Massilia paldalensis sp. nov., Isolated from Stream Bank Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Cultivation
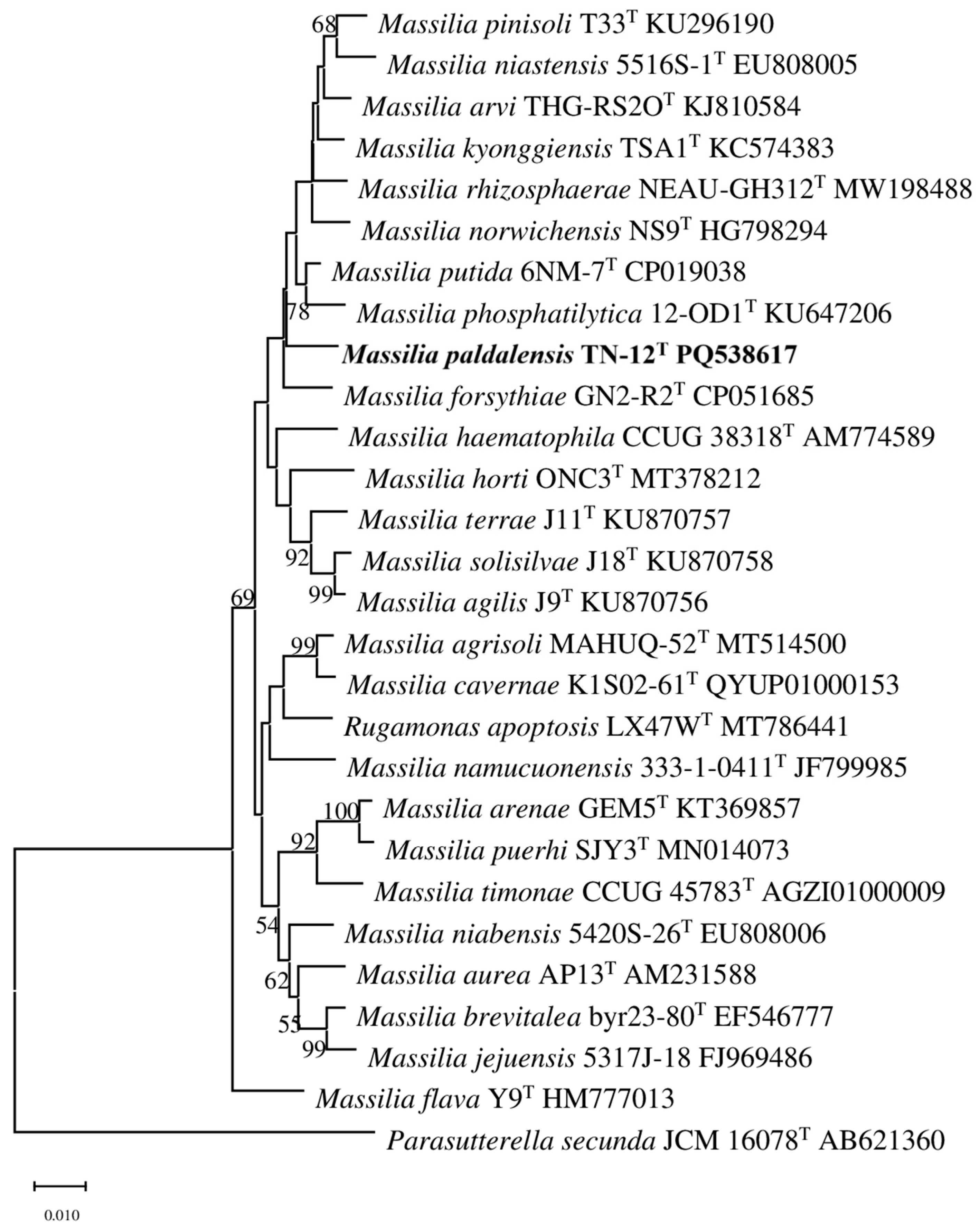
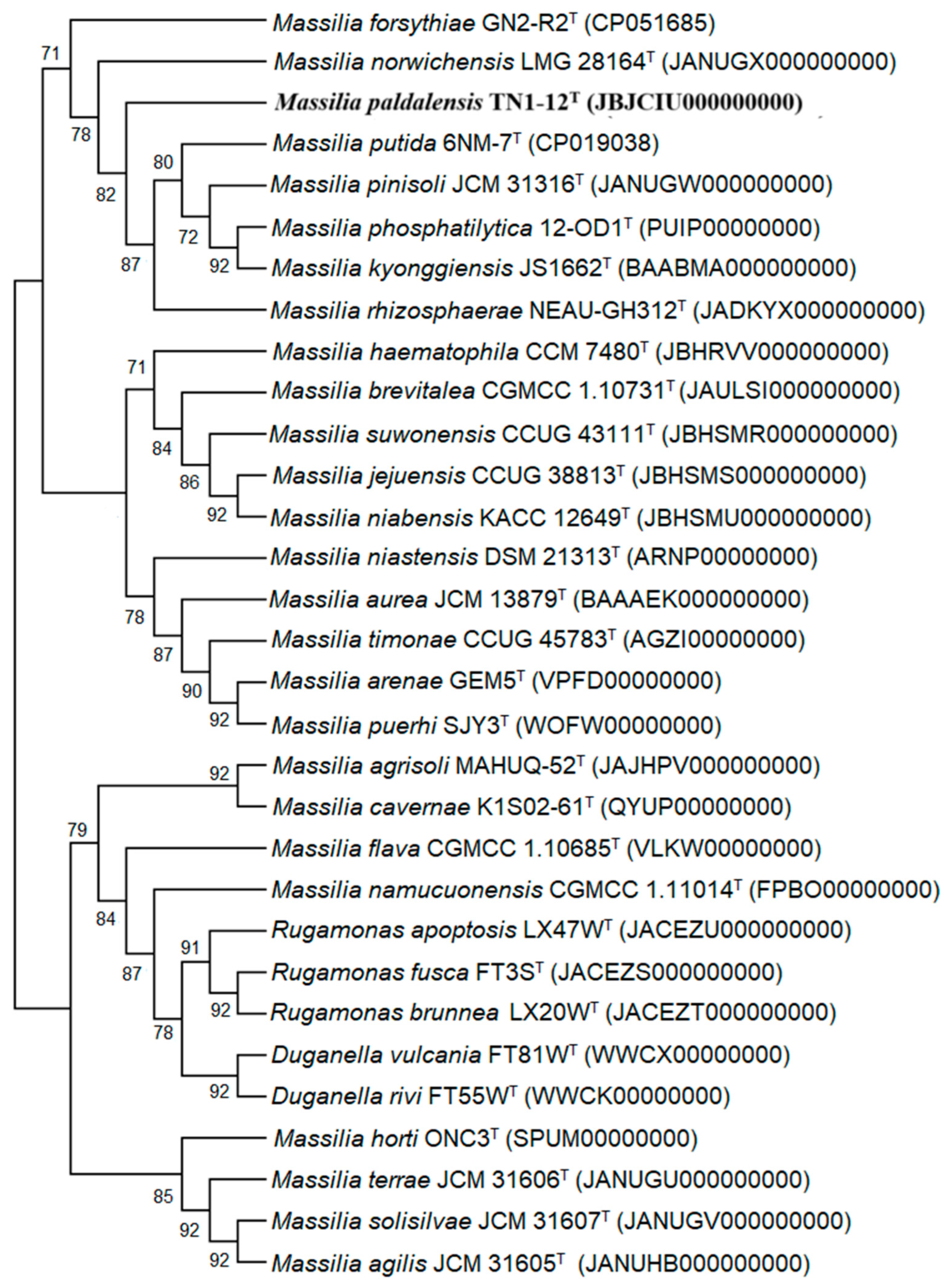
2.2. 16S rRNA Gene, Genome and Phylogenetic Analyses
2.3. Physiology and Chemotaxonomy
3. Results
3.1. 16S rRNA Gene, Genome and Phylogenetic Analyses
3.2. Physiology and Chemotaxonomy
4. Discussion
Description of Massilia paldalensis sp. nov.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- La Scola, B.; Birtles, R.J.; Mallet, M.N.; Raoult, D. Massilia timonae gen. nov., sp. nov., isolated from blood of an immunocompromised patient with cerebellar lesions. J. Clin. Microbiol. 1998, 36, 2847–2852. [Google Scholar] [CrossRef] [PubMed]
- Kämpfer, P.; Lodders, N.; Martin, K.; Falsen, E. Revision of the genus Massilia La Scola et al. 2000, with an emended description of the genus and inclusion of all species of the genus Naxibacter as new combinations, and proposal of Massilia consociata sp. nov. Int. J. Syst. Evol. Microbiol. 2011, 61, 1528–1533. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Yu, X.; Wang, G. Massilia tieshanensis sp. nov., isolated from mining soil. Int. J. Syst. Evol. Microbiol. 2012, 62 Pt 10, 2356–2362. [Google Scholar] [CrossRef]
- Weon, H.-Y.; Kim, B.-Y.; Hong, S.-B.; Jeon, Y.-A.; Koo, B.-S.; Kwon, S.-W.; Stackebrandt, E. Massilia niabensis sp. nov. and Massilia niastensis sp. nov., isolated from air samples. Int. J. Syst. Evol. Microbiol. 2009, 59 Pt 7, 1656–1660. [Google Scholar] [CrossRef]
- Gallego, V.; Sánchez-Porro, C.; García, M.T.; Ventosa, A. Massilia aurea sp. nov., isolated from drinking water. Int. J. Syst. Evol. Microbiol. 2006, 56 Pt 10, 2449–2453. [Google Scholar] [CrossRef]
- Heo, J.; Won, M.; Lee, D.; Han, B.-H.; Hong, S.-B.; Kwon, S.-W. Duganella dendranthematis sp. nov. and Massilia forsythiae sp. nov., isolated from flowers. Int. J. Syst. Evol. Microbiol. 2022, 72, 005487. [Google Scholar] [CrossRef]
- Feng, G.-D.; Yang, S.-Z.; Li, H.-P.; Zhu, H.-H. Massilia putida sp. nov., a dimethyl disulfide-producing bacterium isolated from wolfram mine tailing. Int. J. Syst. Evol. Microbiol. 2016, 66, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Cao, P.; Du, C.; Zhang, X.; Bing, H.; Li, L.; Sun, P.; Xiang, W.; Zhao, J.; Wang, X. Massilia rhizosphaerae sp. nov., a rice-associated rhizobacterium with antibacterial activity against Ralstonia solanacearum. Int. J. Syst. Evol. Microbiol. 2021, 71, 005009. [Google Scholar] [CrossRef]
- Dahal, R.H.; Chaudhary, D.K.; Kim, J. Genome insight and description of antibiotic producing Massilia antibiotica sp. nov., isolated from oil-contaminated soil. Sci. Rep. 2021, 11, 12864. [Google Scholar]
- Kour, D.; Kaur, T.; Devi, R.; Yadav, A.; Singh, M.; Joshi, D. Beneficial microbiomes for bioremediation of diverse contaminated environments for environmental sustainability: Present status and future challenges. Environ. Sci. Pollut. Res. 2021, 28, 24917–24939. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A quality control tool for high throughput sequence data. In Babraham Bioinformatics; Babraham Institute: Babraham, UK, 2010. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Na, S.I.; Kim, Y.O.; Yoon, S.H.; Ha, S.M.; Baek, I.; Chun, J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 2018, 56, 280–285. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Smibert, R.M.; Krieg, N.R. Phenotypic characterization. In Methods for General and Molecular Bacteriology; Gerhardt, P., Murray, R.G.E., Wood, W.A., Krieg, N.R., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 607–654. [Google Scholar]
- Sasser, M. Identification of bacteria by gas chromatography of cellular fatty acids. Tech. Note 1990, 101, 1–6. [Google Scholar]
- Minnikin, D.E.; O’donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Hiraishi, A.; Ueda, Y.; Ishihara, J.; Mori, T. Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J. Gen. Appl. Microbiol. 1996, 42, 457–469. [Google Scholar] [CrossRef]
- Wayne, L.G. International Committee on Systematic Bacteriology: Announcement of the report of the ad hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Zentralblatt Bakteriol. Mikrobiol. Hygiene A 1988, 268, 433–434. [Google Scholar]
- Abou-Shanab, R.A.; van Berkum, P.; Angle, J.S. Heavy metal resistance and genotypic analysis of metal resistance genes in gram-positive and gram-negative bacteria present in Ni-rich serpentine soil and in the rhizosphere of Alyssum murale. Chemosphere 2007, 68, 360–367. [Google Scholar] [CrossRef] [PubMed]
| Characterictics | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Isolation source | Soil | Wolfram mine tailing [7] | Flowers [6] | Rhizosphere soil of rice | Drinking water [5] | Air [4] |
| Media | R2A, TSA, NA, LB | R2A, PYE [7] | R2A [6] | R2A, NA, ISP2 | PCA, R2A, TSB, NA [5] | R2A, NA [4] |
| Oxidase | + | − | − | + | + | + |
| Catalase | + | + | + | ND | + | + |
| Urea | − | − | − | − | − | − |
| NaCl tolerance (%, w/v) | 0–2 | 0–1 | 0–1 | 0–5 | 0–2 | 0–1 |
| Temperature for growth | 10–40 | 20–37 | 10–30 | 10–40 | 10–30 | 10–35 |
| pH (optimum) | 5–8 (7) | 6–8 (7) | 4.5–8.5 (7) | 4–8 (7) | 4.5–9 (7.5) | 6.5–9 (7) |
| Hydrolysis of: | ||||||
| Casein | + | − | − | ND | + | − |
| Starch | + | + | + | − | + | + |
| Gelatin | + | − | + | + | − | − |
| Esculin | + | − | + | − | + | − |
| Tween 80 | + | − | − | − | − | + |
| Assimilation of: | ||||||
| d-Glucose | + | + | + | + | + | − |
| l-Arabinose | + | + | + | − | − | − |
| d-Mannose | + | + | − | + | + | − |
| d-Mannitol | − | − | − | − | − | − |
| N-acetyl-glucosamine | − | + | − | + | − | − |
| Maltose | + | + | + | + | + | − |
| Potassium gluconate | + | + | − | + | − | − |
| Capric acid | + | − | − | + | − | − |
| Adipic acid | − | − | − | + | + | − |
| Malic acid | + | − | + | − | + | + |
| Trisodium citrate | + | − | − | − | + | − |
| Phenylacetic acid | − | − | − | + | − | − |
| Enzyme activities: | ||||||
| Lipase (C14) | − | − | − | + | − | − |
| Cystine arylamidase | − | − | − | + | − | − |
| Trypsin | − | − | − | + | + | − |
| α-chymotrypsin | + | + | − | + | − | − |
| α-galactosidase | − | + | + | − | − | − |
| β-galactosidase | − | + | + | − | − | + |
| β-glucuronidase | − | − | − | − | − | + |
| β-glucosidase | + | + | + | + | + | − |
| N-acetyl- β-glucosaminidase | − | + | − | + | + | − |
| α-mannosidase | − | − | − | − | + | − |
| DNA G+C content (mol%) | 66.7 | 66.8 [7] | 66.5 [6] | 66.3 | 66.0 [5] | 67.8 [4] |
| Fatty Acid | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 10:0 | − | − | − | − | 0.62 | 0.56 |
| 10:0 3OH | 6.09 | 3.69 | 4.83 | − | − | 5.73 |
| 12:0 | 3.73 | 1.45 | 2.27 | − | − | 8.32 |
| 12:0 3OH | − | 0.88 | − | − | 0.5 | − |
| 14:0 | 1.79 | 3.52 | 2.68 | − | 1.33 | − |
| 14:0 2OH | 3.27 | 2.31 | 2.93 | − | − | − |
| 14:0 iso | − | − | − | 0.7 | − | − |
| 15:0 iso | − | − | − | − | 0.89 | − |
| 15:0 anteiso | − | − | − | − | 14.83 | − |
| 16:0 | 31.50 | 26.21 | 30.82 | 39.7 | 7.26 | 22.55 |
| 16:0 iso | − | − | − | 1.4 | 16.46 | − |
| 17:0 anteiso | − | − | − | − | 17.22 | − |
| 17:0 iso | − | − | − | − | 2.49 | − |
| 17:0 cyclo | 18.82 | 6.69 | − | − | − | − |
| 17:1 w8c | − | − | − | − | 0.59 | − |
| 18:0 | − | − | − | − | 0.56 | − |
| 18:1 w9c | − | − | − | − | 7.86 | − |
| 18:1 w7c 11-methyl | − | 1.31 | − | − | − | − |
| Summed Feature 3 * | 23.85 | 26.37 | 46.50 | 47.9 | 3.16 | 51.10 |
| Summed Feature 8 ** | 10.41 | 26.06 | 9.37 | 9.8 | 25.08 | 10.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, N.T.A.; Kim, J. Massilia paldalensis sp. nov., Isolated from Stream Bank Soil. Diversity 2025, 17, 327. https://doi.org/10.3390/d17050327
Nguyen NTA, Kim J. Massilia paldalensis sp. nov., Isolated from Stream Bank Soil. Diversity. 2025; 17(5):327. https://doi.org/10.3390/d17050327
Chicago/Turabian StyleNguyen, Nhi Thi Ai, and Jaisoo Kim. 2025. "Massilia paldalensis sp. nov., Isolated from Stream Bank Soil" Diversity 17, no. 5: 327. https://doi.org/10.3390/d17050327
APA StyleNguyen, N. T. A., & Kim, J. (2025). Massilia paldalensis sp. nov., Isolated from Stream Bank Soil. Diversity, 17(5), 327. https://doi.org/10.3390/d17050327







