Species Composition and Phylogenetic Diversity of Acetic Acid Bacteria Communities in Homemade Vinegars
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and 16S-23S rDNA ITS Amplicon Metagenomic Analysis
2.2. Taxonomic Diversity of AAB
3. Results and Discussion
3.1. Species Diversity Detected with 16S-23S rRNA ITS Sequencing
3.2. Phylogenetic Diversity Detected with 16S-23S rRNA ITS Sequencing
3.3. Non-Acetic Acid Bacteria
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lynch, K.M.; Zannini, E.; Wilkinson, S.; Daenen, L.; Arendt, E.K. Physiology of Acetic Acid Bacteria and Their Role in Vinegar and Fermented Beverages. Compr. Rev. Food Sci. Food Saf. 2019, 18, 587–625. [Google Scholar] [CrossRef]
- Sengun, I.Y.; Karabiyikli, S. Importance of Acetic Acid Bacteria in Food Industry. Food Control 2011, 22, 647–656. [Google Scholar] [CrossRef]
- Saichana, N.; Matsushita, K.; Adachi, O.; Frébort, I.; Frebortova, J. Acetic Acid Bacteria: A Group of Bacteria with Versatile Biotechnological Applications. Biotechnol. Adv. 2015, 33, 1260–1271. [Google Scholar] [CrossRef]
- Jozala, A.F.; de Lencastre-Novaes, L.C.; Lopes, A.M.; de Carvalho Santos-Ebinuma, V.; Mazzola, P.G.; Pessoa, A., Jr.; Grotto, D.; Gerenutti, M.; Chaud, M.V. Bacterial Nanocellulose Production and Application: A 10-Year Overview. Appl. Microbiol. Biotechnol. 2016, 100, 2063–2072. [Google Scholar] [CrossRef]
- Trček, J.; Dogsa, I.; Accetto, T.; Stopar, D. Acetan and Acetan-like Polysaccharides: Genetics, Biosynthesis, Structure, and Viscoelasticity. Polymers 2021, 13, 815. [Google Scholar] [CrossRef]
- Ho, C.W.; Lazim, A.M.; Fazry, S.; Zaki, U.K.H.H.; Lim, S.J. Varieties, Production, Composition and Health Benefits of Vinegars: A Review. Food Chem. 2017, 221, 1621–1630. [Google Scholar] [CrossRef]
- Gao, Q.; Song, Y.; Liang, Y.; Li, Y.; Chang, Y.; Ma, R.; Cao, X.; Wang, S. Dynamics of Physicochemical Properties, Functional Compounds and Antioxidant Capacity during Spontaneous Fermentation of Lycium ruthenicum Murr. (Qinghai–Tibet Plateau) Natural Vinegar. Foods 2022, 11, 1344. [Google Scholar] [CrossRef]
- Lisa, S.; Paolo, G. Vinegars of the World. In Vinegars of the World; Springer: Milano, Italy, 2009; pp. 1–16. [Google Scholar]
- Özdemir, G.B.; Özdemir, N.; Ertekin-Filiz, B.; Gökırmaklı, Ç.; Kök-Taş, T.; Budak, N.H. Volatile Aroma Compounds and Bioactive Compounds of Hawthorn Vinegar Produced from Hawthorn Fruit (Crataegus tanacetifolia (Lam.) Pers.). J. Food Biochem. 2022, 46, e13676. [Google Scholar] [CrossRef]
- Mas, A.; Torija, M.J.; del Carmen García-Parrilla, M.C.; Troncoso, A.M. Acetic Acid Bacteria and the Production and Quality of Wine Vinegar. Sci. World J. 2014, 2014, 394671. [Google Scholar] [CrossRef]
- Yun, J.; Zhao, F.; Zhang, W.; Yan, H.; Zhao, F.; Ai, D. Monitoring the Microbial Community Succession and Diversity of Liangzhou Fumigated Vinegar during Solid-State Fermentation with next-Generation Sequencing. Ann. Microbiol. 2019, 69, 279–289. [Google Scholar] [CrossRef]
- Kou, R.; Li, M.; Xing, J.; He, Y.; Wang, H.; Fan, X. Exploring of Seasonal Dynamics of Microbial Community in Multispecies Fermentation of Shanxi Mature Vinegar. J. Biosci. Bioeng. 2022, 133, 375–381. [Google Scholar] [CrossRef]
- Li, L.; Li, N.; Fu, J.; Liu, J.; Wen, X.P.; Cao, H.; Xu, H.; Zhang, Y.; Cao, R. Synthesis of an Autochthonous Microbial Community by Analyzing the Core Microorganisms Responsible for the Critical Flavor of Bran Vinegar. Food Res. Int. 2024, 175, 113742. [Google Scholar] [CrossRef]
- Ribič, A.; Trček, J. Customized 16S-23S rDNA ITS Amplicon Metagenomics for Acetic Acid Bacteria Species Identification in Vinegars and Kombuchas. Microorganisms 2024, 12, 1023. [Google Scholar] [CrossRef]
- Li, S.; Li, P.; Feng, F.; Luo, L.X. Microbial Diversity and Their Roles in the Vinegar Fermentation Process. Appl. Microbiol. Biotechnol. 2015, 99, 4997–5024. [Google Scholar] [CrossRef]
- Kim, B.R.; Shin, J.; Guevarra, R.B.; Lee, J.H.; Kim, D.W.; Seol, K.H.; Lee, J.H.; Kim, H.B.; Isaacson, R.E. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef]
- Gullo, M.; Caggia, C.; De Vero, L.; Giudici, P. Characterization of Acetic Acid Bacteria in “Traditional Balsamic Vinegar”. Int. J. Food Microbiol. 2006, 106, 209–212. [Google Scholar] [CrossRef]
- Trček, J.; Mahnič, A.; Rupnik, M. Diversity of the Microbiota Involved in Wine and Organic Apple Cider Submerged Vinegar Production as Revealed by DHPLC Analysis and Next-Generation Sequencing. Int. J. Food Microbiol. 2016, 223, 57–62. [Google Scholar] [CrossRef]
- Trcek, J. Quick Identification of Acetic Acid Bacteria Based on Nucleotide Sequences of the 16S-23S rDNA Internal Transcribed Spacer Region and of the PQQ-Dependent Alcohol Dehydrogenase Gene. Syst. Appl. Microbiol. 2005, 28, 735–745. [Google Scholar] [CrossRef]
- Modin, O.; Liébana, R.; Saheb-Alam, S.; Wilén, B.M.; Suarez, C.; Hermansson, M.; Persson, F. Hill-Based Dissimilarity Indices and Null Models for Analysis of Microbial Community Assembly. Microbiome 2020, 8, 132. [Google Scholar] [CrossRef]
- Alberdi, A.; Gilbert, M.T.P. A Guide to the Application of Hill Numbers to DNA-Based Diversity Analyses. Mol. Ecol. Resour. 2019, 19, 804–817. [Google Scholar] [CrossRef]
- Feranchuk, S.; Belkova, N.; Potapova, U.; Kuzmin, D.; Belikov, S. Evaluating the Use of Diversity Indices to Distinguish between Microbial Communities with Different Traits. Res. Microbiol. 2018, 169, 254–261. [Google Scholar] [CrossRef]
- Haegeman, B.; Hamelin, J.; Moriarty, J.; Neal, P.; Dushoff, J.; Weitz, J.S. Robust Estimation of Microbial Diversity in Theory and in Practice. ISME J. 2013, 7, 1092–1101. [Google Scholar] [CrossRef]
- Ott, A.; Quintela-Baluja, M.; Zealand, A.M.; O’Donnell, G.; Haniffah, M.R.M.; Graham, D.W. Improved Quantitative Microbiome Profiling for Environmental Antibiotic Resistance Surveillance. Environ. Microbiome 2021, 16, 21. [Google Scholar] [CrossRef]
- Chao, A.; Chiu, C.H.; Jost, L. Unifying Species Diversity, Phylogenetic Diversity, Functional Diversity, and Related Similarity and Differentiation Measures through Hill Numbers. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 297–324. [Google Scholar] [CrossRef]
- Chao, A.; Gotelli, N.J.; Hsieh, T.C.; Sander, E.L.; Ma, K.H.; Colwell, R.K.; Ellison, A.M. Rarefaction and Extrapolation with Hill Numbers: A Framework for Sampling and Estimation in Species Diversity Studies. Ecol. Monogr. 2014, 84, 45–67. [Google Scholar] [CrossRef]
- Li, D. hillR: Taxonomic, Functional, and Phylogenetic Diversity and Similarity through Hill Numbers. J. Open Source Softw. 2018, 3, 1041. [Google Scholar] [CrossRef]
- Chao, A.; Henderson, P.A.; Chiu, C.H.; Moyes, F.; Hu, K.H.; Dornelas, M.; Magurran, A.E. Measuring Temporal Change in Alpha Diversity: A Framework Integrating Taxonomic, Phylogenetic and Functional Diversity and the INEXT.3D Standardization. Methods Ecol. Evol. 2021, 12, 1926–1940. [Google Scholar] [CrossRef]
- Karničnik, B.; Accetto, T.; Fanedl, L.; Jugović, I.; Trček, J. Isolation and Characterization of Komagataeibacter piraceti sp. nov. and Novacetimonas labruscae sp. nov.: Two Novel Microaerobic Cellulose-Producing Acetic Acid Bacteria from Vinegars. Microorganisms 2025, 13, 456. [Google Scholar] [CrossRef]
- Chochevska, M.; Jančovska Seniceva, E.; Veličkovska, S.K.; Naumova-Leţia, G.; Mirčeski, V.; Rocha, J.M.F.; Esatbeyoglu, T. Electrochemical Determination of Antioxidant Capacity of Traditional Homemade Fruit Vinegars Produced with Double Spontaneous Fermentation. Microorganisms 2021, 9, 1946. [Google Scholar] [CrossRef]
- Marič, L.; Cleenwerck, I.; Accetto, T.; Vandamme, P.; Trček, J. Description of Komagataeibacter melaceti sp. nov. and Komagataeibacter melomenusus sp. nov. Isolated from Apple Cider Vinegar. Microorganisms 2020, 8, 1178. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D.; van den Brand, T. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. 2022. Available online: https://cran.r-project.org/web/packages/ggplot2/index.html (accessed on 10 June 2025).
- Raivo, K. pheatmap: Pretty Heatmaps. 2019. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 10 June 2025).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Posit Team. RStudio: Integrated Development Environment for R; Posit Software; PBC: Boston, MA, USA, 2023; Available online: https://posit.co/products/open-source/rstudio (accessed on 15 April 2025).
- Huang, T.; Lu, Z.M.; Peng, M.Y.; Chai, L.J.; Zhang, X.J.; Shi, J.S.; Li, Q.; Xu, Z.H. Constructing a Defined Starter for Multispecies Vinegar Fermentation via Evaluation of the Vitality and Dominance of Functional Microbes in an Autochthonous Starter. Appl. Environ. Microbiol. 2022, 88, e02175-21. [Google Scholar] [CrossRef]
- Kong, H.; Kim, S.H.; Jeong, W.S.; Kim, S.Y.; Yeo, S.H. Microbiome and Volatile Metabolic Profile of Acetic Acid Fermentation Using Multiple Starters for Traditional Grain Vinegar. Fermentation 2023, 9, 423. [Google Scholar] [CrossRef]
- Gullo, M.; De Vero, L.; Giudici, P. Succession of Selected Strains of Acetobacter pasteurianus and Other Acetic Acid Bacteria in Traditional Balsamic Vinegar. Appl. Environ. Microbiol. 2009, 75, 2585–2589. [Google Scholar] [CrossRef]
- Hua, S.; Wang, Y.; Wang, L.; Zhou, Q.; Li, Z.; Liu, P.; Wang, K.; Zhu, Y.; Han, D.; Yu, Y. Regulatory Mechanisms of Acetic Acid, Ethanol and High Temperature Tolerances of Acetic Acid Bacteria during Vinegar Production. Microb. Cell Factories 2024, 23, 324. [Google Scholar] [CrossRef]
- He, Q.; Wang, S.; Feng, K.; Michaletz, S.T.; Hou, W.; Zhang, W.; Li, F.; Zhang, Y.; Wang, D.; Peng, X.; et al. High Speciation Rate of Niche Specialists in Hot Springs. ISME J. 2023, 17, 1303–1314. [Google Scholar] [CrossRef]
- Jost, L. Entropy and Diversity. Oikos 2006, 113, 363–375. [Google Scholar] [CrossRef]
- Loganathan, P.; Nair, S. Swaminathania salitolerans gen. nov., sp. nov., a Salt-Tolerant, Nitrogen-Fixing and Phosphate-Solubilizing Bacterium from Wild Rice (Porteresia coarctata Tateoka). Int. J. Syst. Evol. Microbiol. 2004, 54, 1185–1190. [Google Scholar] [CrossRef]
- Chao, A.; Chiu, C.H.; Jost, L. Phylogenetic Diversity Measures Based on Hill Numbers. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 3599–3609. [Google Scholar] [CrossRef]
- Xu, W.; Huang, Z.; Zhang, X.; Li, Q.; Lu, Z.; Shi, J.; Xu, Z.; Ma, Y. Monitoring the Microbial Community during Solid-State Acetic Acid Fermentation of Zhenjiang Aromatic Vinegar. Food Microbiol. 2011, 28, 1175–1181. [Google Scholar] [CrossRef]
- Hidalgo, C.; Mateo, E.; Mas, A.; Torija, M.J. Identification of Yeast and Acetic Acid Bacteria Isolated from the Fermentation and Acetification of Persimmon (Diospyros kaki). Food Microbiol. 2012, 30, 98–104. [Google Scholar] [CrossRef]
- de Bello, F.; Šmilauer, P.; Diniz-Filho, J.A.F.; Carmona, C.P.; Lososová, Z.; Herben, T.; Götzenberger, L. Decoupling Phylogenetic and Functional Diversity to Reveal Hidden Signals in Community Assembly. Methods Ecol. Evol. 2017, 8, 1200–1211. [Google Scholar] [CrossRef]
- Sterniša, M.; Purgatorio, C.; Paparella, A.; Mraz, J.; Smole Možina, S. Combination of Rosemary Extract and Buffered Vinegar Inhibits Pseudomonas and Shewanella Growth in Common Carp (Cyprinus carpio). J. Sci. Food Agric. 2020, 100, 2305–2312. [Google Scholar] [CrossRef]
- Carson, C.F.; Ash, O.; Chakera, A. In Vitro Data Support the Investigation of Vinegar as an Antimicrobial Agent for PD-Associated Pseudomonas Exit Site Infections. Nephrology 2017, 22, 179–181. [Google Scholar] [CrossRef]
- Kim, K.Y.; Jordan, D.; Krishnan, H.B. Rahnella aquatilis, a Bacterium Isolated from Soybean Rhizosphere, Can Solubilize Hydroxyapatite. FEMS Microbiol. Lett. 1997, 153, 273–277. [Google Scholar] [CrossRef]
- Asselin, J.E.; Eikemo, H.; Perminow, J.; Nordskog, B.; Brurberg, M.B.; Beer, S.V. Rahnella spp. Are Commonly Isolated from Onion (Allium cepa) Bulbs and Are Weakly Pathogenic. J. Appl. Microbiol. 2019, 127, 812–824. [Google Scholar] [CrossRef]
- Doonan, J.; Denman, S.; Pachebat, J.A.; McDonald, J.E. Genomic Analysis of Bacteria in the Acute Oak Decline Pathobiome. Microb. Genom. 2019, 5, e000240. [Google Scholar] [CrossRef]
- Moradi-Amirabad, Y.; Rahimian, H.; Babaeizad, V.; Denman, S. Brenneria spp. and Rahnella victoriana Associated with Acute Oak Decline Symptoms on Oak and Hornbeam in Iran. For. Pathol. 2019, 49, e12535. [Google Scholar] [CrossRef]
- Brady, C.; Denman, S.; Kirk, S.; Venter, S.; Rodríguez-Palenzuela, P.; Coutinho, T. Description of Gibbsiella quercinecans gen. nov., sp. nov., Associated with Acute Oak Decline. Syst. Appl. Microbiol. 2010, 33, 444–450. [Google Scholar] [CrossRef]
- Brady, C.; Hunter, G.; Kirk, S.; Arnold, D.; Denman, S. Gibbsiella greigii sp. nov., a Novel Species Associated with Oak Decline in the USA. Syst. Appl. Microbiol. 2014, 37, 417–422. [Google Scholar] [CrossRef]
- Jan-Roblero, J.; Cruz-Maya, J.A.; Guerrero Barajas, C.C. Kosakonia. In Beneficial Microbes in Agro-Ecology: Bacteria and Fungi; Academic Press: Cambridge, MA, USA, 2020; pp. 213–231. [Google Scholar] [CrossRef]
- Kämpfer, P.; McInroy, J.A.; Doijad, S.; Chakraborty, T.; Glaeser, S.P. Kosakonia pseudosacchari sp. nov., an Endophyte of Zea mays. Syst. Appl. Microbiol. 2016, 39, 1–7. [Google Scholar] [CrossRef]
- Krawczyk, K.; Borodynko-Filas, N. Kosakonia cowanii as the New Bacterial Pathogen Affecting Soybean (Glycine max Willd.). Eur. J. Plant Pathol. 2020, 157, 173–183. [Google Scholar] [CrossRef]
- Fatema, K.; Mahmud, N.U.; Gupta, D.R.; Siddiqui, M.N.; Sakif, T.I.; Sarker, A.; Sharpe, A.G.; Islam, T. Enhancing Rice Growth and Yield with Weed Endophytic Bacteria Alcaligenes faecalis and Metabacillus indicus under Reduced Chemical Fertilization. PLoS ONE 2024, 19, e0296547. [Google Scholar] [CrossRef]
- Yin, Z.; Wang, X.; Hu, Y.; Zhang, J.; Li, H.; Cui, Y.; Zhao, D.; Dong, X.; Zhang, X.; Liu, K.; et al. Metabacillus dongyingensis sp. nov. Is Represented by the Plant Growth-Promoting Bacterium BY2G20 Isolated from Saline-Alkaline Soil and Enhances the Growth of Zea mays L. under Salt Stress. mSystems 2022, 7, e0142621. [Google Scholar] [CrossRef]
- Tracz, D.M.; Gilmour, M.W.; Mabon, P.; Beniac, D.R.; Hoang, L.; Kibsey, P.; Van Domselaar, G.; Tabor, H.; Westmacott, G.R.; Corbett, C.R.; et al. Tatumella saanichensis sp. nov., Isolated from a Cystic Fibrosis Patient. Int. J. Syst. Evol. Microbiol. 2015, 65, 1959–1966. [Google Scholar] [CrossRef]
- Hollis, D.G.; Hickman, F.W.; Fanning, G.R.; Farmer, J.J.; Weaver, R.E.; Brenner, D.J. Tatumella ptyseos gen. nov., sp. nov., a Member of the Family Enterobacteriaceae Found in Clinical Specimens. J. Clin. Microbiol. 1981, 14, 79–88. [Google Scholar] [CrossRef]
- Oh, H.M.L.; Tay, L. Bacteraemia Caused by Rahnella aquatilis: Report of Two Cases and Review. Scand. J. Infect. Dis. 1995, 27, 79–80. [Google Scholar] [CrossRef]
- Berinson, B.; Bellon, E.; Christner, M.; Both, A.; Aepfelbacher, M.; Rohde, H. Identification of Kosakonia cowanii as a Rare Cause of Acute Cholecystitis: Case Report and Review of the Literature. BMC Infect. Dis. 2020, 20, 366. [Google Scholar] [CrossRef]
- Blazewicz, S.J.; Barnard, R.L.; Daly, R.A.; Firestone, M.K. Evaluating rRNA as an Indicator of Microbial Activity in Environmental Communities: Limitations and Uses. ISME J. 2013, 7, 2061–2068. [Google Scholar] [CrossRef]
- Matange, K.; Tuck, J.M.; Keung, A.J. DNA Stability: A Central Design Consideration for DNA Data Storage Systems. Nat. Commun. 2021, 12, 1358. [Google Scholar] [CrossRef]
- Vaz-Moreira, I.; Nunes, O.C.; Manaia, C.M. Diversity and Antibiotic Resistance in Pseudomonas spp. from Drinking Water. Sci. Total Environ. 2012, 426, 366–374. [Google Scholar] [CrossRef]
- Cepec, E.; Trček, J. Antimicrobial Resistance of Acetobacter and Komagataeibacter Species Originating from Vinegars. Int. J. Environ. Res. Public Health 2022, 19, 463. [Google Scholar] [CrossRef]
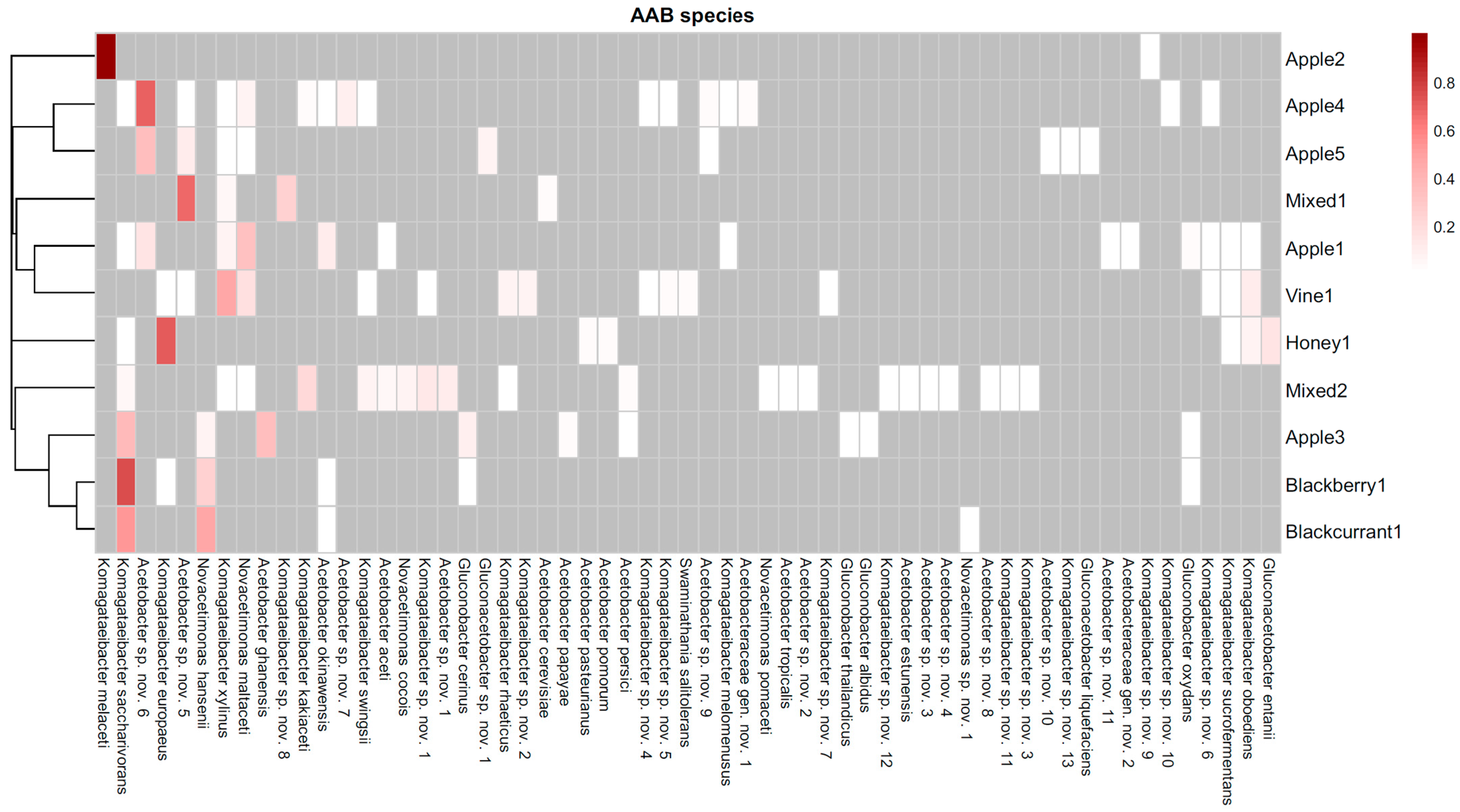

| No. | Sample | Origin | Raw Material | Type of Production | Type of Vessel | pH |
|---|---|---|---|---|---|---|
| 1 | Mixed1 | Lovrenc na Pohorju | Mixed fruit | Spontaneously | Plastic barrel | 3.6 |
| 2 | Blackcurrant1 | Lovrenc na Pohorju | Blackcurrant | Back-slopping | Glass vessel | 2.9 |
| 3 | Apple1 | Lovrenc na Pohorju | Apple | Spontaneously | Plastic barrel | 3.4 |
| 4 | Apple2 | Vurberk | Apple | Back-slopping | Plastic barrel | 3.2 |
| 5 | Apple3 | Lovrenc na Pohorju | Apple | Spontaneously | Wooden barrel | 2.9 |
| 6 | Honey1 | Videm pri Ptuju | Honey | Added pre-culture | Stainless steel barrel | 2.8 |
| 7 | Mixed2 | Pesnica pri Mariboru | Mixed fruit | Back-slopping | Plastic barrel | 3.6 |
| 8 | Apple4 | Lovrenc na Pohorju | Apple | Back-slopping | Wooden barrel | 3.2 |
| 9 | Blackberry1 | Lovrenc na Pohorju | Blackberry | Back-slopping | Glass vessel | 2.8 |
| 10 | Apple5 | Lovrenc na Pohorju | Apple | Back-slopping | Wooden barrel | 3.3 |
| 11 | Vine1 | Lovrenc na Pohorju | Wine | Back-slopping | Plastic barrel | 3.2 |
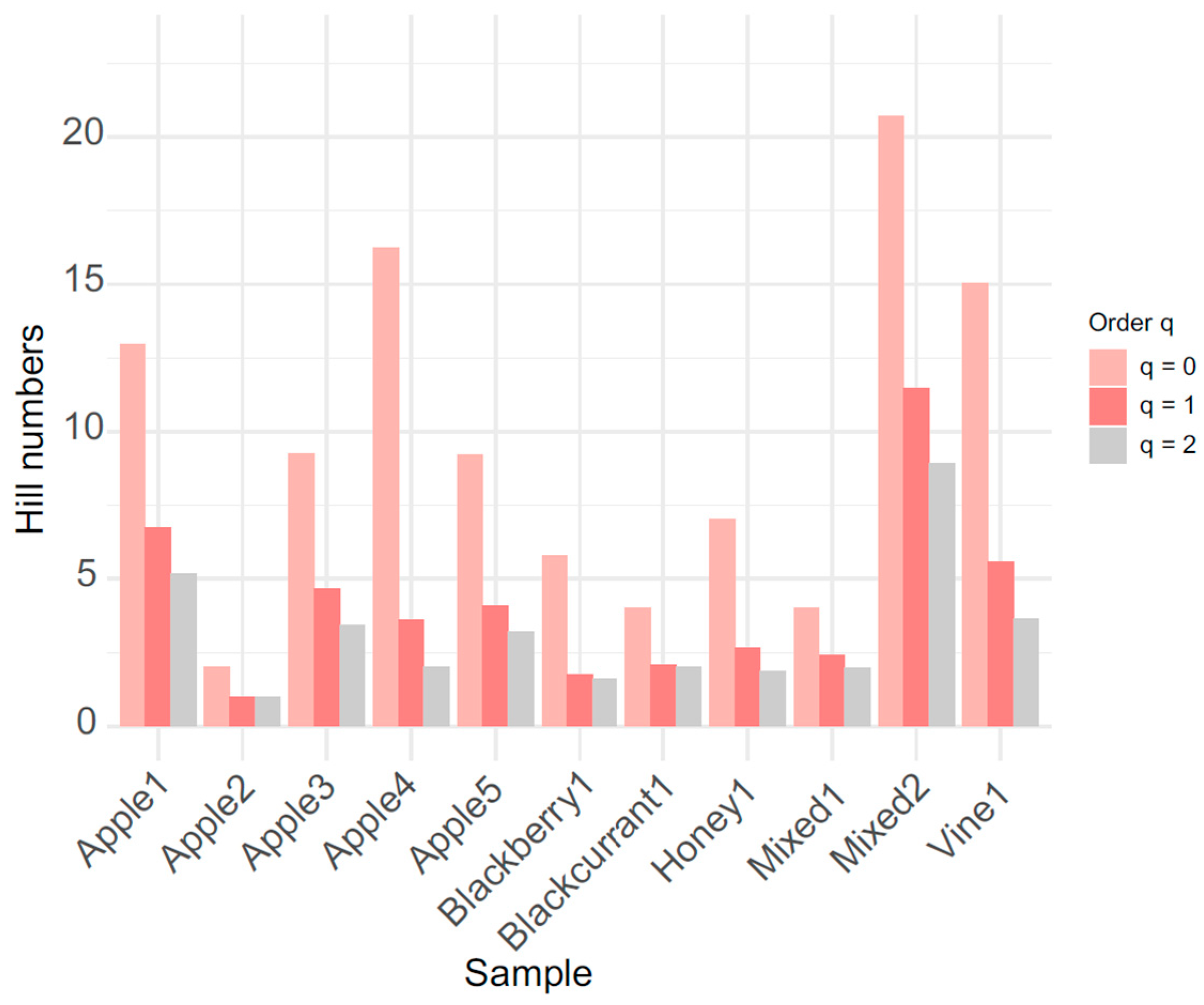
| Sample | q0 | q1 | q2 |
|---|---|---|---|
| Mixed2 | 21 | 11.49 | 8.91 |
| Apple1 | 13 | 6.73 | 5.18 |
| Apple4 | 16 | 3.60 | 2.03 |
| Vine1 | 15 | 5.58 | 3.65 |
| Apple3 | 9 | 4.65 | 3.42 |
| Apple5 | 9 | 4.10 | 3.20 |
| Honey1 | 7 | 2.65 | 1.87 |
| Blackberry1 | 6 | 1.77 | 1.59 |
| Mixed1 | 4 | 2.42 | 1.97 |
| Blackcurrant1 | 4 | 2.08 | 2.02 |
| Apple2 | 2 | 1.01 | 1.00 |
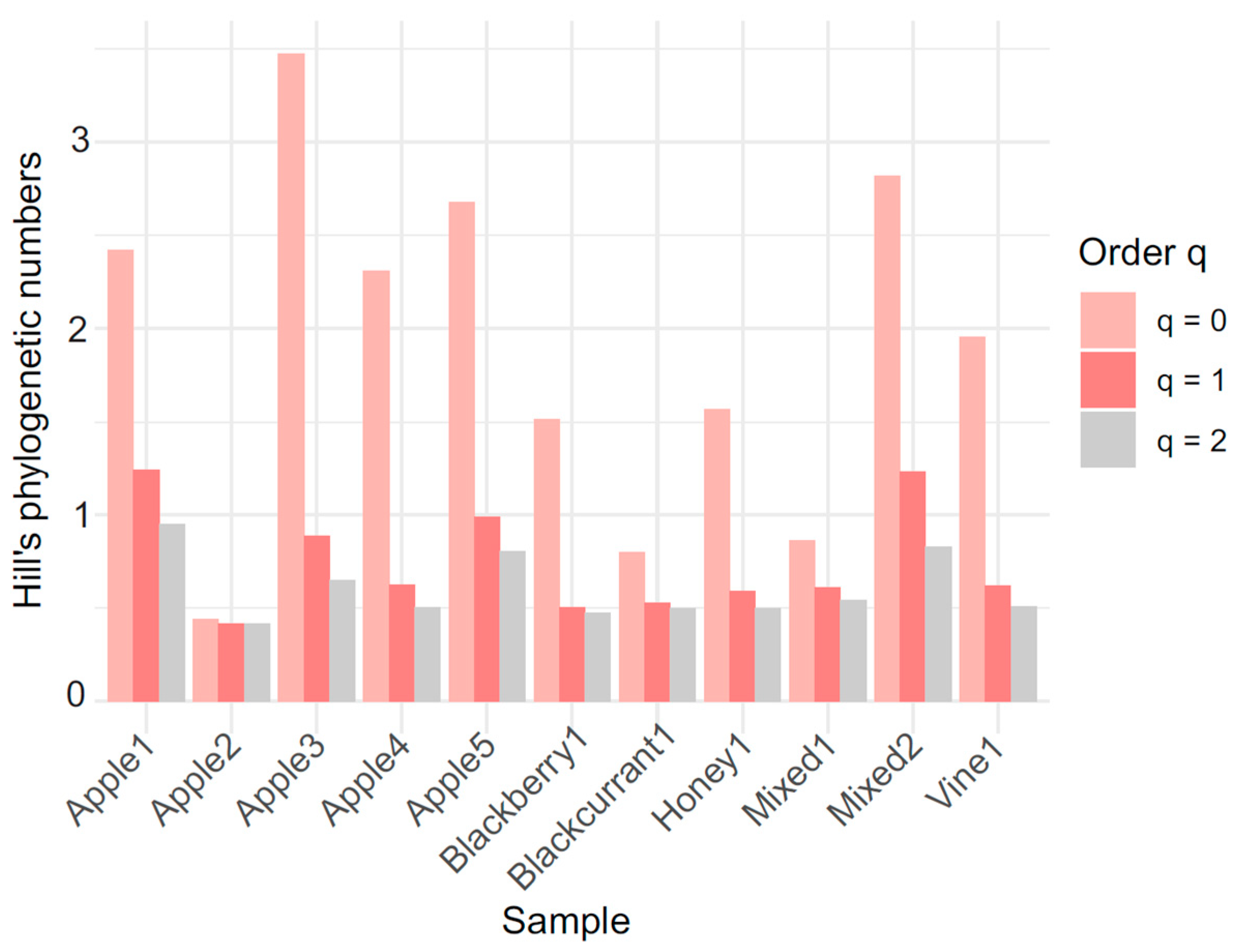
| Sample | q0 | q1 | q2 |
|---|---|---|---|
| Mixed2 | 2.82 | 1.23 | 0.82 |
| Apple1 | 2.42 | 1.24 | 0.94 |
| Apple4 | 2.31 | 0.62 | 0.50 |
| Vine1 | 1.95 | 0.62 | 0.51 |
| Apple3 | 3.47 | 0.89 | 0.65 |
| Apple5 | 2.68 | 0.98 | 0.80 |
| Honey1 | 1.57 | 0.60 | 0.49 |
| Blackberry1 | 1.51 | 0.50 | 0.47 |
| Mixed1 | 0.86 | 0.61 | 0.54 |
| Blackcurrant1 | 0.80 | 0.53 | 0.50 |
| Apple2 | 0.44 | 0.41 | 0.41 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karničnik, B.; Jugović, I.; Janžekovič, F.; Trček, J. Species Composition and Phylogenetic Diversity of Acetic Acid Bacteria Communities in Homemade Vinegars. Diversity 2025, 17, 770. https://doi.org/10.3390/d17110770
Karničnik B, Jugović I, Janžekovič F, Trček J. Species Composition and Phylogenetic Diversity of Acetic Acid Bacteria Communities in Homemade Vinegars. Diversity. 2025; 17(11):770. https://doi.org/10.3390/d17110770
Chicago/Turabian StyleKarničnik, Bernarda, Igor Jugović, Franc Janžekovič, and Janja Trček. 2025. "Species Composition and Phylogenetic Diversity of Acetic Acid Bacteria Communities in Homemade Vinegars" Diversity 17, no. 11: 770. https://doi.org/10.3390/d17110770
APA StyleKarničnik, B., Jugović, I., Janžekovič, F., & Trček, J. (2025). Species Composition and Phylogenetic Diversity of Acetic Acid Bacteria Communities in Homemade Vinegars. Diversity, 17(11), 770. https://doi.org/10.3390/d17110770







