Insights into the Taxonomy of the Genus Chrysastrella (Chrysophyceae), with Establishment of Chrysastrellaceae fam. nov.
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Morphological Observations
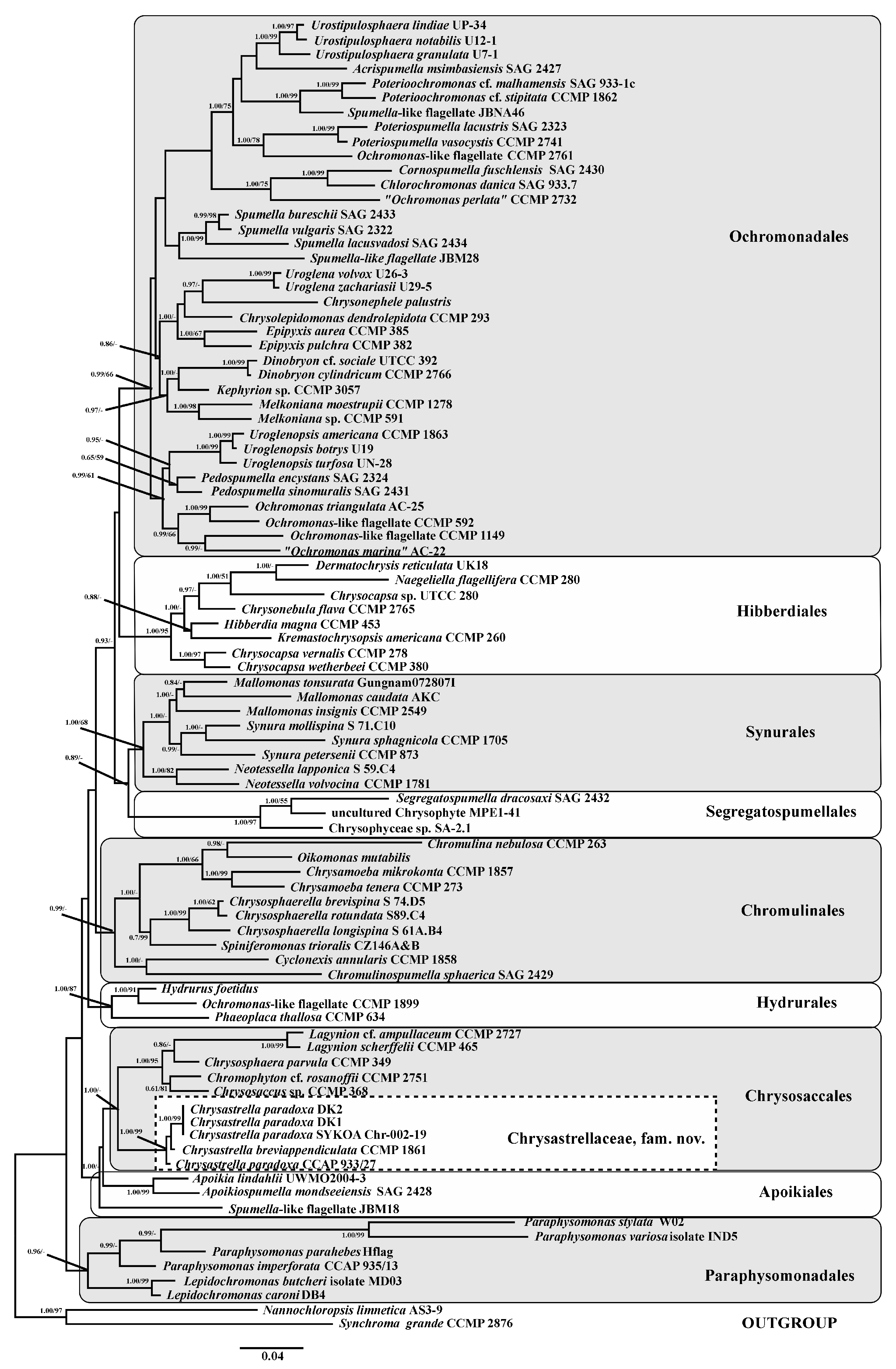
3.2. Phylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andersen, R.A.; Graf, L.; Malakhov, Y.; Yoon, H.S. Rediscovery of the Ochromonas type species Ochromonas triangulata (Chrysophyceae) from its type locality (Lake Veysove, Donetsk region, Ukraine). Phycologia 2017, 56, 591–604. [Google Scholar] [CrossRef]
- Boenigk, J.; Pfandl, K.; Stadler, P.; Chatzinotas, A. High diversity of the ‘Spumella-like’ flagellates: An investigation based on the SSU rRNA gene sequences of isolates from habitats located in six different geographic regions. Environ. Microbiol. 2005, 7, 685–697. [Google Scholar] [CrossRef]
- Pietsch, T.; Arndt, H. Comparison of mixotrophic and heterotrophic chrysomonads of similar size regarding bacterivory and growth rate. Eur. J. Protistol. 2024, 95, 126109. [Google Scholar] [CrossRef]
- Kapustin, D.A.; Iurmanov, A.A.; Kulikovskiy, M.S. A nomenclator of the genus Ochromonas sensu lato (Chrysophyceae). Phytotaxa 2025. accepted. [Google Scholar]
- Chodat, R. Matériaux pour l’histoire des Algues de la Suisse. Bull. Soc. Bot. Genève 1921, 13, 66–114. [Google Scholar]
- Pascher, A. Neue oder wenig bekannte Protisten. XII. Arch. Protististenk. 1924, 48, 492–508. [Google Scholar]
- Pascher, A. Die braune Algenreihe der Chrysophyceen. Arch. Protistenk. 1925, 52, 489–564. [Google Scholar]
- Conrad, W. Recherches sur les flagellates de nos eaux saumâtres. II. Chrysomonadines. Arch. Protistenk. 1926, 56, 167–231. [Google Scholar]
- Deflandre, G. Monographie du genre Trachelomonas Ehr. (Suite et fin). Rev. Gén. Bot. 1927, 39, 73–98. [Google Scholar]
- Deflandre, P. Sur l’abus de l’emploi en paléontologie du nom de genre Trachelomonas. Ann. Protistol. 1934, 4, 151–165. [Google Scholar]
- Rampi, L. Note sur les Chrysostomatacées du dépôt de Crognuolo (Monte Amiata). Bull. Soc. Fr. Microsc. 1939, 8, 15–20. [Google Scholar]
- Zanon, V. Saggio systematica dell Crisostomacee deposito Quaternario di Crisostomacee in Roma (Nota preventiva). Acta Pont. Acad. Sci. 1947, 11, 43–62. [Google Scholar]
- Frenguelli, J. Nuevas especies argentinas del género Chrysastrella (Crysostomataceae). Not. Mus. La Plata 1945, 10, 99–105. [Google Scholar]
- Frenguelli, J. Analisis microscopico de una segunda serie de muestras de la turbera del rio de la Mision, Rio Grande, Tierra del Fuego, extraidos por el Dr. Väinö Auer. Ann. Acad. Sci. Fenn. Ser. A. III Geol.-Geogr. 1953, 34, 1–52. [Google Scholar]
- Frenguelli, J.; Orlando, H.A. Diatomeas y silicoflagelados del sector Antártico Sudamericano. Inst. Antárt. Argent. Publ. 1958, 5, 1–191. [Google Scholar]
- Hibberd, D.J. Observations on cytology and ultrastructure of Ochromonas tuberculatus sp. nov. (Chrysophyceae), with special reference to the discobolocysts. Br. Phycol. J. 1970, 5, 119–143. [Google Scholar] [CrossRef]
- Hibberd, D.J. Ultrastructure of cyst formation in Ochromonas tuberculata (Chrysophyceae). J. Phycol. 1977, 13, 309–320. [Google Scholar] [CrossRef]
- Kapustin, D. Diversity and taxonomy of chrysophytes from the Pasvik State Nature Reserve (Russia). Water 2024, 16, 2990. [Google Scholar] [CrossRef]
- Guillard, R.R.L.; Lorenzen, C.J. Yellow-green algae with Chlorophyllide C1,2. J. Phycol. 1972, 8, 10–14. [Google Scholar] [CrossRef]
- Katana, A.; Kwiatowski, J.; Spalik, K.; Zakryś, B.; Szalacha, E.; Szymańska, H. Phylogenetic position of Koliella (Chlorophyta) as inferred from nuclear and chloroplast small subunit rDNA. J. Phycol. 2001, 37, 443–451. [Google Scholar] [CrossRef]
- Hamby, R.K.; Sims, L.; Issel, L.; Zimmer, E. Direct ribosomal RNA sequencing: Optimization of extraction and sequencing methods for work with higher plants. Plant Mol. Biol. Rep. 1988, 6, 175–192. [Google Scholar] [CrossRef]
- Daugbjerg, N.; Andersen, R.A. Phylogenetic analysis of the rbcL sequences from haptophytes and heterokont algae suggest their chloroplasts are unrelated. Mol. Biol. Evol. 1997, 14, 1242–1251. [Google Scholar] [CrossRef] [PubMed]
- Gusev, E.S.; Čertnerová, D.; Škaloudová, M.; Škaloud, P. Exploring Cryptic Diversity and Distribution Patterns in the Mallomonas kalinae/rasilis Species Complex with a Description of a New Taxon—Mallomonas furtiva sp. nov. J. Eukaryot. Microbiol. 2018, 65, 38–47. [Google Scholar] [CrossRef]
- Martynenko, N.; Gusev, E.; Kapustin, D.; Kulikovskiy, M. A New Cryptic Species of the Genus Mychonastes (Chlorophyceae, Sphaeropleales). Plants 2022, 11, 3363. [Google Scholar] [CrossRef]
- Martynenko, N.; Kezlya, E.; Gusev, E. Description of a New Species of the Genus Cryptomonas (Cryptophyceae: Cryptomonadales), Isolated from Soils in a Tropical Forest. Diversity 2022, 14, 1001. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Doflein, F. Untersuchungen über Chrysomonadinen. I. Ochromonas granularis Dofl. II. Über Chrysamoeba radians Klebs. Arch. Protistenk. 1922, 44, 149–213. [Google Scholar]
- Doflein, F. Untersuchungen über Chrysomonadinen. III. Arten von Chromulina und Ochromonas aus dem badischen Schwarzwald und ihre Cystenbildung. Arch. Protistenk. 1923, 46, 267–327. [Google Scholar]
- Matvienko, A.M. De chrysomonade nova e viciniis Charkov. Bot. Mat. Otd. Spor. Rast. 1949, 6, 21–25. (In Russian) [Google Scholar]
- Matvienko, A.M. Chrysomonadineae paludis Charkoviensis Mochovatoje. Bot. Mat. Otd. Spor. Rast. 1951, 7, 10–18. (In Russian) [Google Scholar]
- Pusztai, M.; Škaloud, P. Elucidating the evolution and diversity of Uroglena-like colonial flagellates (Chrysophyceae): Polyphyletic origin of the morphotype. Eur. J. Phycol. 2019, 54, 404–416. [Google Scholar] [CrossRef]
- Pusztai, M.; Škaloud, P. Species delimitation within the colonial flagellates Uroglena, Uroglenopsis and Urostipulosphaera (Chrysophyceae). Eur. J. Phycol. 2022, 57, 79–95. [Google Scholar] [CrossRef]
- Findenig, B.M.; Antonis Chatzinotas, A.; Boenigk, J. Taxonomic and Ecological Characterization of Stomatocysts of Spumella-Like Flagellates (Chrysophyceae). J. Phycol. 2010, 46, 868–881. [Google Scholar] [CrossRef]
- Jeong, M.; Kim, J.I.; Nam, S.W.; Shin, W. Molecular Phylogeny and Taxonomy of the Genus Spumella (Chrysophyceae) Based on Morphological and Molecular Evidence. Front. Plant Sci. 2021, 12, 758067. [Google Scholar] [CrossRef] [PubMed]
- Jeong, M.; Wang, Y.; Kim, J.I.; Shin, W. Multigene phylogeny reveals a cryptic diversity in the genus Dinobryon (Chrysophyceae) with integrative description of five new species. Front. Plant Sci. 2023, 14, 1150814. [Google Scholar] [CrossRef]
- Kapustin, D.; Sterlyagova, I.; Patova, E. Morphology of Chrysastrella paradoxa stomatocysts from the Subpolar Urals (Russia) with comments on related morphotypes. Phytotaxa 2019, 402, 295–300. [Google Scholar] [CrossRef]
- Kalina, T. Morphologie und Artbegrenzung von Ochromonas crenata Klebs (Chrysomonadales). Acta Univ. Carolinae–Biol. 1964, 2, 149–153. [Google Scholar]
- Bock, C.; Olefeld, J.L.; Vogt, J.C.; Albach, D.C.; Boenigk, J. Phylogenetic and functional diversity of Chrysophyceae in inland waters. Org. Divers. Evol. 2022, 22, 327–341. [Google Scholar] [CrossRef]
- Malavasi, V.; Pusztai, M.; Jadrná, I.; Škvorová, Z.; Škaloud, P. Morphological diversity and phylogeny of the palmelloid chrysophyte genera Chrysotilos and Globulochrysis, gen. nov. Eur. J. Phycol. 2024, 59, 279–289. [Google Scholar] [CrossRef]
- Bourrelly, P. Recherches sur les Chrysophycées. Morphologie, Phylogénie, Systématique. Rev. Algol. Mém. Hors- Sér. 1957, 1, 1–412. [Google Scholar]
- Kristiansen, J.; Škaloud, P. Chrysophyta. In Handbook of the Protists, 2nd ed.; Archibald, J.M., Simpson, A.G.B., Slamovits, C.H., Eds.; Springer International Publishing AG: Cham, Switzerland, 2017; pp. 331–366. [Google Scholar] [CrossRef]
- Andersen, R.A.; Graf, L.; Malakhov, Y.; Kim, H.; Yoon, H.S. Stylococcus aureus and S. brevis sp. nov. (Chrysophyceae) and their phylogenetic relationship to Lagynion. Phycologia 2025, 64, 282–296. [Google Scholar] [CrossRef]
- Malavasi, V.; Pusztai, M.; Jankowska, K.; Zakryś, B.; Škaloud, P. Revisiting Chrysococcus (Chrysophyceae): New phylogenetic evidence and evolutionary implications. Fottea 2025, 25, 121–127. [Google Scholar] [CrossRef]
- Kim, E.; Yubuki, N.; Leander, B.S.; Graham, L.E. Ultrastructure and 18S rDNA Phylogeny of Apoikia lindahlii comb. nov. (Chrysophyceae) and its Epibiontic Protists, Filos agilis gen. et sp. nov. (Bicosoecida) and Nanos amicus gen. et sp. nov. (Bicosoecida). Protist 2010, 161, 177–196. [Google Scholar] [CrossRef]
- Grossmann, L.; Bock, C.; Schweikert, M.; Boenigk, J. Small but Manifold—Hidden Diversity in “Spumella-like Flagellates”. J. Eukaryot. Microbiol. 2016, 63, 419–439. [Google Scholar] [CrossRef]
- Pietsch, T.; Nitsche, F.; Arndt, H. High molecular diversity in the functional group of small bacterivorous non-scaled chrysomonad flagellates. Eur. J. Protistol. 2022, 86, 125915. [Google Scholar] [CrossRef]
- Nicholls, K.H. Chrysococcus furcatus (Dolg.) comb. nov.: A new name for Chrysastrella furcata (Dolg.) Defl. based on the discovery of the vegetative stage. Phycologia 1981, 20, 16–21. [Google Scholar] [CrossRef]
- Kapustin, D.A.; Kapustina, N.V. New records of Chrysococcus furcatus (Chrysophyceae) in Russia. Inland Water Biol. 2018, 11, 384–386. [Google Scholar] [CrossRef]
- Swale, E.M.F.; Belcher, J.H. Ochromonas ostreaeformis nov. sp., a large compressed chrysomonad. New Phytol. 1966, 65, 267–272. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kapustin, D.; Martynenko, N.; Sterlyagova, I.; Iurmanov, A.; Kulikovskiy, M. Insights into the Taxonomy of the Genus Chrysastrella (Chrysophyceae), with Establishment of Chrysastrellaceae fam. nov. Diversity 2025, 17, 824. https://doi.org/10.3390/d17120824
Kapustin D, Martynenko N, Sterlyagova I, Iurmanov A, Kulikovskiy M. Insights into the Taxonomy of the Genus Chrysastrella (Chrysophyceae), with Establishment of Chrysastrellaceae fam. nov. Diversity. 2025; 17(12):824. https://doi.org/10.3390/d17120824
Chicago/Turabian StyleKapustin, Dmitry, Nikita Martynenko, Irina Sterlyagova, Anton Iurmanov, and Maxim Kulikovskiy. 2025. "Insights into the Taxonomy of the Genus Chrysastrella (Chrysophyceae), with Establishment of Chrysastrellaceae fam. nov." Diversity 17, no. 12: 824. https://doi.org/10.3390/d17120824
APA StyleKapustin, D., Martynenko, N., Sterlyagova, I., Iurmanov, A., & Kulikovskiy, M. (2025). Insights into the Taxonomy of the Genus Chrysastrella (Chrysophyceae), with Establishment of Chrysastrellaceae fam. nov. Diversity, 17(12), 824. https://doi.org/10.3390/d17120824






