Mitochondrial Genomes of Distant Fish Hybrids Reveal Maternal Inheritance Patterns and Phylogenetic Relationships
Abstract
1. Introduction
2. Materials and Methods
3. Results
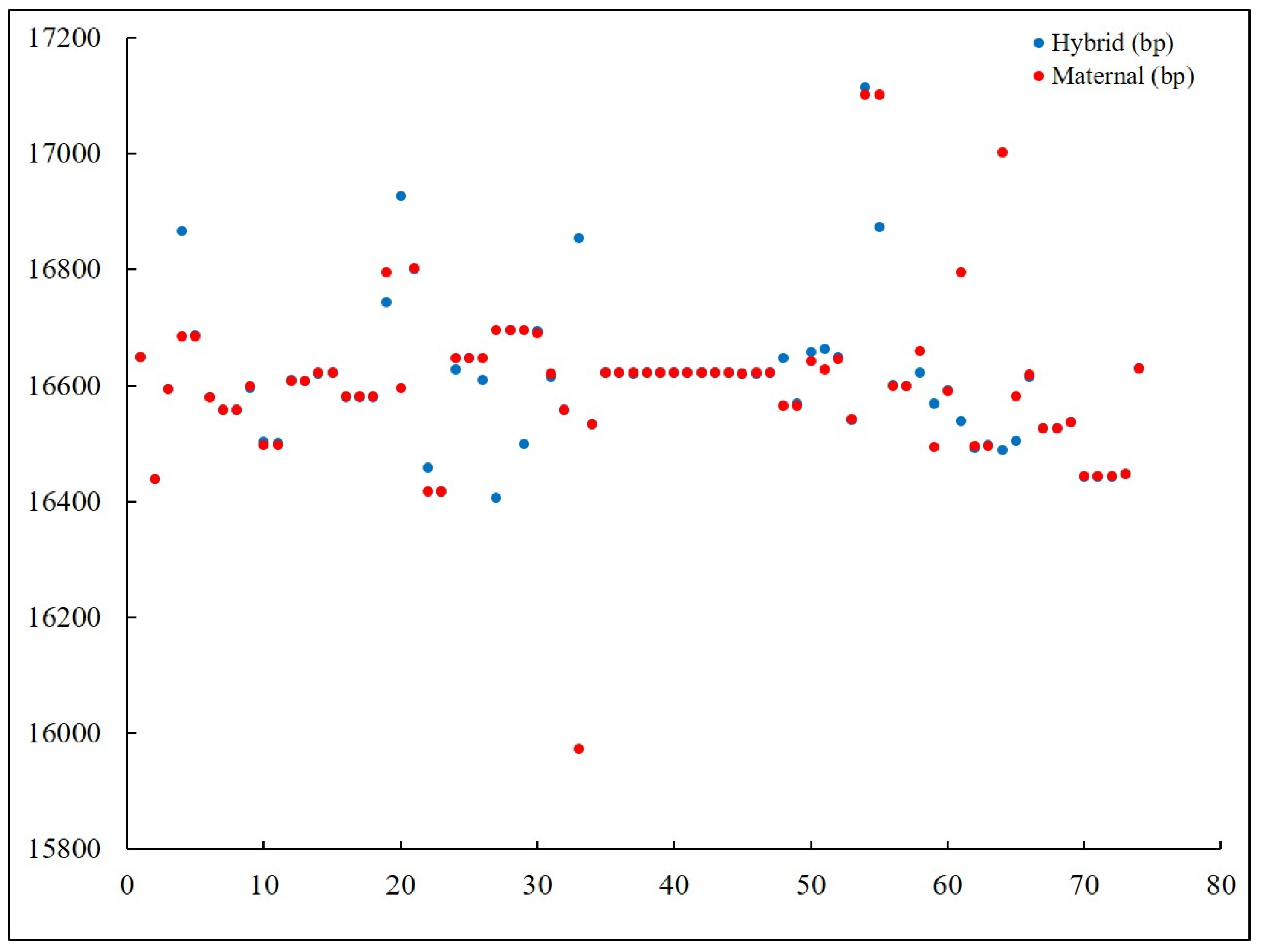
3.1. Distant Hybridization in Fish and Their Genetic Distance
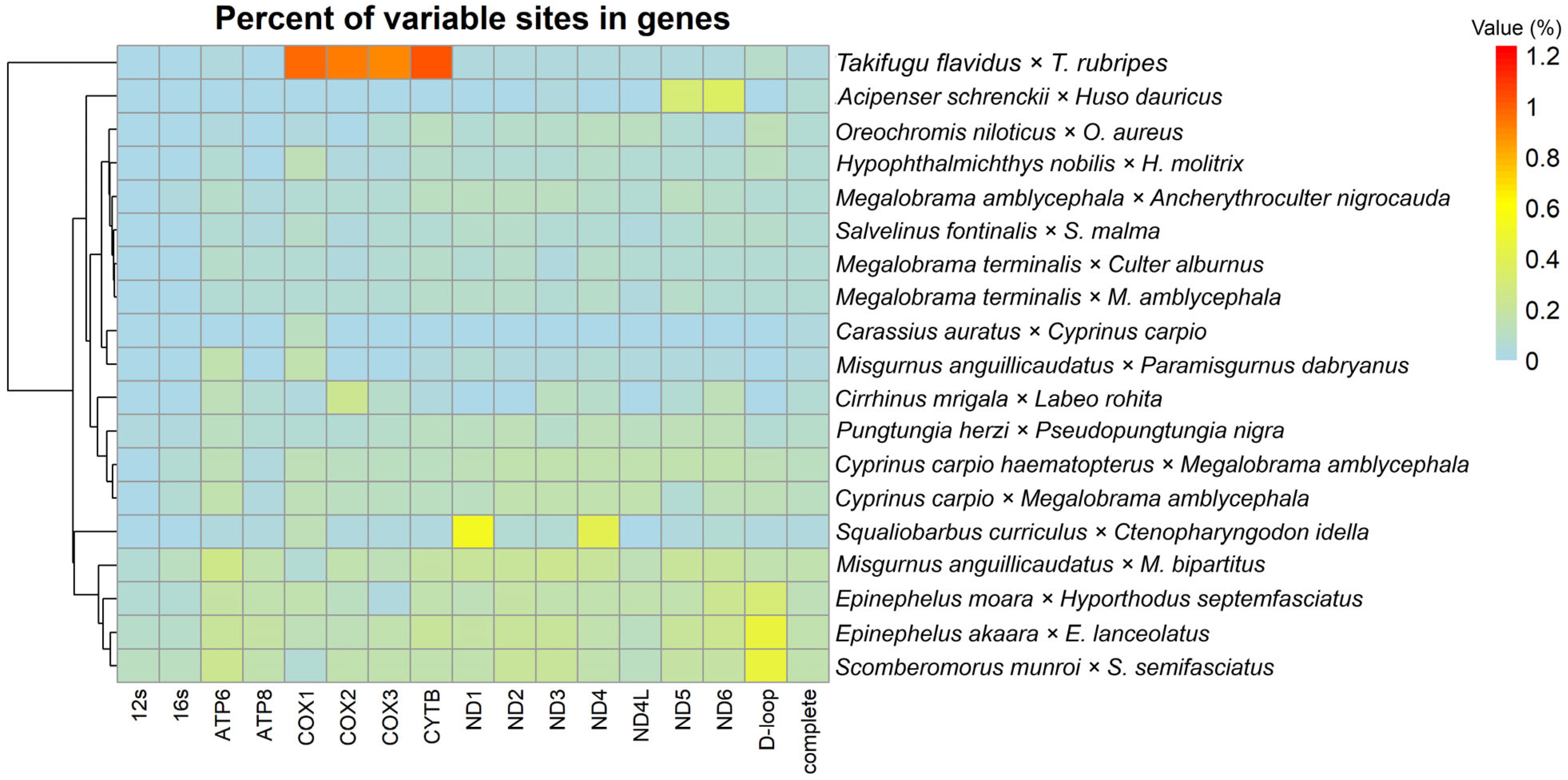
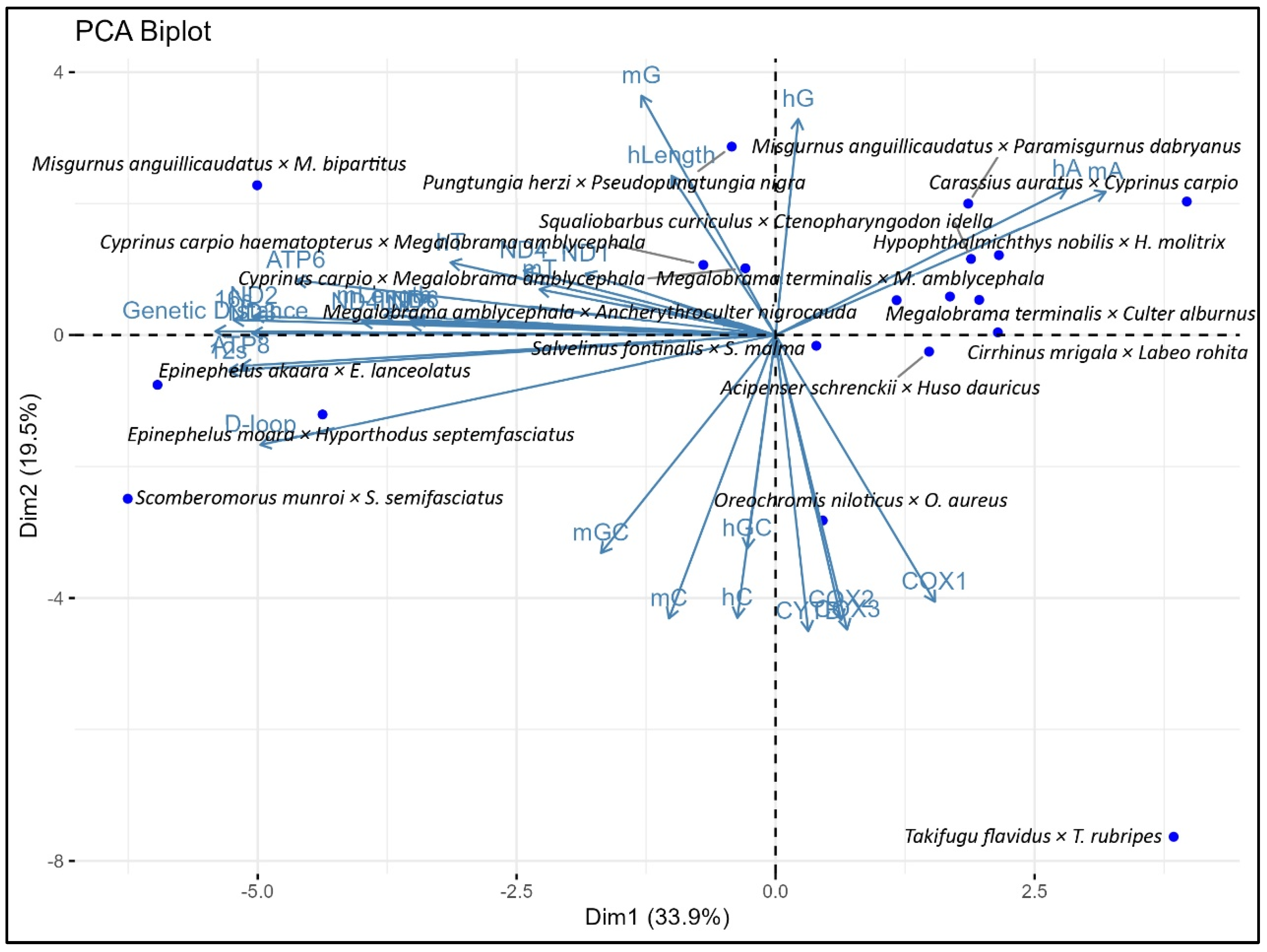
3.2. Association Between Genetic Distance and Genomic Composition and Their Variable Sites
3.3. Purifying Selection Pressure and AT-Skew in Highly Variable PCGs
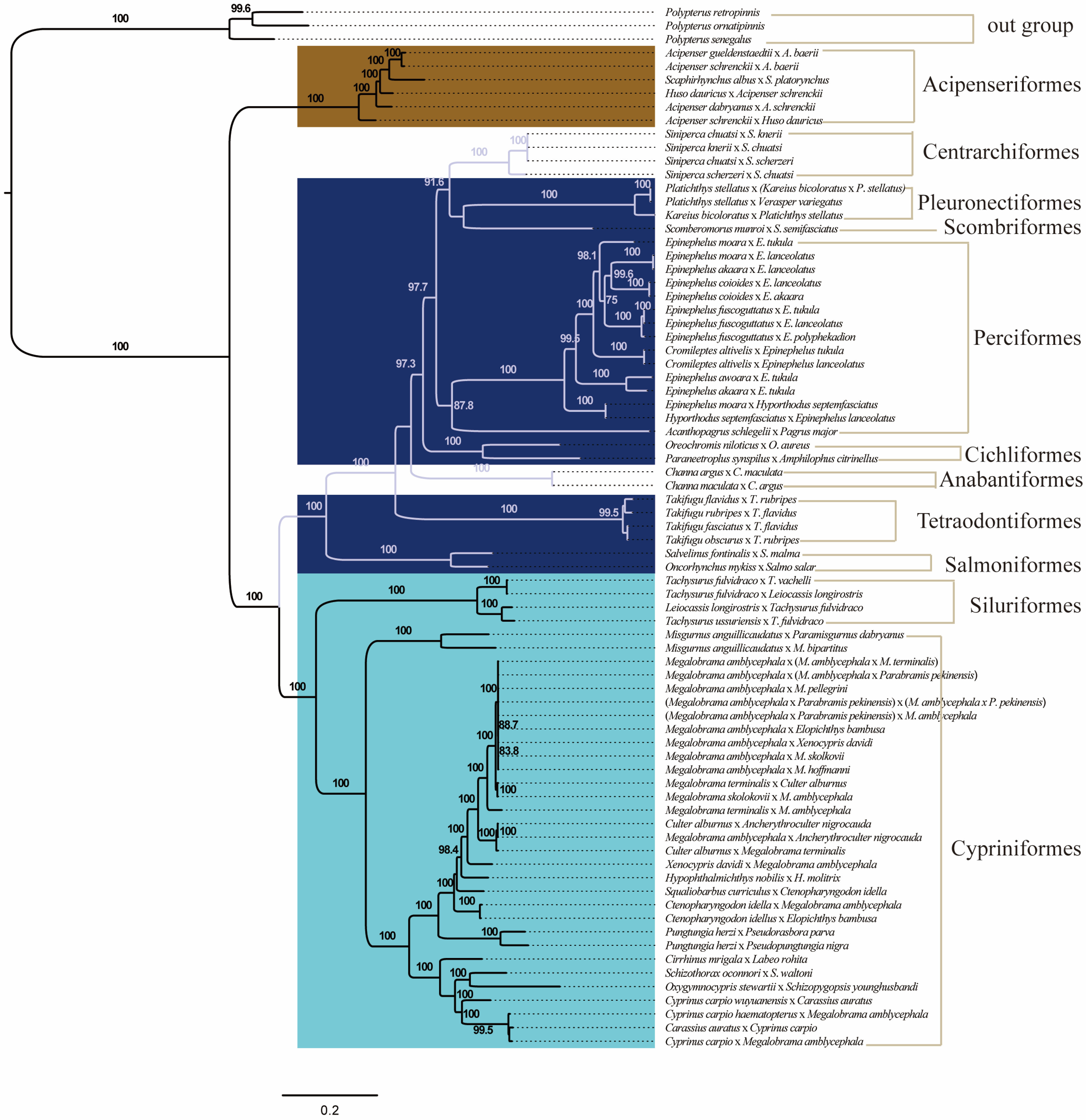
3.4. The Phylogenetic Relationship of Distantly Hybridized Fish
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Q.; Wang, S.; Tang, C.; Tao, M.; Zhang, C.; Zhou, Y.; Qin, Q.; Luo, K.; Wu, C.; Hu, F.; et al. The Research Advances in Distant Hybridization and Gynogenesis in Fish. Rev. Aquac. 2025, 17, e12972. [Google Scholar] [CrossRef]
- Peñalba, J.V.; Runemark, A.; Meier, J.I.; Singh, P.; Wogan, G.O.; Sánchez-Guillén, R.; Mallet, J.; Rometsch, S.J.; Menon, M.; Seehausen, O.; et al. The role of hybridization in species formation and persistence. Cold Spring Harb. Perspect. Biol. 2024, 16, a041445. [Google Scholar] [CrossRef] [PubMed]
- Allendorf, F.W.; Leary, R.F.; Spruell, P.; Wenburg, J.K. The problems with hybrids: Setting conservation guidelines. Trends Ecol. Evol. 2001, 16, 613–622. [Google Scholar] [CrossRef]
- Liu, S.; Wang, S.; Liu, Q.; Zhou, Y.; Zhang, C.; Tao, M.; Luo, K. The summary of fish distant hybridization. In Fish Distant Hybridization; Springer Nature: Singapore, 2022; pp. 325–343. [Google Scholar] [CrossRef]
- Wang, S.; Tang, C.; Tao, M.; Qin, Q.; Zhang, C.; Luo, K.; Zhao, R.; Wang, J.; Ren, L.; Xiao, J.; et al. Establishment and application of distant hybridization technology in fish. Sci. China Life Sci. 2019, 62, 22–45. [Google Scholar] [CrossRef]
- Zbinden, Z.D.; Douglas, M.R.; Chafin, T.K.; Douglas, M.E. A community genomics approach to natural hybridization. Proc. R. Soc. B 2023, 290, 20230768. [Google Scholar] [CrossRef]
- Scribner, K.T.; Page, K.S.; Bartron, M.L. Hybridization in freshwater fishes: A review of case studies and cytonuclear methods of biological inference. Rev. Fish Biol. Fish. 2001, 10, 293–323. [Google Scholar] [CrossRef]
- Ota, K.G. Goldfish as an Experimental Model. In Goldfish Development and Evolution; Springer: Singapore, 2021; pp. 17–44. [Google Scholar] [CrossRef]
- Wang, S.; Ye, X.; Wang, Y.; Chen, Y.; Lin, B.; Yi, Z.; Mao, Z.; Hu, F.; Zhao, R.; Wang, J.; et al. A new type of homodiploid fish derived from the interspecific hybridization of female common carp× male blunt snout bream. Sci. Rep. 2017, 7, 4189. [Google Scholar] [CrossRef]
- Cao, L.; Chen, P.; Hou, X.; Ma, J.; Yang, N.; Lu, Y.; Huang, H. rDNA and mtDNA analysis for the identification of genetic characters in the hybrid grouper derived from hybridization of Cromileptes altivelis (female) × Epinephelus lanceolatus (male). BMC Genom. Data. 2024, 25, 5. [Google Scholar] [CrossRef]
- Qi, Z.; Shi, J.; Yu, Y.; Yin, G.; Zhou, X.; Yu, Y. Paternal Mitochondrial DNA Leakage in Natural Populations of Large-Scale Loach, Paramisgurnus dabryanus. Biology 2024, 13, 604. [Google Scholar] [CrossRef]
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar] [CrossRef]
- Díaz-Arce, N.; Gagnaire, P.A.; Richardson, D.E.; Walter, J.F., III; Arnaud-Haond, S.; Fromentin, J.M.; Brophy, D.; Lutcavage, M.; Addis, P.; Alemany, F.; et al. Unidirectional trans-Atlantic gene flow and a mixed spawning area shape the genetic connectivity of Atlantic bluefin tuna. Mol. Ecol. 2024, 33, e17188. [Google Scholar] [CrossRef]
- Luo, W.; Huang, X.; Xu, X.; Dai, C.; Liu, Q.; Zhu, Y.; Wu, D.; Wang, S.; Liu, Q.; Yang, C. Population genetic characteristics of two crucian carp varieties derived from distant hybridization. Reprod. Breed. 2024, 4, 185–193. [Google Scholar] [CrossRef]
- Arnold, M.L. Natural Hybridization and Evolution; Oxford University Press: Oxford, UK, 1997. [Google Scholar] [CrossRef]
- Long, Z.; Rieseberg, L.H. Documenting homoploid hybrid speciation. Mol. Ecol. 2024, 14, e17412. [Google Scholar] [CrossRef] [PubMed]
- Olave, M.; Nater, A.; Kautt, A.F.; Meyer, A. Early stages of sympatric homoploid hybrid speciation in crater lake cichlid fishes. Nat. Commun. 2022, 13, 5893. [Google Scholar] [CrossRef] [PubMed]
- Seehausen, O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004, 19, 198–207. [Google Scholar] [CrossRef]
- Svardal, H.; Salzburger, W.; Malinsky, M. Genetic variation and hybridization in evolutionary radiations of cichlid fishes. Annu. Rev. Anim. Biosci. 2021, 9, 55–79. [Google Scholar] [CrossRef]
- Huang, Y.; Li, Z.; Li, M.; Zhang, X.; Shi, Q.; Xu, Z. Fish Genomics and Its Application in Disease-Resistance Breeding. Rev. Aquac. 2025, 17, e12973. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, G.; Zhang, Y.; Li, X.; Luo, Z.; Liu, W.; Luo, F.; Liu, H.; Yin, S.; Jiang, J.; et al. Profiling genetic breeding progress in bagrid catfishes. Fishes 2023, 8, 426. [Google Scholar] [CrossRef]
- Bartley, D.M.; Rana, K.; Immink, A.J. The use of inter-specific hybrids in aquaculture and fisheries. Rev. Fish Biol. Fish. 2000, 10, 325–337. [Google Scholar] [CrossRef]
- Diedericks, G.; Maetens, H.; Van Steenberge, M.; Snoeks, J. Testing for hybridization between Nile tilapia (Oreochromis niloticus) and blue spotted tilapia (Oreochromis leucostictus) in the Lake Edward system. J. Great Lakes Res. 2021, 47, 1446–1452. [Google Scholar] [CrossRef]
- Bhujel, R.C. A review of strategies for the management of Nile tilapia (Oreochromis niloticus) broodfish in seed production systems, especially hapa-based systems. Aquaculture 2000, 181, 37–59. [Google Scholar] [CrossRef]
- Kinami, R.; Ineno, T. First evidence of WW superfemale for hybrid sturgeon, the bester (Huso huso × Acipenser ruthenus). Aquaculture 2025, 596, 741808. [Google Scholar] [CrossRef]
- Birstein, V.J.; Doukakis, P.; Sorkin, B.; DeSalle, R. Population aggregation analysis of three caviar-producing species of sturgeons and implications for the species identification of black caviar. Conserv. Biol. 1998, 12, 766–775. [Google Scholar] [CrossRef]
- Curto, M.; Morgado-Santos, M.; Alexandre, C.M.; Alves, M.J.; Gante, H.F.; Gkenas, C.; Medeiros, J.P.; Pinheiro, P.J.; Almeida, P.R.; Magalhães, M.F.; et al. Widespread hybridization between invasive bleak (Alburnus alburnus) and Iberian chub (Squalius spp.): A neglected conservation threat. Fishes 2022, 7, 247. [Google Scholar] [CrossRef]
- Rhymer, J.M.; Simberloff, D. Extinction by hybridization and introgression. Annu. Rev. Ecol. Syst. 1996, 27, 83–109. [Google Scholar] [CrossRef]
- Hirashiki, C.; Kareiva, P.; Marvier, M. Concern over hybridization risks should not preclude conservation interventions. Conserv. Sci. Pract. 2021, 3, e424. [Google Scholar] [CrossRef]
- Perry, W.L.; Feder, J.L.; Lodge, D.M. Implications of hybridization between introduced and resident Orconectes crayfishes. Conserv. Biol. 2001, 15, 1656–1666. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Leary, R.F. Conservation and distribution of genetic variation in a polytypic species, the cutthroat trout. Conserv. Biol. 1988, 2, 170–184. [Google Scholar] [CrossRef]
- Gilg, M.R.; Kerns, E.V.; Gutierrez-Bayona, N.E.; Kooyomjian, C.; Hinojosa, N.A. Dynamic cohort analysis reveals fluctuating patterns of selection within a hybrid zone between the killifish Fundulus heteroclitus and F. grandis. Evol. Biol. 2022, 49, 1–14. [Google Scholar] [CrossRef]
- Barton, N.H.; Hewitt, G.M. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 1985, 16, 113–148. [Google Scholar] [CrossRef]
- Garcia-Vazquez, E.; Moran, P.; Martinez, J.L.; Perez, J.; de Gaudemar, B.; Beall, E. Interspecific hybridization between Atlantic salmon and brown trout introduced in the sub-Antarctic Kerguelen Islands. Aquaculture 2004, 230, 81–88. [Google Scholar] [CrossRef]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. KaKs_Calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.I.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Froese, R.; Pauly, D. (Eds.) FishBase. World Wide Web Electronic Publication. Version (10/2024). 2024. Available online: www.fishbase.org (accessed on 10 October 2024).
- Rahman, M.A.; Lee, S.; Yusoff, F.M.; Rafiquzzaman, S.M. Hybridization and Its Application in Aquaculture. In Sex Control in Aquaculture, 1st ed.; Wang, H., Piferrer, F., Chen, S., Shen, Z., Eds.; Wiley: Hoboken, NJ, USA, 2018; pp. 163–178. [Google Scholar] [CrossRef]
- Zhang, W.Z.; Xiong, X.M.; Zhang, X.J.; Wan, S.M.; Guan, N.N.; Nie, C.H.; Zhao, B.W.; Hsiao, C.D.; Wang, W.M.; Gao, Z.X. Mitochondrial genome variation after hybridization and differences in the first and second generation hybrids of bream fishes. PLoS ONE 2016, 11, e0158915. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Avise, J.C. Molecular Markers, Natural History and Evolution; Springer Science & Business Media: New York, NY, USA, 2012. [Google Scholar] [CrossRef]
- Zhang, Y.; Pan, X.; Yin, Y.; Yang, J.; Wang, X. Fertilization and growth performance in reciprocal hybrids of Dianchi golden-line barbel (Sinocyclocheilus grahami) and domestic common carp (Cyprinus carpio) and crucian carp (Carassius auratus). Aquac. Rep. 2021, 21, 100893. [Google Scholar] [CrossRef]
- Wang, Y.; Yao, J.; Luo, Y.; Tan, H.; Huang, X.; Wang, S.; Qin, Q.; Zhang, C.; Tao, M.; Dabrowski, K.; et al. Two new types of homodiploid fish and polyploid hybrids derived from the distant hybridization of female koi carp and male bighead carp. Mar. Biotechnol. 2021, 23, 628–640. [Google Scholar] [CrossRef]
- Yu, X.; Setyawan, P.; Bastiaansen, J.W.; Liu, L.; Imron, I.; Groenen, M.A.; Komen, H.; Megens, H.J. Genomic analysis of a Nile tilapia strain selected for salinity tolerance shows signatures of selection and hybridization with blue tilapia (Oreochromis aureus). Aquaculture 2022, 560, 738527. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, B.; Zou, Z.; Li, D.; Zhu, J.; Yu, J.; Xiao, W.; Yang, H. Multiple trait comparison and global intestine transcriptional provide new insights into bases of heterosis in hybrid tilapia (Oreochromis niloticus × Oreochromis aureus). Comp. Biochem. Physiol. Part D Genom. Proteom. 2024, 50, 101236. [Google Scholar] [CrossRef] [PubMed]
- Feuerstein, C.A.; Kovach, R.P.; Kruse, C.G.; Jaeger, M.E.; Bell, D.A.; Robinson, Z.L.; Whiteley, A.R. Genetic variation and hybridization determine the outcomes of conservation reintroductions. Conserv. Lett. 2024, 17, e13049. [Google Scholar] [CrossRef]
- Selz, O.M.; Seehausen, O. Interspecific hybridization can generate functional novelty in cichlid fish. Proc. R. Soc. B 2019, 286, 20191621. [Google Scholar] [CrossRef]
- Gjedrem, T.; Robinson, N. Advances by selective breeding for aquatic species: A review. Agric. Sci. 2014, 5, 1152. [Google Scholar] [CrossRef]
- Mastor, N.N.I.; Zuldin, W.H.; Faudzi, N.M.; Rodrigues, K.F.; Ransangan, J. Fish Hybridization: Enhancing Genetic Potential, Ecological Implications and Ethical Perspectives. Int. J. Agric. Biosci. 2025, 14, 374–387. [Google Scholar] [CrossRef]
- Bogár, K.; Stanivuk, J.; Géczi, A.; Fazekas, G.L.; Kovács, B.; Lázár, B.; Molnár, M.; Ardó, L.; Ljubobratović, U.; Kovács, G.; et al. Investigation of Sexes and Fertility Potential of Female Russian Sturgeon (Acipenser gueldenstaedtii) and Male American Paddlefish (Polyodon spathula) Hybrids. Life 2024, 14, 818. [Google Scholar] [CrossRef]
- Parida, S.N.; Parida, C.K.; Rout, A.K.; Kumar, V.; Dhar, S.; Bisai, K.; Behera, B.; Behera, B.K. Mitochondrial Genes and Their Application in Fish Diversity Studies. In Current Trends in Fisheries Biotechnology; Springer Nature: Singapore, 2024; pp. 13–25. [Google Scholar] [CrossRef]
- Minhas, B.F.; Beck, E.A.; Cheng, C.H.C.; Catchen, J. Novel mitochondrial genome rearrangements including duplications and extensive heteroplasmy could underlie temperature adaptations in Antarctic notothenioid fishes. Sci. Rep. 2023, 13, 6939. [Google Scholar] [CrossRef]
- Huang, X.; Jiang, Y.; Yang, T. Genetic variation and phylogeographic patterns of Harpadon nehereus in China offshore inferred from mitochondrial cytochrome b (Cyt b) sequences: With implications for its fishery management. Mar. Biodivers. 2024, 54, 18. [Google Scholar] [CrossRef]
- Kartavtsev, Y.P.; Sharina, S.N.; Saitoh, K.; Imoto, J.M.; Hanzawa, N.; Redin, A.D. Phylogenetic relationships of Russian far eastern flatfish (Pleuronectiformes, Pleuronectidae) based on two mitochondrial gene sequences, Co-1 and Cyt-b, with inferences in order phylogeny using complete mitogenome data. Mitochondrial DNA Part A 2016, 27, 667–678. [Google Scholar] [CrossRef]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. London Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, H.; Cui, J.; Yan, X.; Zhang, X.; Luo, M.; Tang, C.; Ren, L.; Liu, S. Interactions between mitochondrial and nuclear genomes and co-regulation of mitochondrial and nuclear gene expression in reciprocal intergeneric hybrids between Carassius auratus red var. × Cyprinus carpio L. Reprod. Breed. 2021, 1, 213–220. [Google Scholar] [CrossRef]
- Turner, B.J.; Fisher, M.T.; Taylor, D.S.; Davis, W.P.; Jarrett, B.L. Evolution of ‘maleness’ and outcrossing in a population of the self-fertilizing killifish, Kryptolebias marmoratus. Evol. Ecol. Res. 2006, 8, 1475–1486. [Google Scholar]
- Mackiewicz, M.; Tatarenkov, A.; Taylor, D.S.; Turner, B.J.; Avise, J.C. Extensive outcrossing and and rodioecy in a vertebrate species that otherwise reproduces as a self-fertilizing hermaphrodite. Proc. Natl. Acad. Sci. USA 2006, 103, 9924–9928. [Google Scholar] [CrossRef]
- Morgan, J.A.; Macbeth, M.; Broderick, D.; Whatmore, P.; Street, R.; Welch, D.J.; Ovenden, J.R. Hybridisation, paternal leakage and mitochondrial DNA linearization in three anomalous fish (Scombridae). Mitochondrion 2013, 13, 852–861. [Google Scholar] [CrossRef]
- Pouliot-Drouin, A.; Niaison, T.; Breton, S.; Bettinazzi, S. Investigating the role of mitochondrial membrane potential in paternal inheritance of mitochondria. Biol. J. Linn. Soc. 2025, 144, blae050. [Google Scholar] [CrossRef]
- Soroka, M. Doubly uniparental inheritance of mitochondrial DNA in freshwater mussels: History and status of the European species. J. Zool. Syst. Evol. Res. 2020, 58, 598–614. [Google Scholar] [CrossRef]
- Plazzi, F.; Cassano, A.; Passamonti, M. The quest for Doubly Uniparental Inheritance in heterodont bivalves and its detection in Meretrix lamarckii (Veneridae: Meretricinae). J. Zool. Syst. Evol. Res. 2015, 53, 87–94. [Google Scholar] [CrossRef]
- Yang, C.; Dai, C.; Liu, Q.; Zhu, Y.; Huang, X.; Xu, X.; Zhou, Y.; Wang, S.; Liu, Q.; Liu, S. Different ploidy-level hybrids derived from female common carp × male topmouth culter. Aquaculture 2025, 594, 741366. [Google Scholar] [CrossRef]
- Stewart, J.B.; Freyer, C.; Elson, J.L.; Larsson, N.G. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat. Rev. Genet. 2008, 9, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Consuegra, S.; John, E.; Verspoor, E.; de Leaniz, C.G. Patterns of natural selection acting on the mitochondrial genome of a locally adapted fish species. Genet. Sel. Evol. 2015, 47, 58. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.N.; Sultana, S. Codon usage bias and purifying selection identified in Cirrhinus reba mitogenome. J. Adv. Biotechnol. Exp. Ther. 2022, 5, 605–614. [Google Scholar] [CrossRef]
- Tamario, C.; Sunde, J.; Petersson, E.; Tibblin, P.; Forsman, A. Ecological and evolutionary consequences of environmental change and management actions for migrating fish. Front. Ecol. Evol. 2019, 7, 271. [Google Scholar] [CrossRef]
- Vasemägi, A.; Sulku, J.; Bruneaux, M.; Thalmann, O.; Mäkinen, H.; Ozerov, M. Prediction of harmful variants on mitochondrial genes: Test of habitat-dependent and demographic effects in a euryhaline fish. Ecol. Evol. 2017, 7, 3826–3835. [Google Scholar] [CrossRef]
- Pavlova, A.; Gan, H.M.; Lee, Y.P.; Austin, C.M.; Gilligan, D.M.; Lintermans, M.; Sunnucks, P. Purifying selection and genetic drift shaped Pleistocene evolution of the mitochondrial genome in an endangered Australian freshwater fish. Heredity 2017, 118, 466–476. [Google Scholar] [CrossRef]
- Zhao, D.; Guo, Y.; Gao, Y. Natural selection drives the evolution of mitogenomes in Acrossocheilus. PLoS ONE 2022, 17, e0276056. [Google Scholar] [CrossRef]
- Charneski, C.A.; Honti, F.; Bryant, J.M.; Hurst, L.D.; Feil, E.J. Atypical AT skew in Firmicute genomes results from selection and not from mutation. PLoS Genet. 2011, 7, e1002283. [Google Scholar] [CrossRef]
- Brown, W.M.; George, M., Jr.; Wilson, A.C. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 1967–1971. [Google Scholar] [CrossRef]
- Ruiz-Pesini, E.; Mishmar, D.; Brandon, M.; Procaccio, V.; Wallace, D.C. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 2004, 303, 223–226. [Google Scholar] [CrossRef]
- Wallace, D.C.; Chalkia, D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol. 2013, 5, a021220. [Google Scholar] [CrossRef]
- Rasal, K.D.; Kumar, P.V.; Risha, S.; Asgolkar, P.; Harshavarthini, M.; Acharya, A.; Shinde, S.; Dhere, S.; Rasal, A.; Sonwane, A.; et al. Genetic improvement and genomic resources of important cyprinid species: Status and future perspectives for sustainable production. Front. Genet. 2024, 15, 1398084. [Google Scholar] [CrossRef]
- Ceballos-Concha, A.; Asche, F.; Cárdenas-Retamal, R. Salmon Aquaculture in Chile: Production Growth and Socioeconomic Impacts. Rev. Aquac. 2025, 17, e12993. [Google Scholar] [CrossRef]
- Araujo, G.S.; Silva, J.W.A.D.; Cotas, J.; Pereira, L. Fish farming techniques: Current situation and trends. J. Mar. Sci. Eng. 2022, 10, 1598. [Google Scholar] [CrossRef]
- Prabu, E.; Rajagopalsamy, C.B.T.; Ahilan, B.; Jeevagan, I.J.M.A.; Renuhadevi, M.J.A.R. Tilapia–an excellent candidate species for world aquaculture: A review. Annu. Res. Rev. Biol. 2019, 31, 1–14. [Google Scholar] [CrossRef]
- Nguyen, N.H. Genetic improvement for important farmed aquaculture species with a reference to carp, tilapia and prawns in Asia: Achievements, lessons and challenges. Fish Fish. 2016, 17, 483–506. [Google Scholar] [CrossRef]
- Kestemont, P.; Mélard, C.; Held, J.A.; Dabrowski, K. Culture methods of Eurasian perch and yellow perch early life stages. In Biology and Culture of Percid Fishes: Principles and Practices; Springer: Dordrecht, The Netherlands, 2015; pp. 265–293. [Google Scholar] [CrossRef]
- Basurco, B.; Lovatelli, A.; García, B. Current status of Sparidae aquaculture. In Sparidae; Wiley Online Library: Hoboken, NJ, USA, 2011; pp. 1–50. [Google Scholar] [CrossRef]
- Marengo, M.; Durieux, E.D.; Marchand, B.; Francour, P. A review of biology, fisheries and population structure of Dentex dentex (Sparidae). Rev. Fish Biol. Fish. 2014, 24, 1065–1088. [Google Scholar] [CrossRef]
- Rimmer, M.A.; Glamuzina, B. A review of grouper (Family Serranidae: Subfamily Epinephelinae) aquaculture from a sustainability science perspective. Rev. Aquac. 2019, 11, 58–87. [Google Scholar] [CrossRef]
- Li, Y.; Gao, P.; Zhou, K.; Yao, Z.; Sun, Z.; Qin, H.; Lai, Q. Effects of saline and alkaline stresses on the survival, growth, and physiological responses in juvenile mandarin fish (Siniperca chuatsi). Aquaculture 2024, 591, 741143. [Google Scholar] [CrossRef]
- Li, X.Q.; Liu, X.M.; Leng, X.J.; Wang, X.C. Effect of salinity on growth and flesh quality of snakehead Channa argus. Oceanol. Limnol. Sin. 2008, 5, 014. [Google Scholar]
- Bronzi, P.; Chebanov, M.; Michaels, J.T.; Wei, Q.; Rosenthal, H.; Gessner, J. Sturgeon meat and caviar production: Global update 2017. J. Appl. Ichthyol. 2019, 35, 257–266. [Google Scholar] [CrossRef]
- Birstein, V.J.; Doukakis, P.; DeSalle, R. Molecular phylogeny of Acipenseridae: Nonmonophylyof Scaphirhynchinae. Copeia 2002, 2002, 287–301. [Google Scholar] [CrossRef]
- Lobanov, V.P.; Pate, J.; Joyce, A. Sturgeon and paddlefish: Review of research on broodstock and early life stage management. Aquac. Fish. 2023, 9, 871–882. [Google Scholar] [CrossRef]
- Grozea, A.; Furdean, S.; Lalescu, D.; Grozea, I.; Pătruică, S. Analyzing the growth patterns of sterlet (Acipenser ruthenus) vs. Hybrid sturgeon (a. Ruthenus, female x a. Gueldenstaedtii, male) fingerlings reared within a recirculating aquaculture system. Int. Multidiscip. Sci. Geoconf. SGEM 2023, 23, 17–24. [Google Scholar] [CrossRef]


| SL | Scientific Names of Hybrid Fish (Names of Hybrid Fish in the Same Genera Appear as Abbreviations) | Freshwater Fish |
|---|---|---|
| 1 | Acanthopagrus schlegelii × Pagrus major | N |
| 2 | Acipenser dabryanus × A. schrenckii | Y |
| 3 | Acipenser gueldenstaedtii × A. baerii | N |
| 4 | Acipenser schrenckii × A. baerii | Y |
| 5 | Acipenser schrenckii × Huso dauricus | Y |
| 6 | Carassius auratus × Cyprinus carpio | Y |
| 7 | Channa argus × C. maculata | Y |
| 8 | Channa maculata × C. argus | Y |
| 9 | Cirrhinus mrigala × Labeo rohita | Y |
| 10 | Cromileptes altivelis × Epinephelus lanceolatus | N |
| 11 | Cromileptes altivelis × Epinephelus tukula | N |
| 12 | Ctenopharyngodon idella × Megalobrama amblycephala | Y |
| 13 | Ctenopharyngodon idellus × Elopichthys bambusa | Y |
| 14 | Culter alburnus × Ancherythroculter nigrocauda | Y |
| 15 | Culter alburnus × Megalobrama terminalis | Y |
| 16 | Cyprinus carpio haematopterus × Megalobrama amblycephala | Y |
| 17 | Cyprinus carpio wuyuanensis × Carassius auratus | Y |
| 18 | Cyprinus carpio × Megalobrama amblycephala | Y |
| 19 | Epinephelus akaara × E. lanceolatus | N |
| 20 | Epinephelus akaara × E. tukula | N |
| 21 | Epinephelus awoara × Epinephelus tukula | N |
| 22 | Epinephelus coioides × E. akaara | N |
| 23 | Epinephelus coioides × E. lanceolatus | N |
| 24 | Epinephelus fuscoguttatus × E. lanceolatus | N |
| 25 | Epinephelus fuscoguttatus × E. polyphekadion | N |
| 26 | Epinephelus fuscoguttatus × E. tukula | N |
| 27 | Epinephelus moara × E. lanceolatus | N |
| 28 | Epinephelus moara × E. tukula | N |
| 29 | Epinephelus moara × Hyporthodus septemfasciatus | N |
| 30 | Huso dauricus × Acipenser schrenckii | Y |
| 31 | Hypophthalmichthys nobilis × H. molitrix | Y |
| 32 | Hyporthodus septemfasciatus × Epinephelus lanceolatus | N |
| 33 | Kareius bicoloratus × Platichthys stellatus | N |
| 34 | Leiocassis longirostris × Tachysurus fulvidraco | Y |
| 35 | Megalobrama amblycephala × (M. amblycephala × M. terminalis) | Y |
| 36 | Megalobrama amblycephala × (M. amblycephala × Parabramis pekinensis) | Y |
| 37 | Megalobrama amblycephala × Ancherythroculter nigrocauda | Y |
| 38 | Megalobrama amblycephala × Elopichthys bambusa | Y |
| 39 | Megalobrama amblycephala × M. hoffmanni | Y |
| 40 | Megalobrama amblycephala × M. pellegrini | Y |
| 41 | Megalobrama amblycephala × M. skolkovii | Y |
| 42 | (Megalobrama amblycephala × Parabramis pekinensis) × (M. amblycephala × P. pekinensis) | Y |
| 43 | (Megalobrama amblycephala × Parabramis pekinensis) × M. amblycephala | Y |
| 44 | Megalobrama amblycephala × Xenocypris davidi | Y |
| 45 | Megalobrama skolokovii × M. amblycephala | Y |
| 46 | Megalobrama terminalis × Culter alburnus | Y |
| 47 | Megalobrama terminalis × M. amblycephala | Y |
| 48 | Misgurnus anguillicaudatus × M. bipartitus | Y |
| 49 | Misgurnus anguillicaudatus × Paramisgurnus dabryanus | Y |
| 50 | Oncorhynchus mykiss × Salmo salar | N |
| 51 | Oreochromis niloticus × O. aureus | N |
| 52 | Oxygymnocypris stewartii × Schizopygopsis younghusbandi | Y |
| 53 | Paraneetroplus synspilus × Amphilophus citrinellus | Y |
| 54 | Platichthys stellatus × (Kareius bicoloratus × P. stellatus) | N |
| 55 | Platichthys stellatus × Verasper variegatus | N |
| 56 | Pungtungia herzi × Pseudopungtungia nigra | Y |
| 57 | Pungtungia herzi × Pseudorasbora parva | Y |
| 58 | Salvelinus fontinalis × S. malma | Y |
| 59 | Scaphirhynchus albus × S. platorynchus | Y |
| 60 | Schizothorax oconnori × S. waltoni | Y |
| 61 | Scomberomorus munroi × S. semifasciatus | N |
| 62 | Siniperca chuatsi × S. knerii | Y |
| 63 | Siniperca chuatsi × S. scherzeri | Y |
| 64 | Siniperca knerii × S. chuatsi | Y |
| 65 | Siniperca scherzeri × S. chuatsi | Y |
| 66 | Squaliobarbus curriculus × Ctenopharyngodon idella | Y |
| 67 | Tachysurus fulvidraco × Leiocassis longirostris | Y |
| 68 | Tachysurus fulvidraco × T. vachelli | Y |
| 69 | Tachysurus ussuriensis × T. fulvidraco | Y |
| 70 | Takifugu fasciatus × T. flavidus | N |
| 71 | Takifugu flavidus × T. rubripes | N |
| 72 | Takifugu obscurus × T. rubripes | N |
| 73 | Takifugu rubripes × T. flavidus | N |
| 74 | Xenocypris davidi × Megalobrama amblycephala | Y |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Safiul Azam, F.M.; Ao, L.; Lin, C.; Wang, J.; Li, R.; Zou, Y. Mitochondrial Genomes of Distant Fish Hybrids Reveal Maternal Inheritance Patterns and Phylogenetic Relationships. Diversity 2025, 17, 510. https://doi.org/10.3390/d17080510
Chen S, Safiul Azam FM, Ao L, Lin C, Wang J, Li R, Zou Y. Mitochondrial Genomes of Distant Fish Hybrids Reveal Maternal Inheritance Patterns and Phylogenetic Relationships. Diversity. 2025; 17(8):510. https://doi.org/10.3390/d17080510
Chicago/Turabian StyleChen, Shixi, Fardous Mohammad Safiul Azam, Li Ao, Chanchun Lin, Jiahao Wang, Rui Li, and Yuanchao Zou. 2025. "Mitochondrial Genomes of Distant Fish Hybrids Reveal Maternal Inheritance Patterns and Phylogenetic Relationships" Diversity 17, no. 8: 510. https://doi.org/10.3390/d17080510
APA StyleChen, S., Safiul Azam, F. M., Ao, L., Lin, C., Wang, J., Li, R., & Zou, Y. (2025). Mitochondrial Genomes of Distant Fish Hybrids Reveal Maternal Inheritance Patterns and Phylogenetic Relationships. Diversity, 17(8), 510. https://doi.org/10.3390/d17080510







