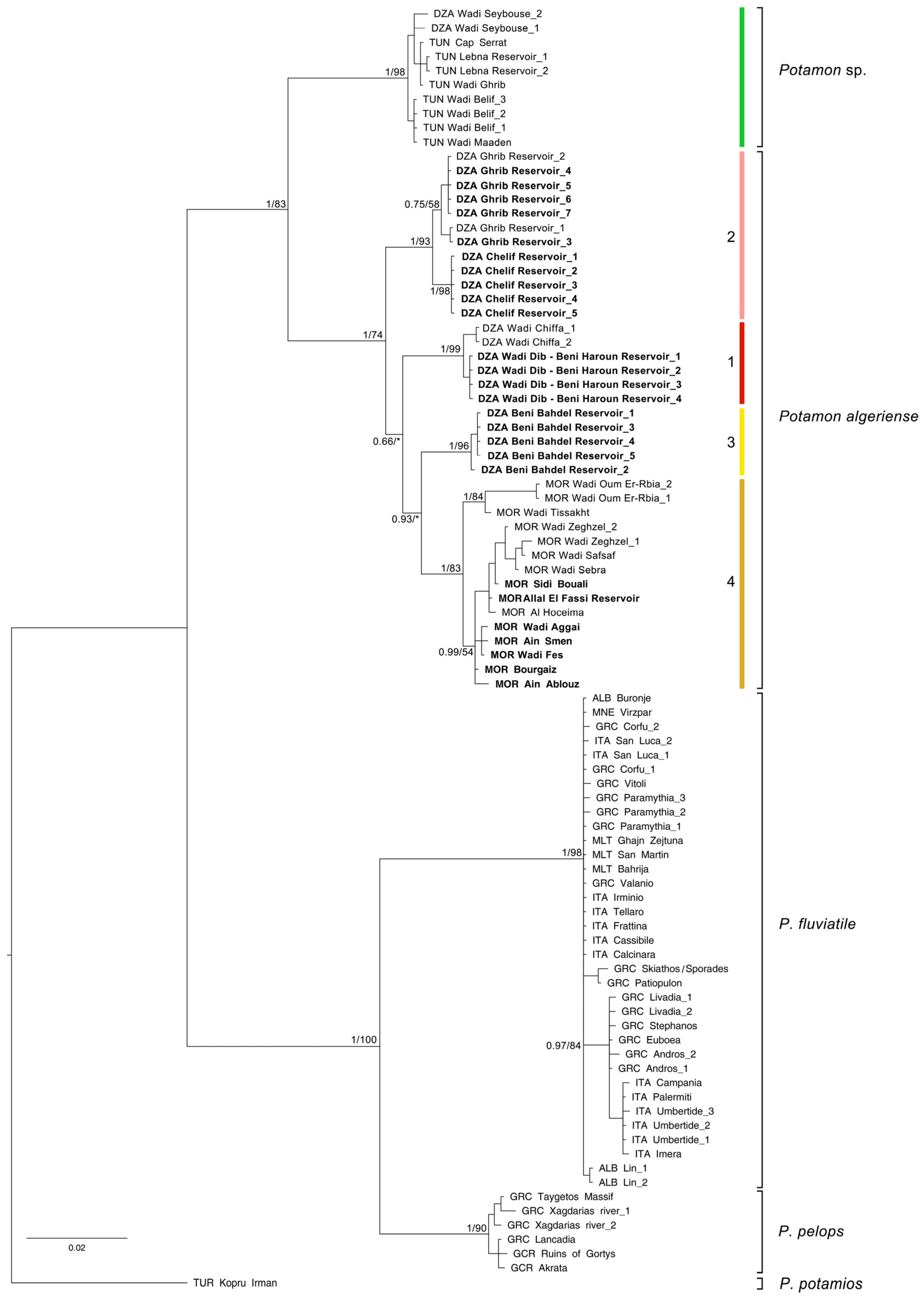
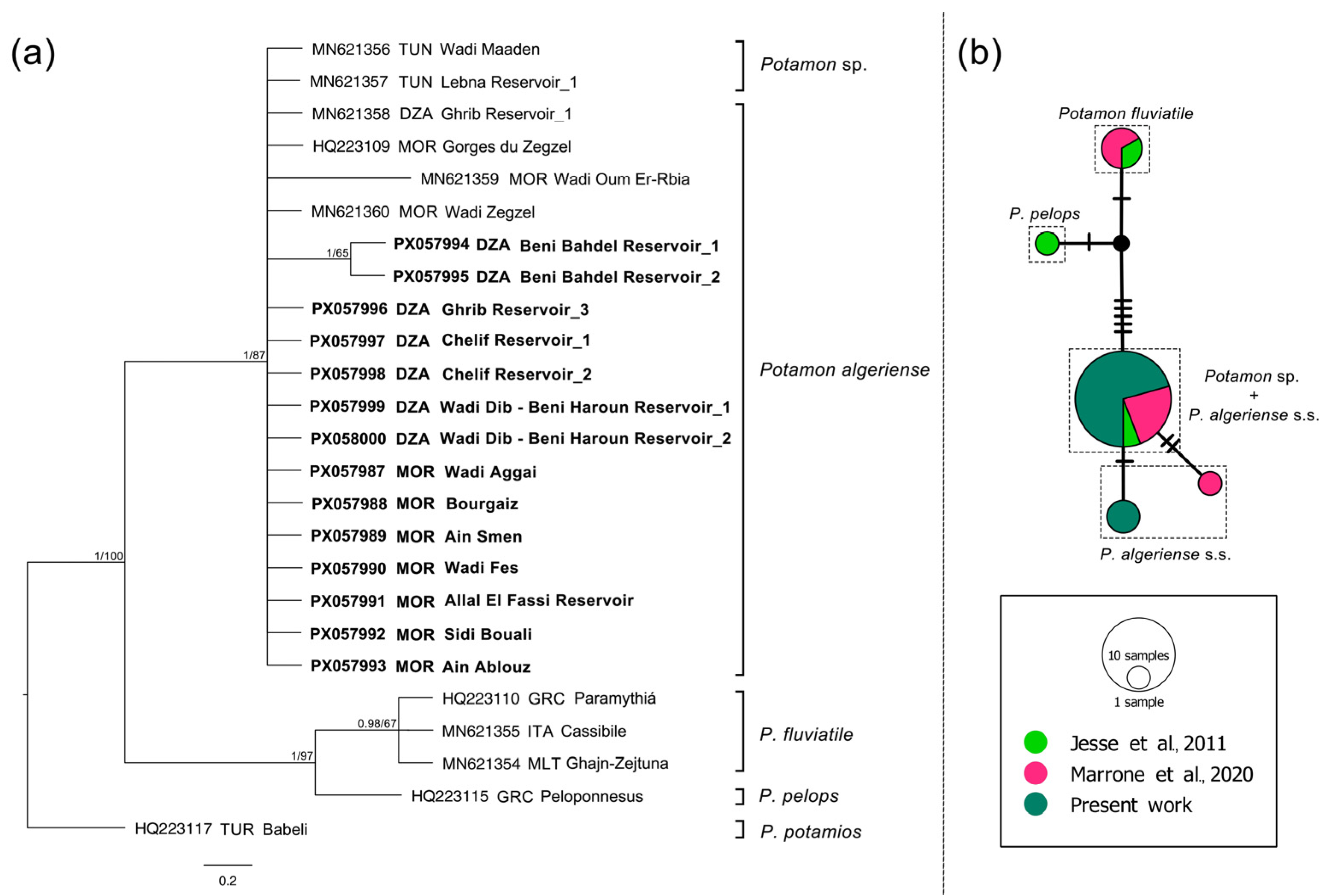
| Taxon | Subclade | Country | Site | Hydrographical Basin | Latitude | Longitude | Date | N | COI | ND1 | 28S | Source |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Potamon algeriense | 1 | Algeria | Wadi Dib—Beni Haroun reservoir | Wadi El Kebir | 36.56619 | 6.29691 | 26/10/2023 | 4 | PX057708-PX057711 | PX099053-PX099056 | PX057999/PX058000/-/- | Present work |
| 1 | Algeria | Wadi Agrioun | Wadi Agrioun | 36.48946 | 5.27753 | (2009-2019) | 1 | - | MZ507394 | - | [17] | |
| 1 | Algeria | Mezouara | Wadi Soummam | 36.64608 | 4.61209 | (2009-2019) | 1 | - | MZ507386 | - | [17] | |
| 1 | Algeria | Wadi Chiffa | Wadi Mazafran | 36.47430 | 2.75030 | 2013 | 2 | MN622982/MN622985 | MN639343/MN639346 | - | [16] | |
| 2 | Algeria | Ghrib reservoir | Wadi Chelif | 36.16140 | 2.56060 | 2015 | 2 | MN622983/MN622984 | MN639344/MN639345 | MN621358/- | [16] | |
| 2 | Algeria | Ghrib reservoir | Wadi Chelif | 36.16189 | 2.56028 | 22/10/2023; 25/11/2023 | 5 | PX057698-PX057702 | PX099043-PX099047 | PX057996/-/-/-/- | Present work | |
| 2 | Algeria | Chelif reservoir | Wadi Chelif | 35.98367 | 0.41444 | 15/10/2023 | 5 | PX057703-PX057707 | PX099048-PX099052 | PX057997/PX057998/-/-/- | Present work | |
| 3 | Algeria | Beni Bahdel reservoir | Wadi Tafna | 34.71183 | −1.50411 | 14/10/2023; 23/11/2023 | 5 | PX057693-PX057697 | PX099038-PX099042 | PX057994/PX057995/-/-/- | Present work | |
| 4 | Morocco | Gorges du Zegzel | Wadi Moulouya | N.A. | N.A. | 2005 | 1 | EU908247§ | EU908268 | HM244838*/HQ223109 | [36,37,38] | |
| 4 | Morocco | Wadi Zegzel | Wadi Moulouya | 34.84490 | −2.34270 | 31/04/19 | 1 | MN622991 | MN639352 | - | [16] | |
| 4 | Morocco | Wadi Zegzel | Wadi Moulouya | 34.84490 | −2.35350 | 31/04/19 | 1 | MN622990 | MN639351 | MN621360 | [16] | |
| 4 | Morocco | Wadi Safsaf | Wadi Moulouya | 34.90760 | −2.63570 | 30/04/2019 | 1 | MN622989 | MN639350 | - | [16] | |
| 4 | Morocco | Wadi Sebra | Wadi Moulouya | 34.88630 | −2.66250 | 30/04/2019 | 1 | MN622988 | MN639349 | - | [16] | |
| 4 | Morocco | Al Hoceima | Wadi Boujbar | 35.23288 | −3.94347 | (2009-2019) | 2 | -/MZ506928 | MZ507390-MZ507391 | MZ507137§-MZ507138§ | [17] | |
| 4 | Morocco | Allal El Fassi reservoir | Wadi Sebou | 33.92011 | −4.67242 | 24/10/2021 | 1 | PX057690 | PX099035 | PX057991 | Present work | |
| 4 | Morocco | Sidi Bouali | Wadi Sebou | 33.77342 | −4.70639 | 25/05/2022 | 1 | PX057691 | PX099036 | PX057992 | Present work | |
| 4 | Morocco | Wadi Aggai | Wadi Sebou | 33.82767 | −4.85142 | 07/03/2021 | 1 | PX057686 | PX099031 | PX057987 | Present work | |
| 4 | Morocco | Ain Smen | Wadi Sebou | 33.96569 | −5.02181 | 13/02/2021 | 1 | PX057688 | PX099033 | PX057989 | Present work | |
| 4 | Morocco | Wadi Fes | Wadi Sebou | 34.04000 | −5.06200 | 23/10/2021 | 1 | PX057689 | PX099034 | PX057990 | Present work | |
| 4 | Morocco | Bourgaiz | Wadi Sebou | 33.92050 | −5.08833 | 04/04/2021 | 1 | PX057687 | PX099032 | PX057988 | Present work | |
| 4 | Morocco | Wadi Laou | Wadi Laou | 35.42216 | −5.11442 | (2009-2019) | 4 | - | MZ507382-MZ507385 | MZ507131§-MZ507134§ | [17] | |
| 4 | Morocco | Ain Ablouz | Wadi Sebou | 33.91158 | −5.13942 | 04/11/2023 | 1 | PX057692 | PX099037 | PX057993 | Present work | |
| 4 | Morocco | Ouazzane | Wadi Loukos | 34.82121 | −5.62973 | (2009-2019) | 1 | - | MZ507392 | MZ507139§ | [17] | |
| 4 | Morocco | Wadi Oum Er-Rbia | Wadi Oum Er-Rbia | 32.43280 | −6.51620 | 10/05/2019 | 1 | MN622987 | MN639348 | MN621359 | [16] | |
| 4 | Morocco | Wadi Tissakht | Wadi Oum Er-Rbia | 31.99694 | −6.71472 | (2009-2019) | 3 | MZ506936/-/MZ506943 | MZ507407/MZ507408/- | MZ507150§/MZ50715§ /MZ507164§ | [17] | |
| 4 | Morocco | Wadi Oum Er-Rbia | Wadi Oum Er-Rbia | 32.29610 | −7.02590 | 05/05/2019 | 1 | MN622986 | MN639347 | - | [16] | |
| 4 | Morocco | Bzou | Wadi Oum Er-Rbia | 32.12131 | −7.03885 | (2009-2019) | 3 | - | MZ507387-MZ507389 | MZ507135§/MZ507136§/- | [17] | |
| Potamon sp. | - | Algeria | Wadi Seybouse | Wadi Seybouse | 36.82750 | 7.75830 | 2013 | 2 | MN622977/MN622972 | MN639338/MN639333 | - | [16] |
| - | Tunisia | Lebna reservoir | Wadi Lebna | 36.74050 | 10.92290 | 2011 | 2 | MN622981/MN622980 | MN639342/MN639341 | MN621357/- | [16] | |
| - | Tunisia | Wadi Joumine | Lake Ichkeul | 37.05187 | 9.67969 | (2009-2019) | 1 | - | MZ507398 | - | [17] | |
| - | Tunisia | Cap Serrat | Ziatine reservoir | 37.20610 | 9.23220 | 2011 | 1 | MN622974 | MN639335 | - | [16] | |
| - | Tunisia | Wadi Béja | Wadi Medjerda | 36.74415 | 9.20691 | (2009-2019) | 1 | - | MZ507395 | MZ507141§ | [17] | |
| - | Tunisia | Wadi Maaden | Wadi El Zouara | 36.91180 | 9.10940 | 2011 | 1 | MN622973 | MN639334 | MN621356 | [16] | |
| - | Tunisia | Wadi Belif | Wadi El Zouara | 37.03170 | 9.09960 | 2011 | 3 | MN622975/MN622976/MN622978 | MN639336/MN639337/MN639339 | - | [16] | |
| - | Tunisia | Firnanah | Wadi Medjerda | 36.65869 | 8.70733 | (2009-2019) | 2 | - | MZ507393/MZ507397 | MZ507140§/- | [17] | |
| - | Tunisia | Wadi Ghrib | Wadi Medjerda | 36.61630 | 8.68590 | 2011 | 1 | MN622979 | MN639340 | - | [16] | |
| - | Tunisia | Hammam Bourguiba | Wadi Bou Namoussa | 36.77354 | 8.56668 | (2009-2019) | 1 | - | MZ507396 | - | [17] |
| Potamon sp. | P. algeriense | P. pelops | P. fluviatile | |
|---|---|---|---|---|
| Potamon sp. | 0.08 | 0.12 | 0.10 | |
| P. algeriense | 0.06 | 0.13 | 0.12 | |
| P. pelops | 0.10 | 0.09 | 0.07 | |
| P. fluviatile | 0.11 | 0.10 | 0.07 |
| Taxon | n | Hn | Hd | π |
|---|---|---|---|---|
| Potamon sp. and P. algeriense | 48 | 26 | 0.962 | 0.03343 |
| Potamon sp. | 10 | 5 | 0.822 | 0.00230 |
| Potamon algeriense | 38 | 21 | 0.950 | 0.02251 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rouabhi, N.; Bouchelouche, D.; Vecchioni, L.; Mabrouki, Y.; Taybi, F.A.; Marrone, F.; Faraone, F.P. The Maghreb as a Hotspot of Diversity for the Freshwater Crab Genus Potamon (Decapoda, Potamidae). Diversity 2025, 17, 562. https://doi.org/10.3390/d17080562
Rouabhi N, Bouchelouche D, Vecchioni L, Mabrouki Y, Taybi FA, Marrone F, Faraone FP. The Maghreb as a Hotspot of Diversity for the Freshwater Crab Genus Potamon (Decapoda, Potamidae). Diversity. 2025; 17(8):562. https://doi.org/10.3390/d17080562
Chicago/Turabian StyleRouabhi, Nesrine, Djaouida Bouchelouche, Luca Vecchioni, Youness Mabrouki, Fouzi Abdelkhaleq Taybi, Federico Marrone, and Francesco Paolo Faraone. 2025. "The Maghreb as a Hotspot of Diversity for the Freshwater Crab Genus Potamon (Decapoda, Potamidae)" Diversity 17, no. 8: 562. https://doi.org/10.3390/d17080562
APA StyleRouabhi, N., Bouchelouche, D., Vecchioni, L., Mabrouki, Y., Taybi, F. A., Marrone, F., & Faraone, F. P. (2025). The Maghreb as a Hotspot of Diversity for the Freshwater Crab Genus Potamon (Decapoda, Potamidae). Diversity, 17(8), 562. https://doi.org/10.3390/d17080562








