Glyphosate-Induced Shifts in Edaphic Microbiota: A Comparative Study of Bacterial and Fungal Responses in Historical Milpa Soils
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites
2.2. Sampling Design
2.3. DNA Isolation and Sequencing
2.4. Bioinformatic Analyses
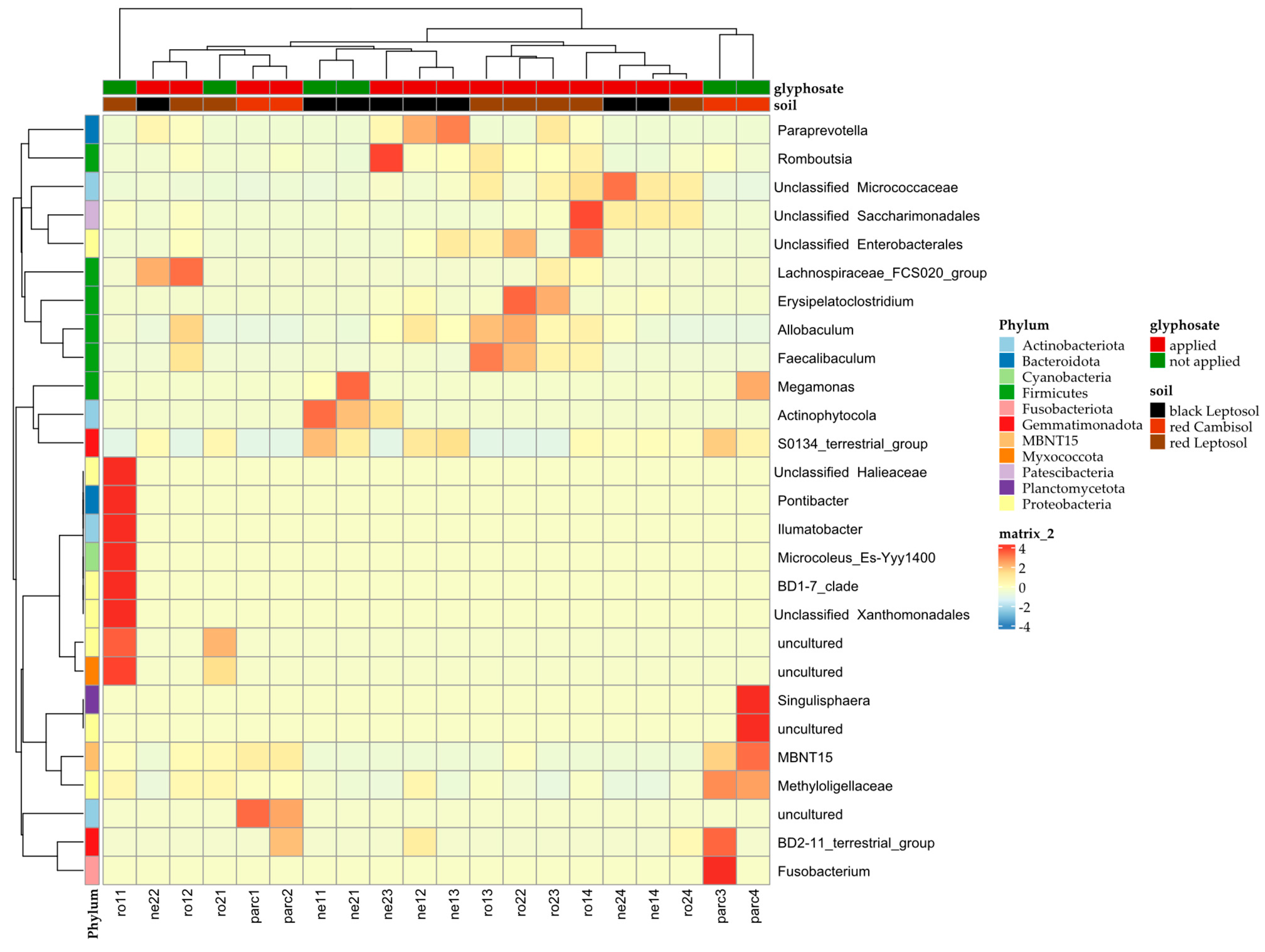
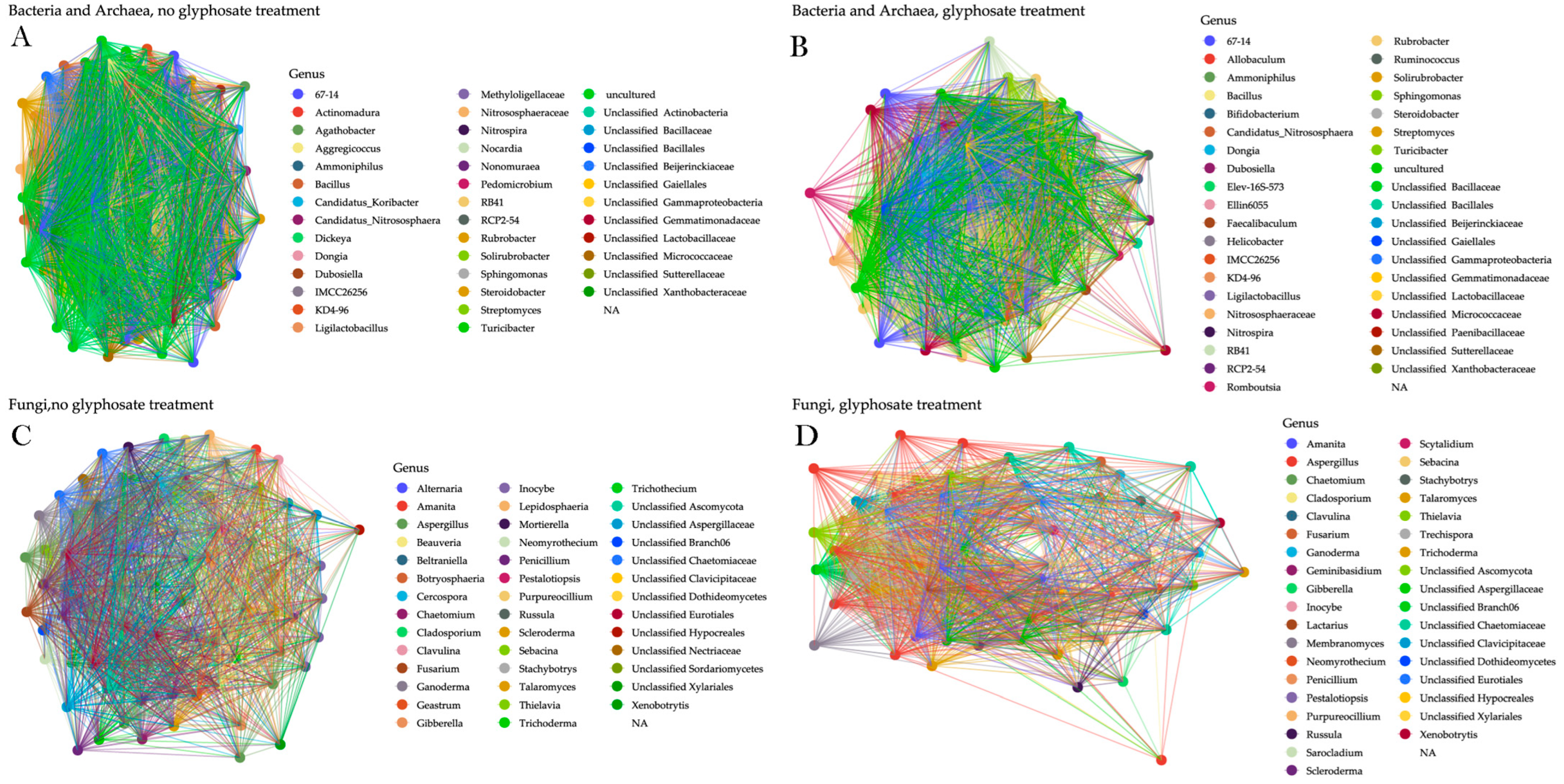
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NMDS | Non-metric multidimensional scaling |
| ASV | Amplicon Sequence Variant |
| ANOSIM | ANalysis Of SIMilarities |
| PermANOVA | Permutational Multivariate Analysis of Variance |
References
- Anthony, M.A.; Bender, S.F.; van der Heijden, M.G.A. Enumerating soil biodiversity. Proc. Natl. Acad. Sci. USA 2023, 120, e2304663120. [Google Scholar] [CrossRef]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; Bodegom, P.M.; Bengtsson-Palme, J.; Anslan, S.; Coelho, L.P.; Harend, H.; et al. Structure and function of the global topsoil microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef]
- Sokol, N.W.; Slessarev, E.; Marschmann, G.L.; Nicolas, A.; Blazewicz, S.J.; Brodie, E.L.; Firestone, M.K.; Foley, M.M.; Hestrin, R.; Hungate, B.A.; et al. Life and death in the soil microbiome: How ecological processes influence biogeochemistry. Nat. Rev. Microbiol. 2022, 20, 415–430. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Philippot, L.; Chenu, C.; Kappler, A.; Rillig, M.C.; Fierer, N. The interplay between microbial communities and soil properties. Nat. Rev. Microbiol. 2024, 22, 226–239. [Google Scholar] [CrossRef]
- Muhammad, M.; Wahab, A.; Waheed, A.; Hakeem, K.R.; Mohamed, H.I.; Basit, A.; Toor, M.D.; Liu, Y.-H.; Li, L.; Li, W.-J. Navigating climate change: Exploring the dynamics between plant–soil microbiomes and their impact on plant growth and productivity. Glob. Change Biol. 2025, 31, e70057. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chi, Y.; Song, S. Important soil microbiota’s effects on plants and soils: A comprehensive 30-year systematic literature review. Front. Microbiol. 2024, 15, 1347745. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez del Río, Á.; Scheu, S.; Rillig, M.C. Soil microbial responses to multiple global change factors as assessed by metagenomics. Nat. Commun. 2025, 16, 5058. [Google Scholar] [CrossRef] [PubMed]
- Arellano-Wattenbarger, G.L.; Córdoba-Agudelo, M.; Rocha, J. Ancestral roots: Exploring microbial communities in traditional agroecosystems for sustainable agriculture. Geoderma Reg. 2025, 41, e00960. [Google Scholar] [CrossRef]
- Terán, C. Milpa, biodiversidad y diversidad cultural. In Biodiversidad Desarrollo Humano en Yucatán; Secretaria de Desarrollo Urbano y Medio Ambiente: Mexico City, Mexico, 2010. [Google Scholar]
- González-Esquivel, C.E.; Briones-Guzmán, C.; Tovar-López, E.; López-Ridaura, S.; Arnés, E.; Camacho-Villa, T.C. Sustainability evaluation of contrasting milpa systems in the Yucatán Peninsula, Mexico. Environ. Dev. Sustain. 2025, 27, 9233–9255. [Google Scholar] [CrossRef]
- Duke, S.O. Glyphosate: Uses other than in glyphosate-resistant crops, mode of action, degradation in plants, and effects on non-target plants and agricultural microbes. In Reviews of Environmental Contamination and Toxicology Volume 255: Glyphosate; Knaak, J.B., Ed.; Springer International Publishing: Cham, Switzerland, 2021; pp. 1–65. [Google Scholar]
- Osten, J.R.; Borges-Ramírez, M.M.; Ruiz-Velazco, N.G.; Helguera, E.; Arellano-Aguilar, O.; Peregrina-Lucano, A.A.; Lozano-Kasten, F. Glyphosate and AMPA in groundwater, surface water, and soils related to different types of crops in Mexico. Bull Environ. Contam. Toxicol. 2025, 114, 44. [Google Scholar] [CrossRef]
- Dill, G.M.; Sammons, R.D.; Feng, P.C.C.; Kohn, F.; Kretzmer, K.; Mehrsheikh, A.; Bleeke, M.; Honegger, J.L.; Farmer, D.; Wright, D.; et al. Glyphosate: Discovery, development, applications, and properties. In Glyphosate Resistance in Crops and Weeds; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010; pp. 1–33. [Google Scholar]
- Leino, L.; Tall, T.; Helander, M.; Saloniemi, I.; Saikkonen, K.; Ruuskanen, S.; Puigbò, P. Classification of the glyphosate target enzyme (5-enolpyruvylshikimate-3-phosphate synthase) for assessing sensitivity of organisms to the herbicide. J. Hazard. Mater. 2021, 408, 124556. [Google Scholar] [CrossRef]
- Hertel, R.; Gibhardt, J.; Martienssen, M.; Kuhn, R.; Commichau, F.M. Molecular mechanisms underlying glyphosate resistance in bacteria. Environ. Microbiol. 2021, 23, 2891–2905. [Google Scholar] [CrossRef]
- Thiour-Mauprivez, C.; Martin-Laurent, F.; Calvayrac, C.; Barthelmebs, L. Effects of herbicide on non-target microorganisms: Towards a new class of biomarkers? Sci. Total Environ. 2019, 684, 314–325. [Google Scholar] [CrossRef] [PubMed]
- van Bruggen, A.H.C.; Finckh, M.R.; He, M.; Ritsema, C.J.; Harkes, P.; Knuth, D.; Geissen, V. Indirect effects of the herbicide glyphosate on plant, animal and human health through its effects on microbial communities. Front. Environ. Sci. 2021, 9, 763917. [Google Scholar] [CrossRef]
- Fazal, A.; Yang, M.; Wang, X.; Lu, Y.; Yao, W.; Luo, F.; Han, M.; Song, Y.; Cai, J.; Yin, T.; et al. Discrepancies in rhizobacterial assembly caused by glyphosate application and herbicide-tolerant soybean Co-expressing GAT and EPSPS. J. Hazard. Mater. 2023, 450, 131053. [Google Scholar] [CrossRef]
- Fazal, A.; Wen, Z.; Yang, M.; Wang, C.; Hao, C.; Lai, X.; Jie, W.; Yang, L.; He, Z.; Yang, H.; et al. Triple-transgenic soybean in conjunction with glyphosate drive patterns in the rhizosphere microbial community assembly. Environ. Pollut. 2023, 335, 122337. [Google Scholar] [CrossRef]
- Yang, M.; Wen, Z.; Hao, C.; Fazal, A.; Liao, Y.; Luo, F.; Yao, W.; Yin, T.; Yang, R.; Qi, J.; et al. Differential assembly and shifts of the rhizosphere bacterial community by a dual transgenic glyphosate-tolerant soybean line with and without glyphosate application. Horticulturae 2021, 7, 374. [Google Scholar] [CrossRef]
- Barriuso, J.; Mellado, R.P. Relative effect of glyphosate on glyphosate-tolerant maize rhizobacterial communities is not altered by soil properties. J. Microbiol. Biotechnol. 2012, 22, 159–165. [Google Scholar] [CrossRef]
- Morales, M.E.; Allegrini, M.; Basualdo, J.; Iocoli, G.A.; Villamil, M.B.; Zabaloy, M.C. Winter cover crop suppression methods influence on sunflower growth and rhizosphere communities. Front. Microbiol. 2024, 15, 1405842. [Google Scholar] [CrossRef] [PubMed]
- Sha, C.; Wang, Z.; Cao, J.; Chen, J.; Shen, C.; Zhang, J.; Wang, Q.; Wang, M. Management of Spartina alterniflora: Assessing the efficacy of plant growth regulators on ecological and microbial dynamics. Sustainability 2024, 16, 7848. [Google Scholar] [CrossRef]
- Estrada-Medina, H.; Bautista, F.; Jiménez-Osornio, J.J.M.; González-Iturbe, J.A.; Aguilar Cordero, W.d.J. Maya and WRB soil classification in yucatan, mexico: Differences and similarities. Int. Sch. Res. Not. 2013, 2013, 634260. [Google Scholar] [CrossRef]
- Estrada-Medina, H.; Ferrer, M.M.; Montañez-Escalante, P.; Pech-Puch, G.; Álvarez-Rivera, O.O. Foliar nutrient contents of tropical tree species under different management and climate conditions. Ecosistemas Recur. Agropecu. 2023, 10, 2. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- May-Mutul, C.G.; López-Garrido, M.A.; O’Connor-Sánchez, A.; Peña-Ramírez, Y.J.; Labrín-Sotomayor, N.Y.; Estrada-Medina, H.; Ferrer, M.M. Hidden tenants: Microbiota of the rhizosphere and phyllosphere of Cordia dodecandra trees in Mayan forests and homegardens. Plants 2022, 11, 3098. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 28 November 2024).
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; May, T.W.; Frøslev, T.G.; Pawlowska, J.; Lindahl, B.; Põldmaa, K.; Truong, C.; et al. The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: Sequences, taxa and classifications reconsidered. Nucleic Acids Res. 2023, 52, D791–D797. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2025. [Google Scholar]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kurtz, Z.D.; Müller, C.L.; Miraldi, E.R.; Littman, D.R.; Blaser, M.J.; Bonneau, R.A. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol. 2015, 11, e1004226. [Google Scholar] [CrossRef]
- Neu, A.T.; Allen, E.E.; Roy, K. Defining and quantifying the core microbiome: Challenges and prospects. Proc. Natl. Acad. Sci. USA 2021, 118, e2104429118. [Google Scholar] [CrossRef] [PubMed]
- Antonov, M.; Csárdi, G.; Horvát, S.; Müller, K.; Nepusz, T.; Noom, D.; Salmon, M.; Traag, V.; Welles, B.F.; Zanini, F. igraph enables fast and robust network analysis across programming languages. arXiv 2023, arXiv:2311.10260. [Google Scholar] [CrossRef]
- Kajihara, K.T.; Hynson, N.A. Networks as tools for defining emergent properties of microbiomes and their stability. Microbiome 2024, 12, 184. [Google Scholar] [CrossRef]
- Estrada-Medina, H.; Canto-Canché, B.B.; De los Santos-Briones, C.; O’Connor-Sánchez, A. Yucatán in black and red: Linking edaphic analysis and pyrosequencing-based assessment of bacterial and fungal community structures in the two main kinds of soil of Yucatán State. Microbiol Res. 2016, 188–189, 23–33. [Google Scholar] [CrossRef] [PubMed]
- López-Ramírez, T.M.; Estrada-Medina, H.; Ferrer, M.M.; O’Connor-Sánchez, A. Divergence in the soil and rhizosphere microbial communities of monoculture and silvopastoral traditional C. dodecandra agroforestry systems in Yucatan, Mexico. Soil Use Manage 2023, 39, 1205–1218. [Google Scholar] [CrossRef]
- Santillán, J.; López-Martínez, R.; Aguilar-Rangel, E.J.; Hernández-García, K.; Vásquez-Murrieta, M.S.; Cram, S.; Alcántara-Hernández, R.J. Microbial diversity and physicochemical characteristics of tropical karst soils in the northeastern Yucatan peninsula, Mexico. Appl. Soil Ecol. 2021, 165, 103969. [Google Scholar] [CrossRef]
- Feng, X.; Tao, Y.; Dai, Z.; Chu, Z.; Wei, Y.; Tao, M.; He, Y.; Chen, H. Effects of transgenic modification on the bacterial communities in different niches of maize under glyphosate toxicity. Environ. Pollut. 2024, 362, 125023. [Google Scholar] [CrossRef]
- Kepler, R.M.; Epp Schmidt, D.J.; Yarwood, S.A.; Cavigelli, M.A.; Reddy, K.N.; Duke, S.O.; Bradley, C.A.; Williams, M.M.; Maula, J.E. Soil microbial communities in diverse agroecosystems exposed to the herbicide glyphosate. Appl. Environ. Microbiol. 2020, 86, e01744-19. [Google Scholar] [CrossRef]
- Lupwayi, N.Z.; Blackshaw, R.E.; Geddes, C.M.; Dunn, R.; Petri, R.M. Multi-year and multi-site effects of recurrent glyphosate applications on the wheat rhizosphere microbiome. Environ. Res. 2022, 215, 114363. [Google Scholar] [CrossRef]
- Almeida, A.; Mitchell, A.L.; Tarkowska, A.; Finn, R.D. Benchmarking taxonomic assignments based on 16S rRNA gene profiling of the microbiota from commonly sampled environments. GigaScience 2018, 7, giy054. [Google Scholar] [CrossRef]
- Sakon, H.; Nagai, F.; Morotomi, M.; Tanaka, R. Sutterella parvirubra sp. nov. and Megamonas funiformis sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2008, 58, 970–975. [Google Scholar] [CrossRef] [PubMed]
- Molteni, C.; Forni, D.; Cagliani, R.; Sironi, M. Comparative genomics reveal a novel phylotaxonomic order in the genus Fusobacterium. Commun. Biol. 2024, 7, 1102. [Google Scholar] [CrossRef]
- Arunrat, N.; Uttarotai, T.; Kongsurakan, P.; Sereenonchai, S.; Hatano, R. Bacterial Community Structure in Soils With Fire-Deposited Charcoal Under Rotational Shifting Cultivation of Upland Rice in Northern Thailand. Ecol. Evol. 2025, 15, e70851. [Google Scholar] [CrossRef] [PubMed]
- Boyarshin, K.S.; Adamova, V.V.; Wentao, Z.; Obuhova, O.Y.; Kolkova, M.V.; Nesterenko, V.A.; Bespalova, O.S.; Kluyeva, V.V.; Degtyareva, K.A.; Kurkina, Y.N.; et al. The Effect of Long-Term Agricultural Use on the Bacterial Microbiota of Chernozems of the Forest-Steppe Zone. Diversity 2023, 15, 191. [Google Scholar] [CrossRef]
- Kong, D.; Xu, L.; Dai, M.; Ye, Z.; Ma, B.; Tan, X. Deciphering the functional assembly of microbial communities driven by heavy metals in the tidal soils of Hangzhou Bay. Environ. Pollut. 2024, 360, 124671. [Google Scholar] [CrossRef] [PubMed]
- Nedashkovskaya, O.I.; Kim, S.B. Pontibacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–4. [Google Scholar]
- Dai, J.; Dai, W.; Qiu, C.; Yang, Z.; Zhang, Y.; Zhou, M.; Zhang, L.; Fang, C.; Gao, Q.; Yang, Q.; et al. Unraveling adaptation of Pontibacter korlensis to radiation and infertility in desert through complete genome and comparative transcriptomic analysis. Sci. Rep. 2015, 5, 10929. [Google Scholar] [CrossRef]
- Xu, L.; Zeng, X.-C.; Nie, Y.; Luo, X.; Zhou, E.; Zhou, L.; Pan, Y.; Li, W. Pontibacter diazotrophicus sp. nov., a Novel Nitrogen-Fixing Bacterium of the Family Cytophagaceae. PLoS ONE 2014, 9, e92294. [Google Scholar] [CrossRef]
- Liu, X.; Du, Z.; Zhou, T.; Li, B.; Wang, J.; Wang, J.; Zhu, L. Evaluation of agricultural soil health after applying atrazine in maize-planted fields based on the response of soil microbes. Appl. Soil Ecol. 2024, 193, 105157. [Google Scholar] [CrossRef]
- Xiao, J.; Lan, S.; Farías, M.E.; Qian, L.; Xia, L.; Song, S.; Wu, L. The living forms of Microcoleus vaginatus and their contributions to the aggregate structure of biocrusts. FEMS Microbiol. Ecol. 2023, 99, fiad040. [Google Scholar] [CrossRef]
- Kim, J.-R.; Yeon, S.-H.; Kim, H.-S.; Ahn, Y.-J. Larvicidal activity against Plutella xylostella of cordycepin from the fruiting body of Cordyceps militaris. Pest Manage Sci. 2002, 58, 713–717. [Google Scholar] [CrossRef]
- Lezama-Gutiérrez, R.; Molina-Ochoa, J.; Chávez-Flores, O.; Ángel-Sahagún, C.A.; Skoda, S.R.; Reyes-Martínez, G.; Barba-Reynoso, M.; Rebolledo-Domínguez, O.; Ruíz-Aguilar, G.M.L.; Foster, J.E. Use of the entomopathogenic fungi Metarhizium anisopliae, Cordyceps bassiana and Isaria fumosorosea to control Diaphorina citri (Hemiptera: Psyllidae) in Persian lime under field conditions. Int. J. Trop. Insect Sci. 2012, 32, 39–44. [Google Scholar] [CrossRef]
- Avery, P.B.; Duren, E.B.; Qureshi, J.A.; Adair, R.C.; Adair, M.M.; Cave, R.D. Field efficacy of Cordyceps javanica, white oil and spinetoram for the management of the Asian citrus psyllid, Diaphorina citri. Insects 2021, 12, 824. [Google Scholar] [CrossRef]
- Pipke, R.; Amrhein, N. Isolation and characterization of a mutant of Arthrobacter sp. strain GLP-1 which utilizes the herbicide glyphosate as its sole source of phosphorus and nitrogen. Appl. Environ. Microbiol. 1988, 54, 2868–2870. [Google Scholar] [CrossRef]
- Bazot, S.; Lebeau, T. Simultaneous mineralization of glyphosate and diuron by a consortium of three bacteria as free- and/or immobilized-cells formulations. Appl. Microbiol. Biotechnol. 2008, 77, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.G.; Dewhirst, F.E.; Tully, J.G.; Paster, B.J.; Yan, L.; Taylor, N.S.; Collins, M.J.; Gorelick, P.L.; Ward, J.M. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J. Clin. Microbiol. 1994, 32, 1238–1245. [Google Scholar] [CrossRef]
- Chang, D.-H.; Rhee, M.-S.; Ahn, S.; Bang, B.-H.; Oh, J.E.; Lee, H.K.; Kim, B.-C. Faecalibaculum rodentium gen. nov., sp. nov., isolated from the faeces of a laboratory mouse. Antonie Van Leeuwenhoek 2015, 108, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- Gerritsen, J.; Fuentes, S.; Grievink, W.; van Niftrik, L.; Tindall, B.J.; Timmerman, H.M.; Rijkers, G.T.; Smidt, H. Characterization of Romboutsia ilealis gen. nov., sp. nov., isolated from the gastro-intestinal tract of a rat, and proposal for the reclassification of five closely related members of the genus Clostridium into the genera Romboutsia gen. nov., Intestinibacter gen. nov., Terrisporobacter gen. nov. and Asaccharospora gen. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 1600–1616. [Google Scholar] [CrossRef] [PubMed]
- Morotomi, M.; Nagai, F.; Sakon, H.; Tanaka, R. Paraprevotella clara gen. nov., sp. nov. and Paraprevotella xylaniphila sp. nov., members of the family ‘Prevotellaceae’ isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2009, 59, 1895–1900. [Google Scholar] [CrossRef]
- Gerritsen, J.; Umanets, A.; Staneva, I.; Hornung, B.; Ritari, J.; Paulin, L.; Rijkers, G.T.; de Vos, W.M.; Smidt, H. Romboutsia hominis sp. nov., the first human gut-derived representative of the genus Romboutsia, isolated from ileostoma effluent. Int. J. Syst. Evol. Microbiol. 2018, 68, 3479–3486. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, K.; Wang, J.; Zeng, Q.; Liu, K.; Zheng, S.; Chen, Y.; Yao, J. Microbial Disruptions in Inflammatory Bowel Disease: A Comparative Analysis. Int. J. Gen. Med. 2024, 17, 1355–1367. [Google Scholar] [CrossRef]
- van Muijlwijk, G.H.; Rice, T.A.; Flavell, R.A.; Palm, N.W.; de Zoete, M.R. Allobaculum mucilyticum sp. nov. and Allobaculum fili sp. nov., isolated from the human intestinal tract. Int. J. Syst. Evol. Microbiol. 2023, 73. [Google Scholar] [CrossRef] [PubMed]
- Seo, B.; Jeon, K.; Baek, I.; Lee, Y.M.; Baek, K.; Ko, G. Faecalibacillus intestinalis gen. nov., sp. nov. and Faecalibacillus faecis sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2019, 69, 2120–2128. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, C.; Appenzeller, B.; Burkart, L.; Coeurdassier, M.; Scheifler, R.; Raoul, F.; Driget, V.; Powolny, T.; Gagnaison, C.; Rieffel, D.; et al. Pervasive exposure of wild small mammals to legacy and currently used pesticide mixtures in arable landscapes. Sci. Rep. 2022, 12, 15904. [Google Scholar] [CrossRef]
- la Cecilia, D.; Maggi, F. Analysis of glyphosate degradation in a soil microcosm. Environ. Pollut. 2018, 233, 201–207. [Google Scholar] [CrossRef]
- Dickinson, C.H. Gliomastix Guéguen; Commonwealth Mycological Institute: Kew, UK, 1968; Volume 115. [Google Scholar]
- Afzal Khan, S.; Hamayun, M.; Kim, H.-Y.; Yoon, H.-J.; Lee, I.-J.; Kim, J.-G. Gibberellin production and plant growth promotion by a newly isolated strain of Gliomastix murorum. World J. Microbiol. Biotechnol. 2009, 25, 829–833. [Google Scholar] [CrossRef]
- Zhao, J.; Shan, T.; Huang, Y.; Liu, X.; Gao, X.; Wang, M.; Jiang, W.; Zhou, L. Chemical composition and in vitro antimicrobial activity of the volatile oils from Gliomastix murorum and Pichia guilliermondii, two endophytic fungi in Paris polyphylla var. yunnanensis. Nat. Prod. Commun. 2009, 4, 1491–1496. [Google Scholar] [CrossRef]
- de Errasti, A.; Novas, M.V.; Carmarán, C.C. Plant-fungal association in trees: Insights into changes in ecological strategies of Peroneutypa scoparia (Diatrypaceae). Flora Morphol. Distrib. Funct. Ecol. Plants 2014, 209, 704–710. [Google Scholar] [CrossRef]
- Ghaedi, M.; Bolboli, Z.; Mostowfizadeh-Ghalamfarsa, R. First report of Peroneutypa scoparia associated with canker disease on Ficus carica in northern Iran. New Dis. Rep. 2023, 48, e12201. [Google Scholar] [CrossRef]
- Zhan, H.; Feng, Y.; Fan, X.; Chen, S. Recent advances in glyphosate biodegradation. Appl. Microbiol. Biotechnol. 2018, 102, 5033–5043. [Google Scholar] [CrossRef] [PubMed]
- Correa, L.O.; Bezerra, A.F.M.; Honorato, L.R.S.; Cortez, A.C.A.; Souza, J.V.B.; Souza, E.S. Amazonian soil fungi are efficient degraders of glyphosate herbicide; novel isolates of Penicillium, Aspergillus, and Trichoderma. Braz. J. Biol. 2023, 83. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Kumar, V.; Gill, J.P.K.; Datta, S.; Singh, S.; Dhaka, V.; Kapoor, D.; Wani, A.B.; Dhanjal, D.S.; Kumar, M.; et al. Herbicide Glyphosate: Toxicity and Microbial Degradation. Int. J. Environ. Res. Public Health 2020, 17, 7519. [Google Scholar] [CrossRef] [PubMed]


| Soil Type | pH | EC | OC | N | P | Na | K | Ca | AD | Sand | Silt | Clay |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (dS/m) | (%) | (mg/kg) | (cmol/kg) | g/cm3 | (%) | |||||||
| BL | 7.51 | 0.50 | 2.79 | 0.16 | 20.17 | 0.13 | 0.09 | 40.68 | 0.92 | 67.48 | 16.00 | 16.52 |
| RL | 7.24 | 0.52 | 1.50 | 0.13 | 2.37 | 0.15 | 0.11 | 19.23 | 0.98 | 45.48 | 24.00 | 30.52 |
| RC | 6.92 | 0.30 | 2.86 | 0.39 | 8.30 | 0.03 | 0.25 | 6.82 | 1.14 | 37.82 | 16.10 | 46.09 |
| Group | Samples | Degree | Betweenness | Local Transitivity | Global Transitivity | Modularity (Greedy) |
|---|---|---|---|---|---|---|
| AB | APP | 0.010 | 18.121 | 0.752 | 0.751 | 0.109 |
| AB | NAP | 0.009 | 13.355 | 0.789 | 0.789 | 0.087 |
| FU | APP | 0.013 | 15.412 | 0.754 | 0.757 | 0.124 |
| FU | NAP | 0.014 | 8.933 | 0.791 | 0.788 | 0.122 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ocaña-Ek, M.A.; García-Romero, A.d.C.; Álvarez-Rivera, O.O.; Tzec-Gamboa, M.d.C.; Estrada-Medina, H.; Ferrer, M.M. Glyphosate-Induced Shifts in Edaphic Microbiota: A Comparative Study of Bacterial and Fungal Responses in Historical Milpa Soils. Diversity 2025, 17, 803. https://doi.org/10.3390/d17110803
Ocaña-Ek MA, García-Romero AdC, Álvarez-Rivera OO, Tzec-Gamboa MdC, Estrada-Medina H, Ferrer MM. Glyphosate-Induced Shifts in Edaphic Microbiota: A Comparative Study of Bacterial and Fungal Responses in Historical Milpa Soils. Diversity. 2025; 17(11):803. https://doi.org/10.3390/d17110803
Chicago/Turabian StyleOcaña-Ek, María Alejandra, Anell del Carmen García-Romero, Oscar Omar Álvarez-Rivera, Magnolia del Carmen Tzec-Gamboa, Héctor Estrada-Medina, and Miriam M. Ferrer. 2025. "Glyphosate-Induced Shifts in Edaphic Microbiota: A Comparative Study of Bacterial and Fungal Responses in Historical Milpa Soils" Diversity 17, no. 11: 803. https://doi.org/10.3390/d17110803
APA StyleOcaña-Ek, M. A., García-Romero, A. d. C., Álvarez-Rivera, O. O., Tzec-Gamboa, M. d. C., Estrada-Medina, H., & Ferrer, M. M. (2025). Glyphosate-Induced Shifts in Edaphic Microbiota: A Comparative Study of Bacterial and Fungal Responses in Historical Milpa Soils. Diversity, 17(11), 803. https://doi.org/10.3390/d17110803







