Mitogenomic Insights into Adaptive Evolution of African Ground Squirrels in Arid Environments
Abstract
1. Introduction
2. Materials and Methods
2.1. Mitogenome Assembly and Structural Characterization
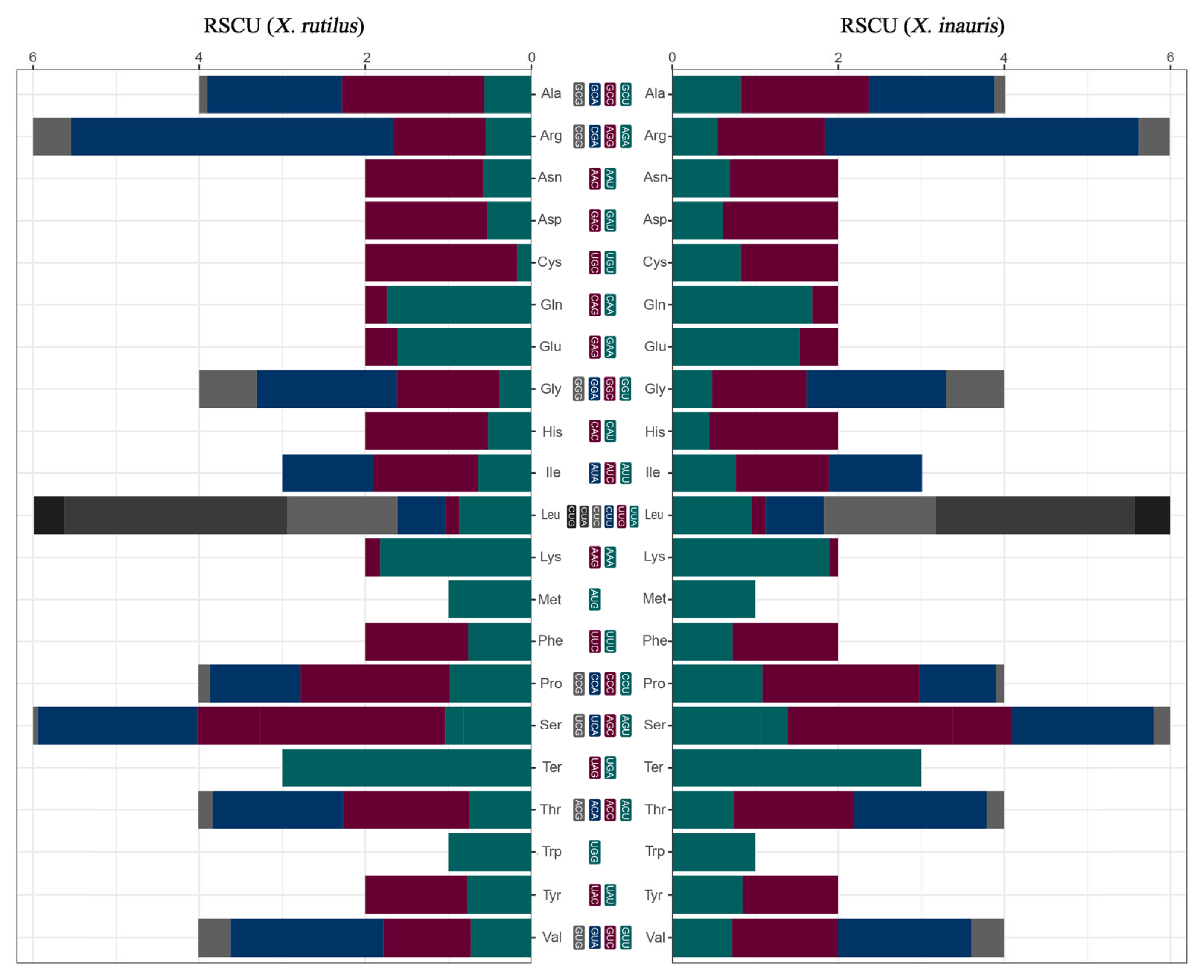
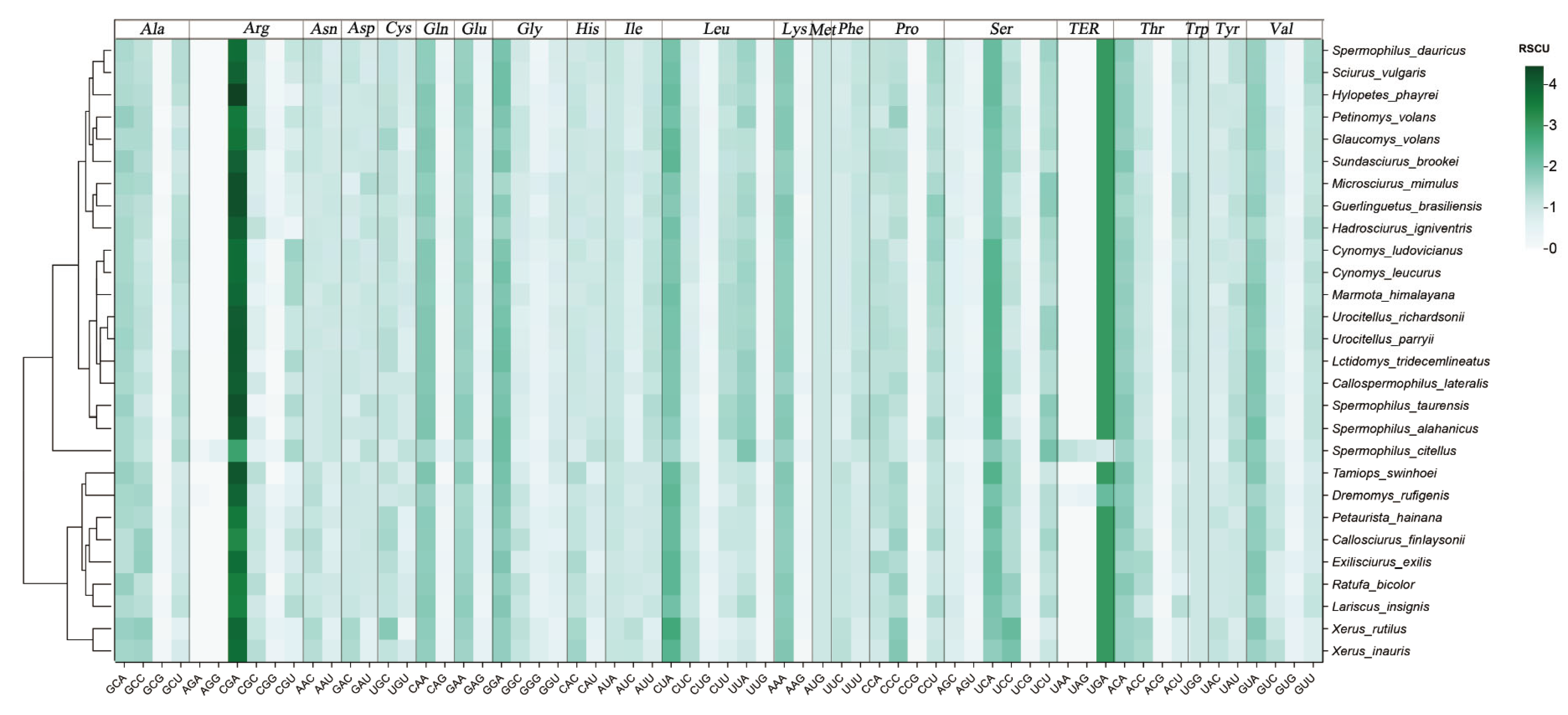
2.2. Relative Synonymous Codon Usage (RSCU)
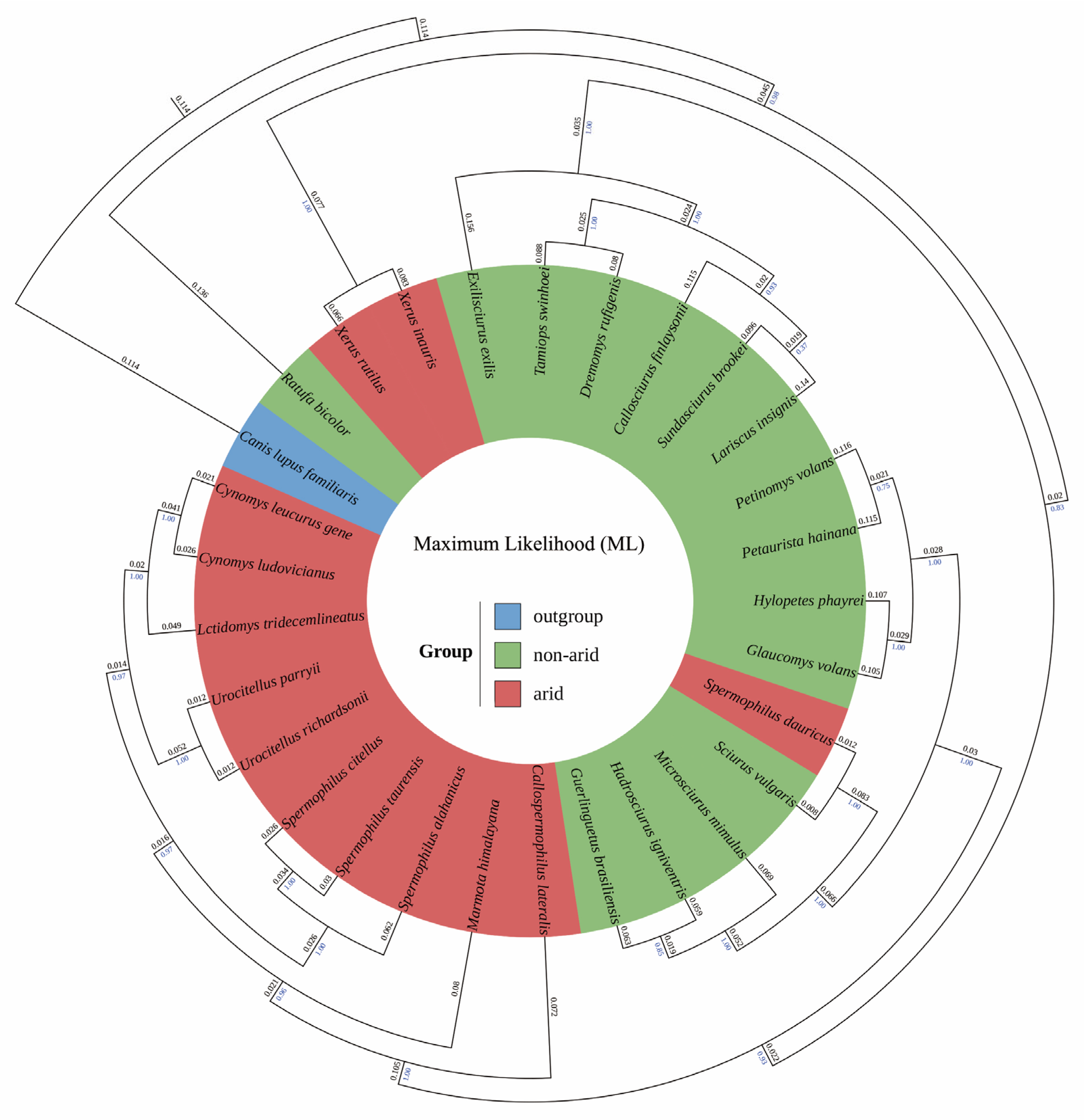
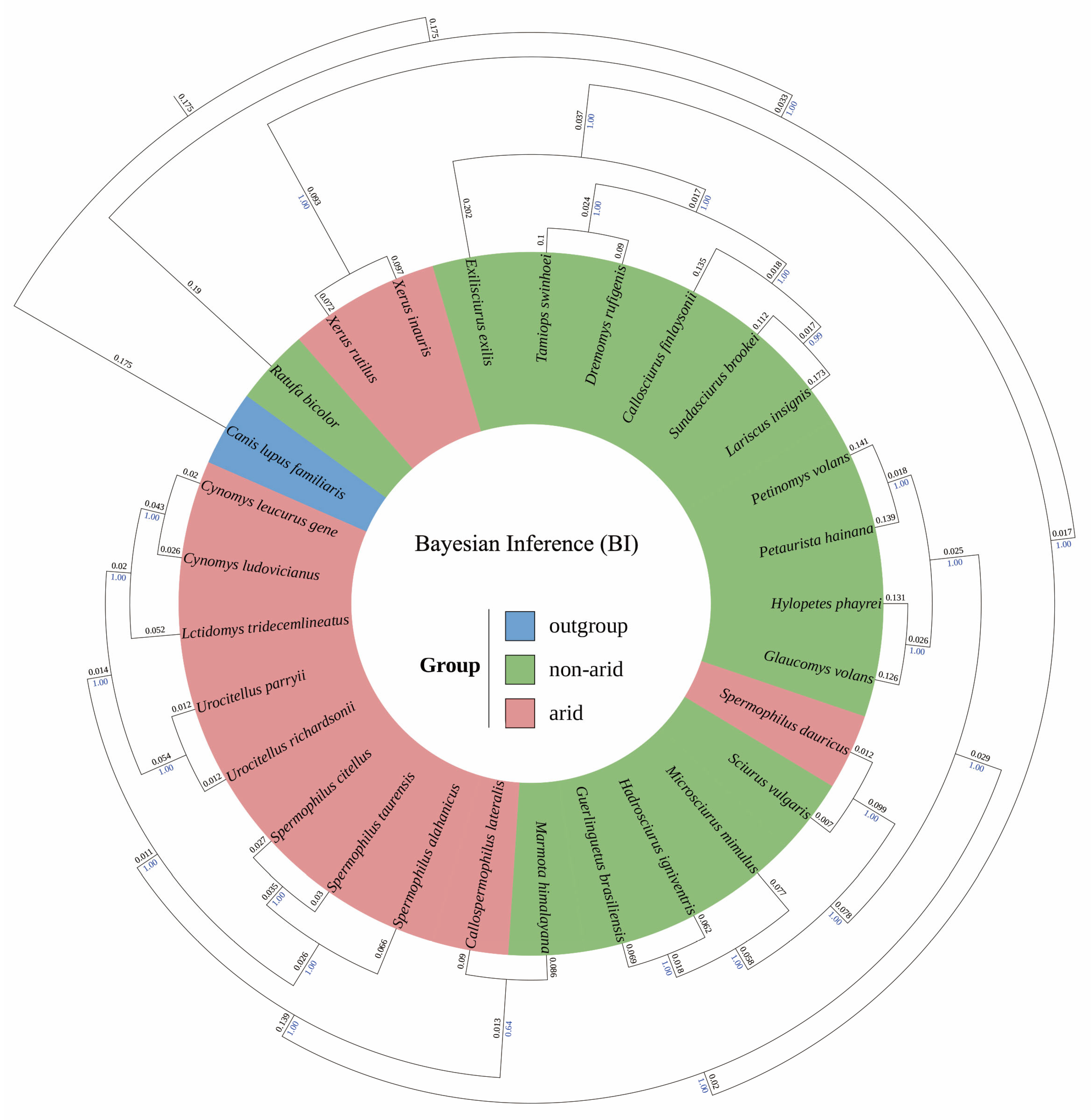
2.3. Phylogenetic Reconstruction
2.4. Positive Selection Analysis
3. Results
3.1. Mitochondrial Genome Annotation
3.2. Relative Synonymous Codon Usage (RSCU)
3.3. Phylogenetic Reconstruction
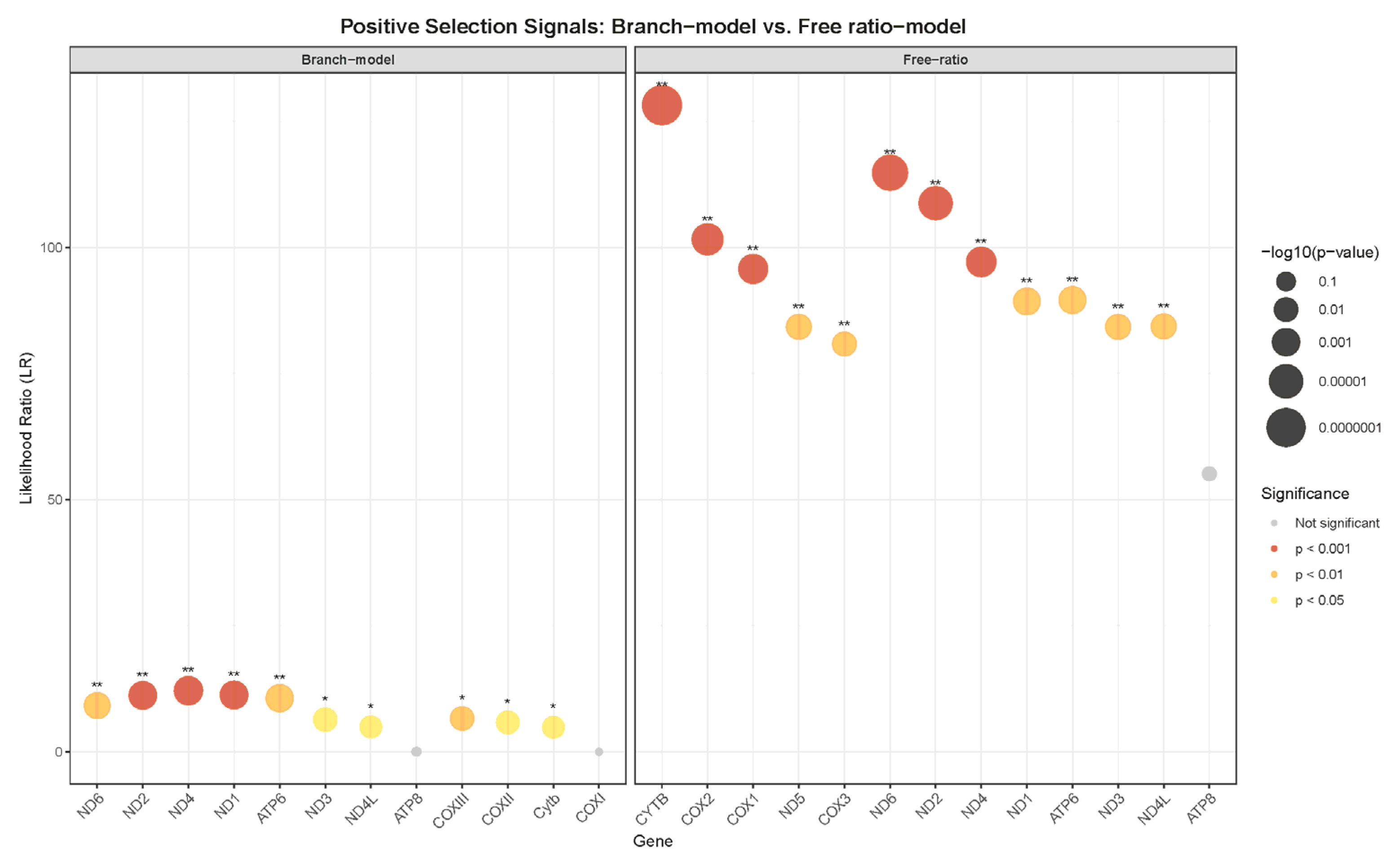
3.4. Selection Pressure Analysis
4. Discussions
4.1. Mitochondrial Genome Annotation and Structural Specificity
4.2. Codon Usage Bias and Metabolic Adaptation
4.3. Phylogenetic Concordance and Biogeographic Processes
4.4. Positive Selection and Arid Adaptation
4.5. Limitations and Methodological Considerations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heske, E.J. Squirrels of the world. J. Mammal. 2013, 94, 957–958. [Google Scholar] [CrossRef][Green Version]
- Whatton, J.F.; Steele, M.A.; Koprowski, J.L.; Thorington, R.W.J. Squirrels of the World; Johns Hopkins University Press: Baltimore, MD, USA, 2012. [Google Scholar]
- Santicchia, F.; Dantzer, B.; van Kesteren, F.; Palme, R.; Martinoli, A.; Ferrari, N.; Wauters, L.A. Stress in biological invasions: Introduced invasive grey squirrels increase physiological stress in native eurasian red squirrels. J. Anim. Ecol. 2018, 87, 1342–1352. [Google Scholar] [CrossRef]
- Marinelli, L. North American tree squirrels. J. Mammal. 2004, 85, 168–169. [Google Scholar] [CrossRef]
- Zhang, B.; Shi, Z.; Chen, X.; Hou, X.; Wang, J.; Li, J.; Chang, G. Seed dispersal of three sympatric oak species by forest rodents in the south slope of Qinling Mountains, China. Acta Ecol. Sin. 2016, 36, 6750–6757. [Google Scholar] [CrossRef]
- Schwimmer, H.; Haim, A. Physiological adaptations of small mammals to desert ecosystems. Integr. Zool. 2009, 4, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Neves, C.; Pillay, N. Kidney form and function vary along an aridity gradient in the African striped mouse, genus Rhabdomys. Mamm. Biol. 2025. [Google Scholar] [CrossRef]
- Herron, M.D.; Castoe, T.A.; Parkinson, C.L. Sciurid phylogeny and the paraphyly of holarctic ground squirrels (Spermophilus). Mol. Phylogenetics Evol. 2004, 31, 1015–1030. [Google Scholar] [CrossRef] [PubMed]
- Mercer, J.M.; Roth, V.L. The effects of cenozoic global change on squirrel phylogeny. Science 2003, 299, 1568–1572. [Google Scholar] [CrossRef]
- Meredith, R.W.; Janečka, J.E.; Gatesy, J.; Ryder, O.A.; Fisher, C.A.; Teeling, E.C.; Goodbla, A.; Eizirik, E.; Simão, T.L.L.; Stadler, T.; et al. Impacts of the cretaceous terrestrial revolution and kpg extinction on mammal diversification. Science 2011, 334, 521–524. [Google Scholar] [CrossRef]
- Favre, A.; Päckert, M.; Pauls, S.U.; Jähnig, S.C.; Uhl, D.; Michalak, I.; Muellner-Riehl, A.N. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. Camb. Philos. Soc. 2015, 90, 236–253. [Google Scholar] [CrossRef]
- Steppan, S.J.; Storz, B.L.; Hoffmann, R.S. Nuclear DNA phylogeny of the squirrels (mammalia: Rodentia) and the evolution of arboreality from c-myc and rag1. Mol. Phylogenet. Evol. 2004, 30, 703–719. [Google Scholar] [CrossRef]
- Li, Q.; Cheng, F.; Jackson, S.M.; Helgen, K.M.; Song, W.Y.; Liu, S.Y.; Sanamxay, D.; Li, S.; Li, F.; Xiong, Y.; et al. Phylogenetic and morphological significance of an overlooked flying squirrel (pteromyini, rodentia) from the eastern himalayas with the description of a new genus. Zool. Res. 2021, 42, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Regan, M.D.; Chiang, E.; Liu, Y.; Tonelli, M.; Verdoorn, K.M.; Gugel, S.R.; Suen, G.; Carey, H.V.; Assadi-Porter, F.M. Nitrogen recycling via gut symbionts increases in ground squirrels over the hibernation season. Science 2022, 375, 460–463. [Google Scholar] [CrossRef]
- Nouri, Z.; Zhang, X.Y.; Khakisahneh, S.; Degen, A.A.; Wang, D.H. The microbiota-gut-kidney axis mediates host osmoregulation in a small desert mammal. npj Biofilms Microbiomes 2022, 8, 16. [Google Scholar] [CrossRef]
- Ventura-Rojas, P.D.; González-Romero, A.; Moreno, C.E.; Sosa, V.J. Effect of rainfall, temperature and climate change on the ecology of the rodents of arid zones: A review. Mammal Rev. 2025, 55, e12372. [Google Scholar] [CrossRef]
- Olenev, G.V.; Grigorkina, E.B. Evolutionary ecological analysis of adaptation strategies of rodent populations under extreme conditions. Russ. J. Ecol. 2016, 47, 486–492. [Google Scholar] [CrossRef]
- Tian, R.; Yin, D.; Liu, Y.; Seim, I.; Xu, S.; Yang, G. Adaptive evolution of energy metabolism-related genes in hypoxia-tolerant mammals. Front. Genet. 2017, 8, 205. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Wang, D.-H. Influencing factors on basal metabolic rate in mammals. Acta Theriol. Sin. 2002, 22, 53–60. [Google Scholar]
- Hill, G.E. Mitonuclear Compensatory Coevolution. Trends Genet. 2020, 36, 403–414. [Google Scholar] [CrossRef]
- Gershoni, M.; Levin, L.; Ovadia, O.; Toiw, Y.; Shani, N.; Dadon, S.; Barzilai, N.; Bergman, A.; Atzmon, G.; Wainstein, J.; et al. Disrupting mitochondrial–nuclear coevolution affects oxphos complex i integrity and impacts human health. Genome Biol. Evol. 2014, 6, 2665–2680. [Google Scholar] [CrossRef]
- Wu, H.; Guang, X.; Al-Fageeh, M.B.; Cao, J.; Pan, S.; Zhou, H.; Zhang, L.; Abutarboush, M.H.; Xing, Y.; Xie, Z.; et al. Camelid genomes reveal evolution and adaptation to desert environments. Nat. Commun. 2014, 5, 5188. [Google Scholar] [CrossRef]
- Wang, S.; Ore, M.J.; Mikkelsen, E.K.; Lee-Yaw, J.; Toews, D.P.L.; Rohwer, S.; Irwin, D. Signatures of mitonuclear coevolution in a warbler species complex. Nat. Commun. 2021, 12, 4279. [Google Scholar] [CrossRef]
- Lee, W.; Zamudio-Ochoa, A.; Buchel, G.; Podlesniy, P.; Marti Gutierrez, N.; Puigròs, M.; Calderon, A.; Tang, H.-Y.; Li, L.; Mikhalchenko, A.; et al. Molecular basis for maternal inheritance of human mitochondrial DNA. Nat. Genet. 2023, 55, 1632–1639. [Google Scholar] [CrossRef]
- Shen, J.; Lyu, X.; Xu, X.; Wang, Z.; Zhang, Y.; Wang, C.; Munaiz, E.D.; Zhang, M.; Havey, M.J.; Shou, W. A nuclear-encoded endonuclease governs the paternal transmission of mitochondria in Cucumis plants. Nat. Commun. 2025, 16, 4266. [Google Scholar] [CrossRef]
- Wang, X.; Shang, Y.; Wu, X.; Wei, Q.; Zhou, S.; Sun, G.; Mei, X.; Dong, Y.; Sha, W.; Zhang, H. Divergent evolution of mitogenomics in Cetartiodactyla niche adaptation. Org. Divers. Evol. 2023, 23, 243–259. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. Novoplasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Wu, X.; Xing, Y.; Wang, X.; Shang, Y.; Chen, Y.; Wang, L.; Han, M.; Sha, W.; Zhang, H. Mitogenomic resolution of phylogenetic conflicts and adaptive signatures in feliform carnivorans. Heredity 2025, 134, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Donath, A.; Jühling, F.; Al-Arab, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. Organellargenomedraw (ogdraw) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. Worldclim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- IUCN. The IUCN Red List of Threatened Species, Version 2022-2; IUCN: Gland, The Switzerland, 2022. [Google Scholar]
- GBIF Occurrence Download. Available online: https://doi.org/10.1016/j.gecco.2020.e01406 (accessed on 3 May 2025).
- Charif, D.; Lobry, J.R. SeqinR 1.0-2: A contributed package to the r project for statistical computing devoted to biological sequences retrieval and analysis. In Structural Approaches to Sequence Evolution; Bastolla, U., Porto, M., Roman, H.E., Vendruscolo, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 207–232. [Google Scholar] [CrossRef]
- Orešič, M.; Shalloway, D. Specific correlations between relative synonymous codon usage and protein secondary Structure11edited by g. Von heijne. J. Mol. Biol. 1998, 281, 31–48. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. Iq-tree 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. Paml 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Bonora, M.; Patergnani, S.; Rimessi, A.; De Marchi, E.; Suski, J.M.; Bononi, A.; Giorgi, C.; Marchi, S.; Missiroli, S.; Poletti, F.; et al. Atp synthesis and storage. Purinergic Signal. 2012, 8, 343–357. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, J.; Min, Z.; Wang, J.; Hong, X.; Pei, X.; Rao, Z.; Xu, X. Structure of atp synthase from an early photosynthetic bacterium chloroflexus aurantiacus. Proc. Natl. Acad. Sci. USA 2025, 122, e2425824122. [Google Scholar] [CrossRef]
- Jonckheere, A.I.; Smeitink, J.A.; Rodenburg, R.J. Mitochondrial atp synthase: Architecture, function and pathology. J. Inherit. Metab. Dis. 2012, 35, 211–225. [Google Scholar] [CrossRef]
- Pavesi, A. Asymmetric evolution in viral overlapping genes is a source of selective protein adaptation. Virology 2019, 532, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, A.E.; Anatskaya, O.V. DNA helix: The importance of being at-rich. Mamm. Genome 2017, 28, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Groucutt, H.S.; White, T.S.; Scerri, E.M.L.; Andrieux, E.; Clark-Wilson, R.; Breeze, P.S.; Armitage, S.J.; Stewart, M.; Drake, N.; Louys, J.; et al. Multiple hominin dispersals into Southwest Asia over the past 400,000 years. Nature 2021, 597, 376–380. [Google Scholar] [CrossRef]
- Abe-Ouchi, A.; Saito, F.; Kawamura, K.; Raymo, M.E.; Okuno, J.i.; Takahashi, K.; Blatter, H. Insolation-driven 100,000-year glacial cycles and hysteresis of ice-sheet volume. Nature 2013, 500, 190–193. [Google Scholar] [CrossRef]
- Elias, S.A.; Short, S.K.; Nelson, C.H.; Birks, H.H. Life and times of the Bering land bridge. Nature 1996, 382, 60–63. [Google Scholar] [CrossRef]
- Steinke, S.; Kienast, M.; Hanebuth, T. On the significance of sea-level variations and shelf paleo-morphology in governing sedimentation in the southern South China Sea during the last deglaciation. Mar. Geol. 2003, 201, 179–206. [Google Scholar] [CrossRef]
- Hu, Z.; Yang, L.; Zhang, M.; Tang, H.; Huang, Y.; Su, Y.; Ding, Y.; Li, C.; Wang, M.; Zhou, Y.; et al. A novel protein Cytb-187aa encoded by the mitochondrial gene Cytb modulates mammalian early development. Cell Metab. 2024, 36, 1586–1597.e7. [Google Scholar] [CrossRef] [PubMed]
- Mahendrarajah, T.A.; Moody, E.R.R.; Schrempf, D.; Szánthó, L.L.; Dombrowski, N.; Davín, A.A.; Pisani, D.; Donoghue, P.C.J.; Szöllősi, G.J.; Williams, T.A.; et al. Atp synthase evolution on a cross-braced dated tree of life. Nat. Commun. 2023, 14, 7456. [Google Scholar] [CrossRef] [PubMed]
- Manee, M.M.; Al-Shomrani, B.M.; Alqahtani, F.H. Mitochondrial DNA of the arabian camel camelus dromedarius. Animals 2024, 14, 2460. [Google Scholar] [CrossRef]
- Epps, C.; Carpenter, L.; Wehausen, J. Genetic relatedness of the preble’s meadow jumping mouse (zapus hudsonius preblei) to nearby subspecies of z. Hudsonius as inferred from variation in cranial morphology, mitochondrial DNA and microsatellite DNA: Implications for taxonomy and conservation. Anim. Conserv. 2005, 8, 329–346. [Google Scholar] [CrossRef]

| Species | Accession Number | AT% | AT_Skew | GC% | GC_Skew | Group |
|---|---|---|---|---|---|---|
| Callospermophilus lateralis | NC_031210.1 | 63.19 | 0.022 | 36.81 | −0.317 | Arid |
| Cynomys leucurus | NC_026705.1 | 62.97 | 0.019 | 37.03 | −0.308 | |
| Cynomys ludovicianus | NC_026706.1 | 62.63 | 0.020 | 37.37 | −0.302 | |
| Lctidomys tridecemlineatus | NC_027278.1 | 63.68 | 0.008 | 36.32 | −0.298 | |
| Spermophilus alashanicus | NC_071768.1 | 64.6 | 0.008 | 35.4 | −0.300 | |
| Spermophilus citellus | NC_059784.1 | 64.59 | 0.015 | 35.41 | −0.302 | |
| Spermophilus dauricus | NC_027283.1 | 63.12 | 0.017 | 36.88 | −0.321 | |
| Spermophilus taurensis | NC_079844.1 | 64.63 | 0.011 | 35.37 | −0.299 | |
| Urocitellus parryii | NC_059785.1 | 63.31 | 0.014 | 36.69 | −0.302 | |
| Urocitellus richardsonii | NC_031209.1 | 63.21 | 0.013 | 36.79 | −0.300 | |
| X. inauris | Supplementary Materials File S1 | 58.18 | 0.075 | 41.82 | −0.361 | |
| X. rutilus | Supplementary Materials File S1 | 57.58 | 0.105 | 42.42 | −0.376 | |
| Lariscus insignis | NC_030070.1 | 61.08 | 0.031 | 38.92 | −0.327 | Non-Arid |
| Hadrosciurus igniventris | NC_050027.1 | 61.94 | 0.011 | 38.06 | −0.312 | |
| Hylopetes phayrei | NC_026443.1 | 62.63 | 0.032 | 37.37 | −0.322 | |
| Glaucomys volans | NC_050026.1 | 62.39 | 0.028 | 37.61 | −0.332 | |
| Guerlinguetus brasiliensis | NC_050010.1 | 62.58 | 0.004 | 37.42 | −0.307 | |
| Exilisciurus exilis | NC_030072.1 | 59.69 | 0.061 | 40.31 | −0.325 | |
| Callosciurus finlaysonii | NC_035817.1 | 60.6 | 0.036 | 39.4 | −0.338 | |
| Dremomys rufigenis | NC_026442.1 | 60.89 | 0.065 | 39.11 | −0.358 | |
| Marmota himalayana | NC_018367.1 | 63.48 | 0.013 | 36.52 | −0.301 | |
| Microsciurus mimulus | NC_050020.1 | 62.2 | 0.006 | 37.8 | −0.303 | |
| Petaurista hainana | NC_023089.1 | 60.38 | 0.052 | 39.62 | −0.328 | |
| Pteromys volans | NC_019612.1 | 62.57 | 0.029 | 37.43 | −0.328 | |
| Ratufa bicolor | NC_023780.1 | 60.32 | 0.051 | 39.68 | −0.336 | |
| Sciurus vulgaris | NC_002369.1 | 62.97 | 0.020 | 37.03 | −0.322 | |
| Sundasciurus brookei | NC_035812.1 | 61.98 | 0.045 | 38.02 | −0.336 | |
| Tamiops swinhoei | NC_026875.1 | 61.3 | 0.065 | 38.7 | −0.361 |
| Gene | Nucleotide Positions | Size (bp) | Strand | Codon | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Xi | Xr | Xi | Xr | ||||||||
| Start | End | Start | End | Xi | Xr | Xi/Xr | Initiation | Amber | Initiation | Amber | |
| tRNA-Phe | 1 | 68 | 1 | 68 | 68 | 68 | + | ||||
| 12SRNA | 69 | 1041 | 69 | 1042 | 973 | 974 | + | ||||
| tRNA-Val | 1042 | 1109 | 1043 | 1110 | 68 | 68 | + | ||||
| 16SRNA | 1110 | 2684 | 1111 | 2688 | 1575 | 1578 | + | ||||
| tRNA-Leu | 2685 | 2759 | 2689 | 2763 | 75 | 75 | + | ||||
| ND1 | 2764 | 3719 | 2768 | 3723 | 956 | 956 | + | ATG | TA | ATG | TA |
| tRNA-Ile | 3720 | 3787 | 3724 | 3792 | 68 | 69 | + | ||||
| tRNA-Gln | 3785 | 3856 | 3790 | 3861 | 72 | 72 | - | ||||
| tRNA-Met | 3862 | 3930 | 3866 | 3934 | 69 | 69 | + | ||||
| ND2 | 3931 | 4972 | 3935 | 4976 | 1042 | 1042 | + | ATC | T | ATC | T |
| tRNA-Trp | 4973 | 5042 | 4977 | 5045 | 70 | 69 | + | ||||
| tRNA-Ala | 5046 | 5114 | 5049 | 5117 | 69 | 69 | - | ||||
| tRNA-Asn | 5132 | 5204 | 5136 | 5207 | 73 | 72 | - | ||||
| tRNA-Cys | 5236 | 5302 | 5238 | 5304 | 67 | 67 | - | ||||
| tRNA-Tyr | 5303 | 5368 | 5305 | 5370 | 66 | 66 | - | ||||
| COXI | 5377 | 6918 | 5379 | 6920 | 1542 | 1542 | + | ATG | TAG | ATG | TAA |
| tRNA-Ser | 6921 | 6989 | 6923 | 6991 | 69 | 69 | - | ||||
| tRNA-Asp | 6993 | 7061 | 6995 | 7063 | 69 | 69 | + | ||||
| COXII | 7062 | 7745 | 7064 | 7747 | 684 | 684 | + | ATG | TAA | ATG | TAA |
| tRNA-Lys | 7748 | 7814 | 7750 | 7817 | 67 | 68 | + | ||||
| ATP8 | 7816 | 8022 | 7819 | 8025 | 207 | 207 | + | ATG | TAG | ATG | TAG |
| ATP6 | 8052 | 8656 | 7980 | 8659 | 605 | 680 | + | ATG | TA | ATG | TA |
| COXIII | 8657 | 9440 | 8660 | 9443 | 784 | 784 | + | ATG | T | ATG | T |
| tRNA-Gly | 9441 | 9510 | 9444 | 9514 | 70 | 71 | + | ||||
| ND3 | 9511 | 9857 | 9515 | 9861 | 347 | 347 | + | ATT | TA | ATT | TA |
| tRNA-Arg | 9858 | 9926 | 9862 | 9930 | 69 | 69 | + | ||||
| ND4L | 9928 | 10224 | 9932 | 10,228 | 297 | 297 | + | ATG | TAA | ATG | TAA |
| ND4 | 10,218 | 11,595 | 10,222 | 11,599 | 1378 | 1378 | + | ATG | T | ATG | T |
| tRNA-His | 11,596 | 11664 | 11,,600 | 11,668 | 69 | 69 | + | ||||
| tRNA-Ser | 11,665 | 11,724 | 11,669 | 11,728 | 60 | 60 | + | ||||
| tRNA-Leu | 11,725 | 11,794 | 11,729 | 11,798 | 70 | 70 | + | ||||
| ND5 | 11,795 | 13,612 | 11,799 | 13,616 | 1818 | 1818 | + | ATC | TAA | ATC | TAA |
| ND6 | 13,596 | 14,126 | 13,600 | 14,130 | 531 | 531 | - | AGA | T | AGA | T |
| tRNA-Glu | 14,121 | 14,189 | 14,125 | 14,193 | 69 | 69 | - | ||||
| Cytb | 14,193 | 15,332 | 14,197 | 15,336 | 1140 | 1140 | + | ATG | TAA | ATG | TAG |
| tRNA-Thr | 15,333 | 15,400 | 15,337 | 15,405 | 68 | 69 | + | ||||
| tRNA-Pro | 15,405 | 15,472 | 15,410 | 15,478 | 68 | 69 | - | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, Y.; Wang, X.; Chen, Y.; Shang, Y.; Cai, H.; Wang, L.; Wu, X. Mitogenomic Insights into Adaptive Evolution of African Ground Squirrels in Arid Environments. Diversity 2025, 17, 538. https://doi.org/10.3390/d17080538
Xing Y, Wang X, Chen Y, Shang Y, Cai H, Wang L, Wu X. Mitogenomic Insights into Adaptive Evolution of African Ground Squirrels in Arid Environments. Diversity. 2025; 17(8):538. https://doi.org/10.3390/d17080538
Chicago/Turabian StyleXing, Yamin, Xibao Wang, Yao Chen, Yongquan Shang, Haotian Cai, Liangkai Wang, and Xiaoyang Wu. 2025. "Mitogenomic Insights into Adaptive Evolution of African Ground Squirrels in Arid Environments" Diversity 17, no. 8: 538. https://doi.org/10.3390/d17080538
APA StyleXing, Y., Wang, X., Chen, Y., Shang, Y., Cai, H., Wang, L., & Wu, X. (2025). Mitogenomic Insights into Adaptive Evolution of African Ground Squirrels in Arid Environments. Diversity, 17(8), 538. https://doi.org/10.3390/d17080538






