Establishment of a COI Haplotype Baseline and Genetic Diversity Evaluation of Vespa soror (Hymenoptera: Vespidae) in Southern China Based on Mitochondrial Gene Sequences
Abstract
1. Introduction
2. Materials and Methods
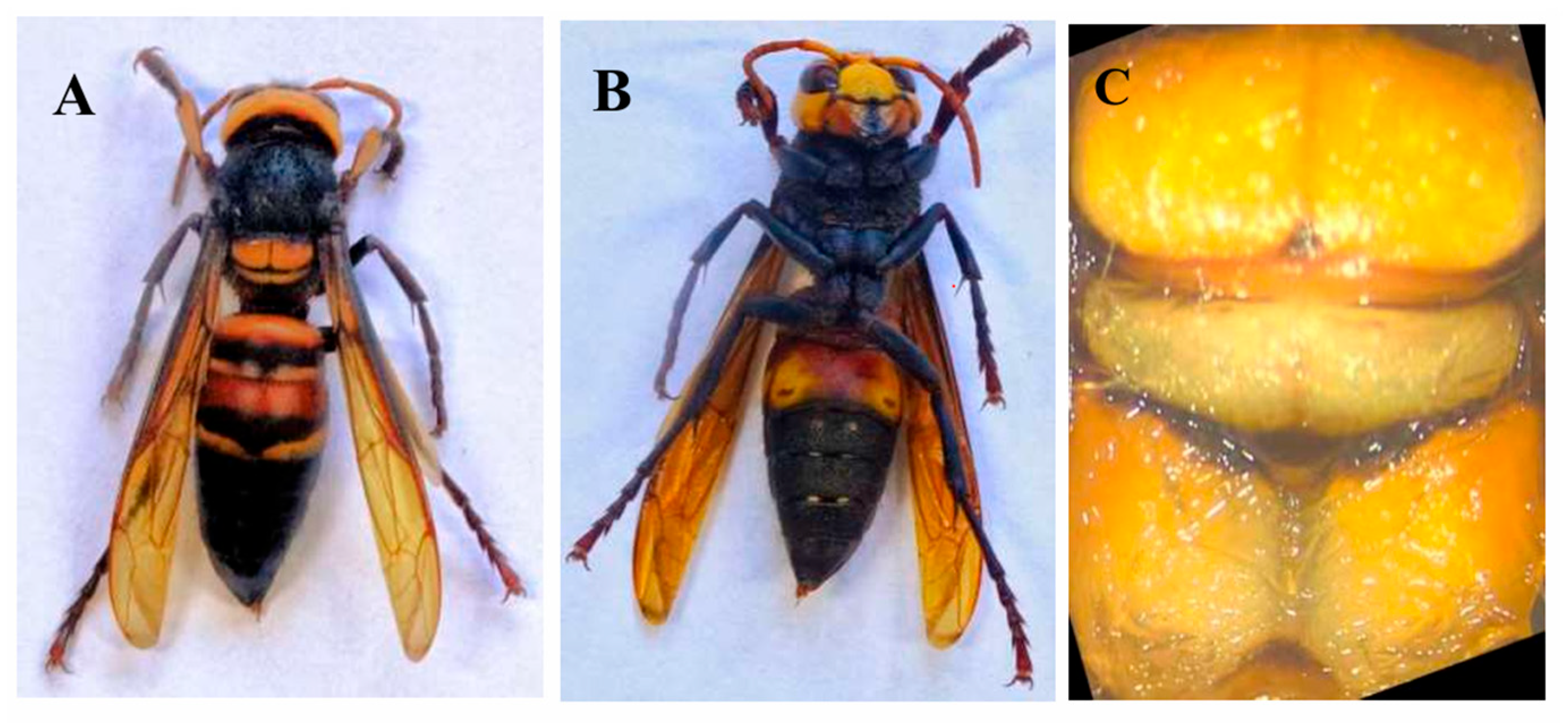
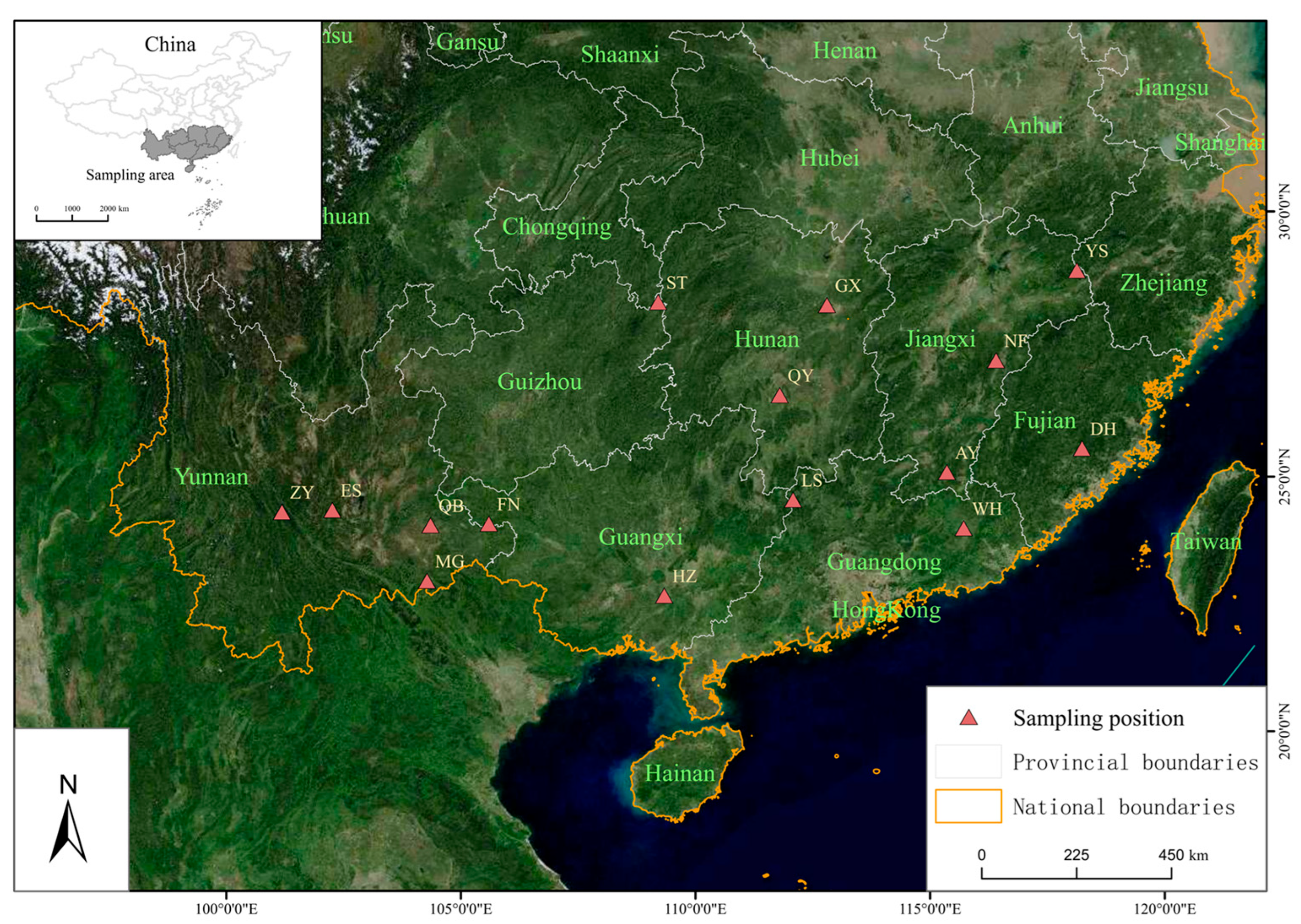
2.1. Sample Collection
2.2. DNA Extraction
2.3. Amplification and Sequencing of the COI Gene
2.4. Data Analysis
3. Results
3.1. Analysis of the Sequence Characteristics of the Mitochondrial COI Gene
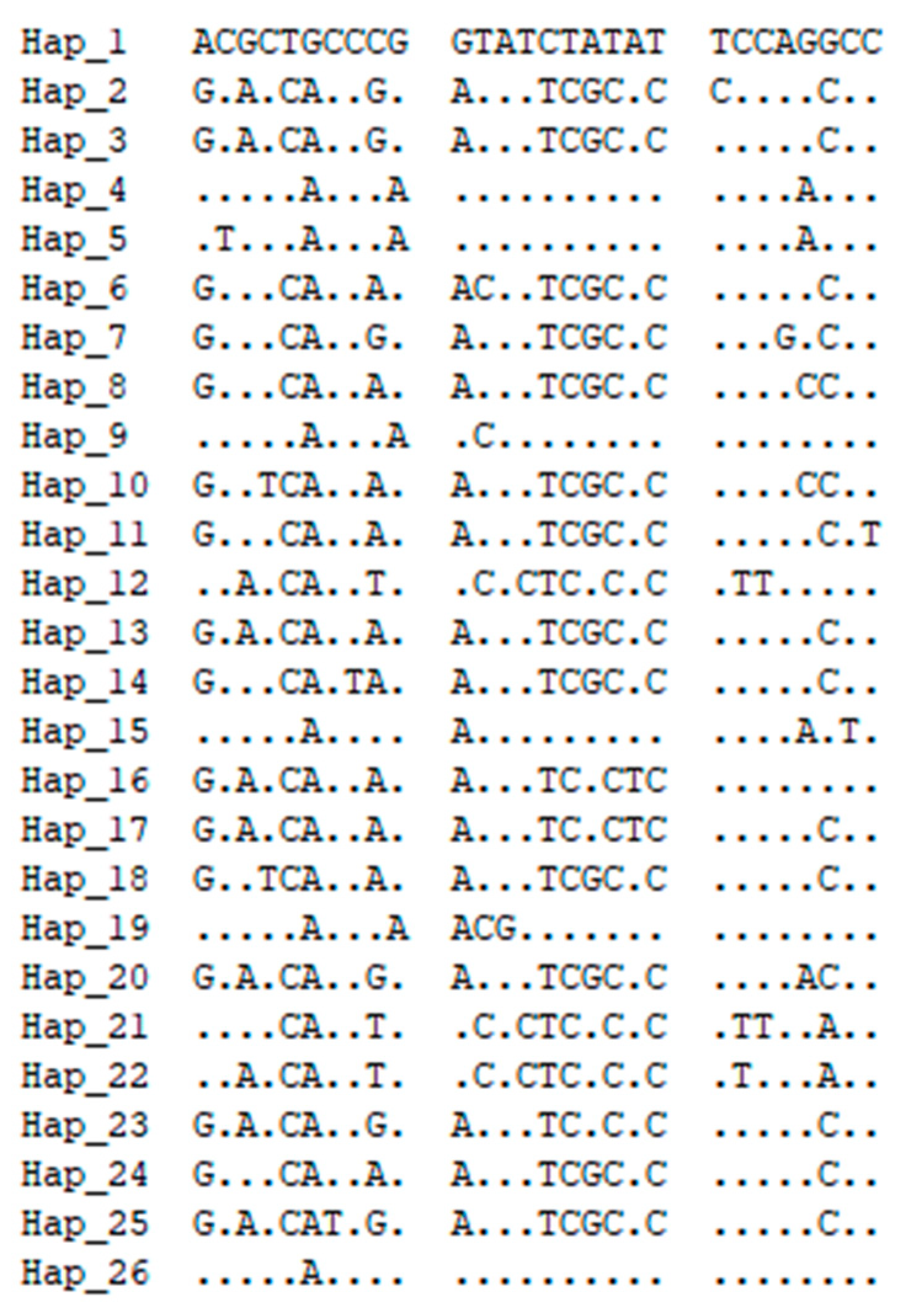
3.2. Haplotype and Genetic Diversity Analysis Based on the Mitochondrial COI Gene
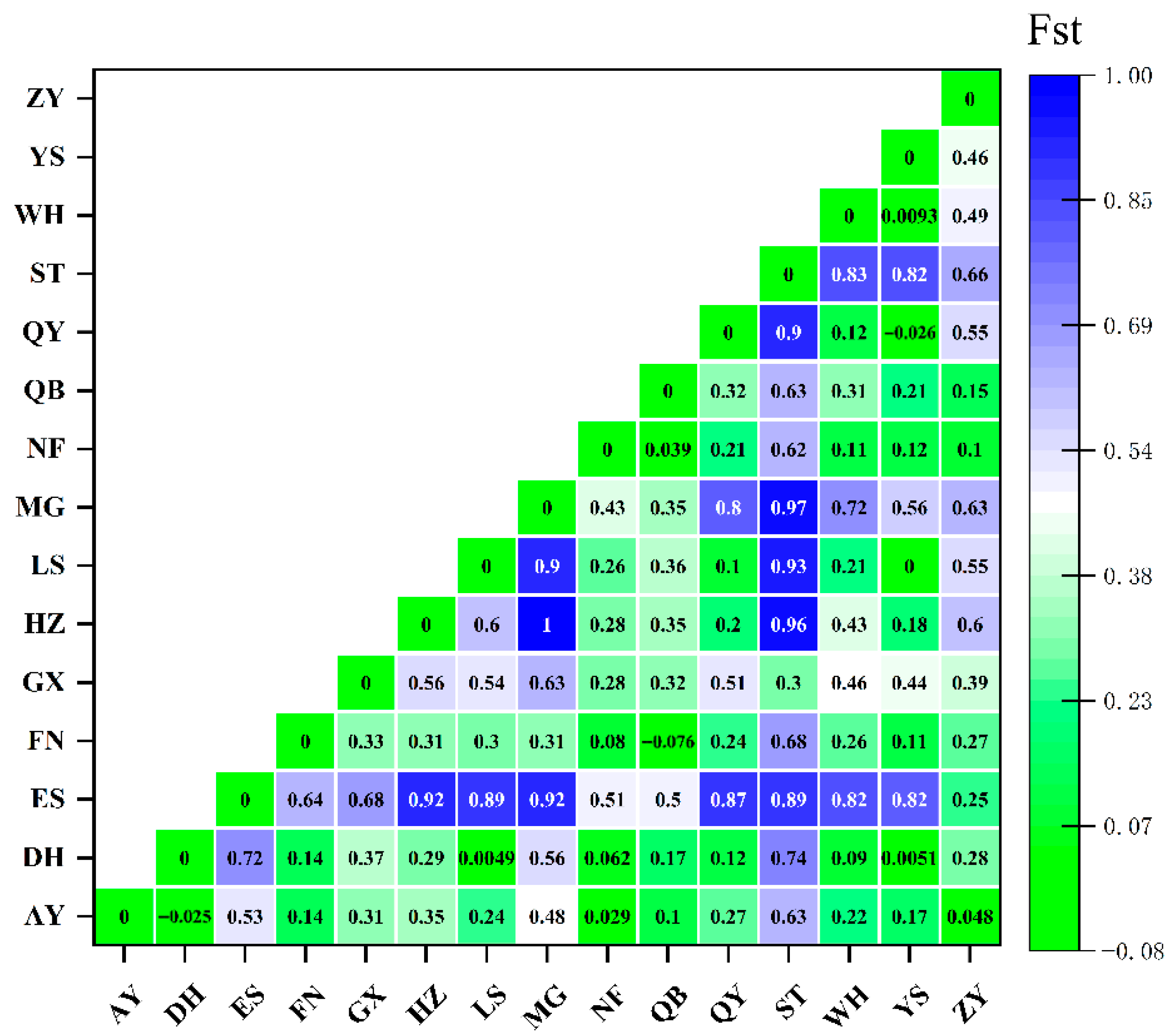
3.3. Genetic Differentiation Analysis of the COI Gene
3.4. Tajima’s D and Fu’s Fs Analysis of Populations
3.5. Haplotype TCS Network Graph Based on the COI Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| COI | cytochrome c oxidase subunit I |
| Cytb | cytochrome b |
| mtDNA | mitochondrial DNA |
| V. soror | Vespa soror |
| V. velutina | Vespa velutina |
| A. cerana | Apis cerana |
| DNA | Deoxyribonucleic Acid |
| PCR | Polymerase Chain Reaction |
| ddH2O | double-distilled water |
| Blast | Basic Local Alignment Search Tool |
| NCBI | National Center for Biotechnology Information |
| A, T, C, G | Adenine, Thymine, Cytosine, Guanine |
| AMOVA | Analysis of Molecular Variance |
| Fst | Fixation Index |
| Nm | Number of migrants per generation |
| Hap | Haplotype |
| Hd | Haplotype Diversity |
| Tajima’s D | Tajima’s D statistic |
| Fu’s Fs | Fu’s Fs statistic |
Appendix A
References
- Carpenter, J.; Kojima, J.-I.; Villemant, C. Phylogeny of hornets: A total evidence approach (Hymenoptera, Vespidae, Vespinae, Vespa). J. Hymenopt. Res. 2013, 32, 1. [Google Scholar] [CrossRef]
- Dehghani, R.; Kassiri, H.; Mazaheri-Tehrani, A.; Hesam, M.; Valazadi, N.; Mohammadzadeh, M. A study on habitats and behavioral characteristics of hornet wasp (Hymenoptera: Vespidae: Vespa orientalis), an important medical-health pest. Biomed. Res. 2019, 30, 61–66. [Google Scholar] [CrossRef]
- Taylor, B.A.; Tembrock, L.R.; Sankovitz, M.; Wilson, T.M.; Looney, C.; Takahashi, J.; Gilligan, T.M.; Smith-Pardo, A.H.; Harpur, B.A. Population genomics of the invasive Northern Giant Hornet Vespa mandarinia in North America and across its native range. Sci. Rep. 2024, 14, 10803. [Google Scholar] [CrossRef] [PubMed]
- Mattila, H.R.; Otis, G.W.; Billen, J.; Nguyen, L.T.; Shimano, S. Comparison of the external morphology of the sternal glands for hornets in the genus Vespa. Biology 2022, 11, 245. [Google Scholar] [CrossRef]
- Kim, C.-J.; Choi, M.B. First discovery of Vespa velutina nigrithorax du Buysson (Hymenoptera: Vespidae), an invasive hornet in the feces of the yellow-throated marten in South Korea. Insects 2021, 12, 296. [Google Scholar] [CrossRef]
- Jeong, H.; Kim, J.M.; Kim, B.; Nam, J.-O.; Hahn, D.; Choi, M.B. Nutritional value of the larvae of the alien invasive wasp Vespa velutina nigrithorax and amino acid composition of the larval saliva. Foods 2020, 9, 885. [Google Scholar] [CrossRef]
- Mozhui, L.; Kakati, L.; Kiewhuo, P.; Changkija, S. Traditional knowledge of the utilization of edible insects in Nagaland, North-East India. Foods 2020, 9, 852. [Google Scholar] [CrossRef]
- Kiewhuo, P.; Mozhui, L.; Kakati, L.; Meyer-Rochow, V. Traditional rearing techniques of the edible Asian giant hornet (Vespa mandarinia Smith) and its socio-economic perspective in Nagaland, India. J. Insects Food Feed 2022, 8, 325–336. [Google Scholar] [CrossRef]
- Mattila, H.R.; Nguyen, L.T.; Perrard, A.; Bain, M.; Otis, G.W. Biology of the southern giant hornet, Vespa soror: Nest architecture, morphological differences among castes, and the genetic structure of colonies. Front. Insect Sci. 2023, 3, 1136297. [Google Scholar] [CrossRef]
- Kumar, P.G.; Srinivasan, G. New record of Vespa soror du Buysson (Hymenoptera: Vespidae) from India. Rec. Zool. Surv. India 2009, 109, 111–112. [Google Scholar] [CrossRef]
- Bass, A.; Needham, K.; Bennett, A.M. First record of Vespa crabro Linnaeus (Hymenoptera: Vespidae) in western North America with a review of recorded species of Vespa Linnaeus in Canada. Zootaxa 2022, 5154, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Smith-Pardo, A.H.; Carpenter, J.M.; Kimsey, L. The diversity of hornets in the genus Vespa (Hymenoptera: Vespidae; Vespinae), their importance and interceptions in the United States. Insect Syst. Divers. 2020, 4, 2. [Google Scholar] [CrossRef]
- Arca, M.; Mougel, F.; Guillemaud, T.; Dupas, S.; Rome, Q.; Perrard, A.; Muller, F.; Fossoud, A.; Capdevielle-Dulac, C.; Torres-Leguizamon, M. Reconstructing the invasion and the demographic history of the yellow-legged hornet, Vespa velutina, in Europe. Biol. Invasions 2015, 17, 2357–2371. [Google Scholar] [CrossRef]
- Granato, A.; Negrisolo, E.; Bonomi, J.; Zulian, L.; Cappa, F.; Bortolotti, L.; Mutinelli, F. Recent confirmation of a single haplotype in the Italian population of Vespa velutina. Biol. Invasions 2019, 21, 2811–2817. [Google Scholar] [CrossRef]
- Wang, W.; Ma, C.; Chen, W.; Jin, Z.; Zhao, M.; Zhang, F.; Liu, Z.; Ma, L. Population genetic diversity of mud crab (Scylla paramamosain) from southeast coastal regions of China based on mitochondrial COI gene sequence. Gene 2020, 751, 144763. [Google Scholar] [CrossRef]
- Wang, H.; Wen, Q.; Wang, T.; Ran, F.; Wang, M.; Fan, X.; Wei, S.; Li, Z.; Tan, J. Next-generation sequencing of four mitochondrial genomes of Dolichovespula (Hymenoptera: Vespidae) with a phylogenetic analysis and divergence time estimation of Vespidae. Animals 2022, 12, 3004. [Google Scholar] [CrossRef]
- Chen, P.-Y.; Wei, S.-J.; Liu, J.-X. The mitochondrial genome of the Vespa mandarinia Smith (Hymenoptera: Vespidae: Vespinae) and a phylogenetic analysis of the Vespoidea. Mitochondrial DNA Part A 2016, 27, 4414–4415. [Google Scholar] [CrossRef]
- Euo, S.-S.; Choi, J.-H.; Choi, M.B.; Kim, I.-K.; Kim, C.-J. The mitochondrial genome of a social wasp, Vespula rufa (Linnaeus, 1758) (Hymenoptera: Vespidae). Mitochondrial DNA Part B 2024, 9, 1024–1028. [Google Scholar] [CrossRef]
- Wu, H.; Qi, S.; Fan, S.; Li, H.; Zhang, Y.; Zhang, Y.; Xu, Q.; Chen, G. Analysis of the Mitochondrial COI Gene and Genetic Diversity of Endangered Goose Breeds. Genes 2024, 15, 1037. [Google Scholar] [CrossRef]
- Xia, L.; Cai, F.; Chen, S.; Cai, Y.; Zhou, K.; Yan, J.; Li, P. Phylogenetic Analysis and Genetic Structure of Schlegel’s Japanese Gecko (Gekko japonicus) from China Based on Mitochondrial DNA Sequences. Genes 2022, 14, 18. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Chapman, N.C.; Raffiudin, R.; Putra, R.E.; Roberts, J.; Widjaya, C.; Buchmann, G.; Holmes, M.; Gloag, R. Loss of mitochondrial diversity in invasive populations of Asian honey bees, Apis cerana (Hymenoptera: Apidae), in the Austral-Pacific. Austral Entomol. 2022, 61, 97–103. [Google Scholar] [CrossRef]
- Wang, M.M.; Yin, P.K.; Tang, Y.N.; Yang, Z.Z.; Xiao, H.; Zhang, C.G.; Yang, Y.H.; Yang, D.S. Utility of DNA barcoding for identification of common Vespa species (Hymenoptera: Vespidae) from Yunnan, China. Entomol. Res. 2022, 52, 111–117. [Google Scholar] [CrossRef]
- Kim, J.S.; Jeong, J.S.; Jeong, S.Y.; Kim, M.J.; Kim, I. Complete mitochondrial genome of the black-tailed hornet, Vespa ducalis (Hymenoptera: Vespidae): Genomic comparisons in Vespoidea. Entomol. Res. 2017, 47, 129–136. [Google Scholar] [CrossRef]
- Sertić Kovačević, M.; Baričević, A.; Kružić, P.; Maurić Maljković, M.; Hamer, B. Barcoding (COI) Sea Cucumber Holothuria mammata Distribution Analysis: Adriatic Rare or Common Species? Genes 2023, 14, 2059. [Google Scholar] [CrossRef]
- Takeuchi, T.; Takahashi, R.; Kiyoshi, T.; Nakamura, M.; Minoshima, Y.; Takahashi, J. The origin and genetic diversity of the yellow-legged hornet, Vespa velutina introduced in Japan. Insectes Sociaux 2017, 64, 313–320. [Google Scholar] [CrossRef]
- Abd-El-Samie, E.M.; Elkafrawy, I.; Osama, M.; Ageez, A. Molecular phylogeny and identification of the Egyptian wasps (Hymenoptera: Vespidae) based on COI mitochondrial gene sequences. Egypt. J. Biol. Pest Control 2018, 28, 36. [Google Scholar] [CrossRef]
- Zhou, C.j.; Feng, M.x.; Tang, Y.t.; Yang, C.x.; Meng, X.l.; Nie, G.x. Species diversity of freshwater shrimp in Henan Province, China, based on morphological characters and COI mitochondrial gene. Ecol. Evol. 2021, 11, 10502–10514. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Proceedings of the Nucleic Acids Symposium Series; Oxford University Press: Oxford, UK, 1999; pp. 95–98. [Google Scholar]
- Hammer, O. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Bloch, N.A.; Runyan, C.L.; Speshock, J.L.; Lambert, B.D.; Wellmann, K.B.; Tifft, K.; Brady, J.A. Investigating Bovine Blood Prokaryotic Microbial Populations Through 16S V4 Sequencing, qPCR, and dPCR, with a Specific Focus on Hemotrophic Mycoplasma wenyonii. Ruminants 2025, 5, 45. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Zhang, X.; He, Y.; Zhang, W.; Wang, Y.; Liu, X.; Cui, A.; Gong, Y.; Lu, J.; Liu, X.; Huo, X. Development of microsatellite marker system to determine the genetic diversity of experimental chicken, duck, goose, and pigeon populations. BioMed Res. Int. 2021, 2021, 8851888. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D.; Nakagawa, S. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Wei, S.-J.; Shi, M.; Chen, X.-X.; Sharkey, M.J.; van Achterberg, C.; Ye, G.-Y.; He, J.-H. New views on strand asymmetry in insect mitochondrial genomes. PLoS ONE 2010, 5, e12708. [Google Scholar] [CrossRef]
- Cen, X.; Zhang, G.; Liu, H.; Yao, G.; Xiong, P.; He, M.; Liu, W. Analysis of genetic diversity in two different shell colors of the giant triton snail (Charonia tritonis) based on mitochondrial COI sequences. Front. Mar. Sci. 2023, 9, 1066750. [Google Scholar] [CrossRef]
- Wang, S.-H.; Zhang, C.; Shang, M.; Wu, X.-G.; Cheng, Y.-X. Genetic diversity and population structure of native mitten crab (Eriocheir sensu stricto) by microsatellite markers and mitochondrial COI gene sequence. Gene 2019, 693, 101–113. [Google Scholar] [CrossRef]
- Yi, C.H.; Yoon, M.; Kim, J.M.; Kim, I.-H.; Cho, I.-Y.; An, H.S. Genetic analysis and population genetic structure of hard-shelled mussel, Mytilus coruscus Gould 1861 (Mytiloida: Mytilidae) from the coasts of South Korea based on mitochondrial cytochrome oxidase (COI) gene sequences. Genes Genom. 2021, 43, 577–585. [Google Scholar] [CrossRef]
- Budge, G.E.; Hodgetts, J.; Jones, E.P.; Ostojá-Starzewski, J.C.; Hall, J.; Tomkies, V.; Semmence, N.; Brown, M.; Wakefield, M.; Stainton, K. The invasion, provenance and diversity of Vespa velutina Lepeletier (Hymenoptera: Vespidae) in Great Britain. PLoS ONE 2017, 12, e0185172. [Google Scholar] [CrossRef]
- Liu, W.; Duan, Z.; Wang, D.; Zhao, W.; Liu, P. Phylogenetic relationships and genetic differentiation of two Salamandrella species as revealed via COI gene from Northeastern China. PLoS ONE 2024, 19, e0298221. [Google Scholar] [CrossRef]
- Jiang, S.; Li, Z.; Li, J.; Xu, K.; Ye, Y. Analysis of genetic diversity and structure of eight populations of Nerita yoldii along the Coast of China based on mitochondrial COI gene. Animals 2024, 14, 718. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, M.J.; Park, J.S.; Lee, K.H.; Jo, Y.H.; Takahashi, J.-i.; Choi, Y.S.; Kim, I. Tracing the invasion characteristics of the yellow-legged hornet, Vespa velutina nigrithorax (Hymenoptera: Vespidae), in Korea using newly detected variable mitochondrial DNA sequences. J. Asia-Pac. Entomol. 2021, 24, 135–147. [Google Scholar] [CrossRef]
- Wright, S. Evolution in Mendelian populations. Genetics 1931, 16, 97. [Google Scholar] [CrossRef]
- Rousset, F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 1997, 145, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- Herrera, C.; Ferragut, J.F.; Leza, M.; Jurado-Rivera, J.A. Invasion genetics of the yellow-legged hornet Vespa velutina in the Westernmost Mediterranean archipelago. J. Pest Sci. 2024, 97, 645–656. [Google Scholar] [CrossRef]
- Sauvard, D.; Imbault, V.; Darrouzet, É. Flight capacities of yellow-legged hornet (Vespa velutina nigrithorax, Hymenoptera: Vespidae) workers from an invasive population in Europe. PLoS ONE 2018, 13, e0198597. [Google Scholar] [CrossRef] [PubMed]
- Wilson, T.M.; Takahashi, J.; Spichiger, S.-E.; Kim, I.; Van Westendorp, P. First reports of Vespa mandarinia (Hymenoptera: Vespidae) in North America represent two separate maternal lineages in Washington state, United States, and British Columbia, Canada. Ann. Entomol. Soc. Am. 2020, 113, 468–472. [Google Scholar] [CrossRef]
- Tajima, F. The effect of change in population size on DNA polymorphism. Genetics 1989, 123, 597–601. [Google Scholar] [CrossRef]
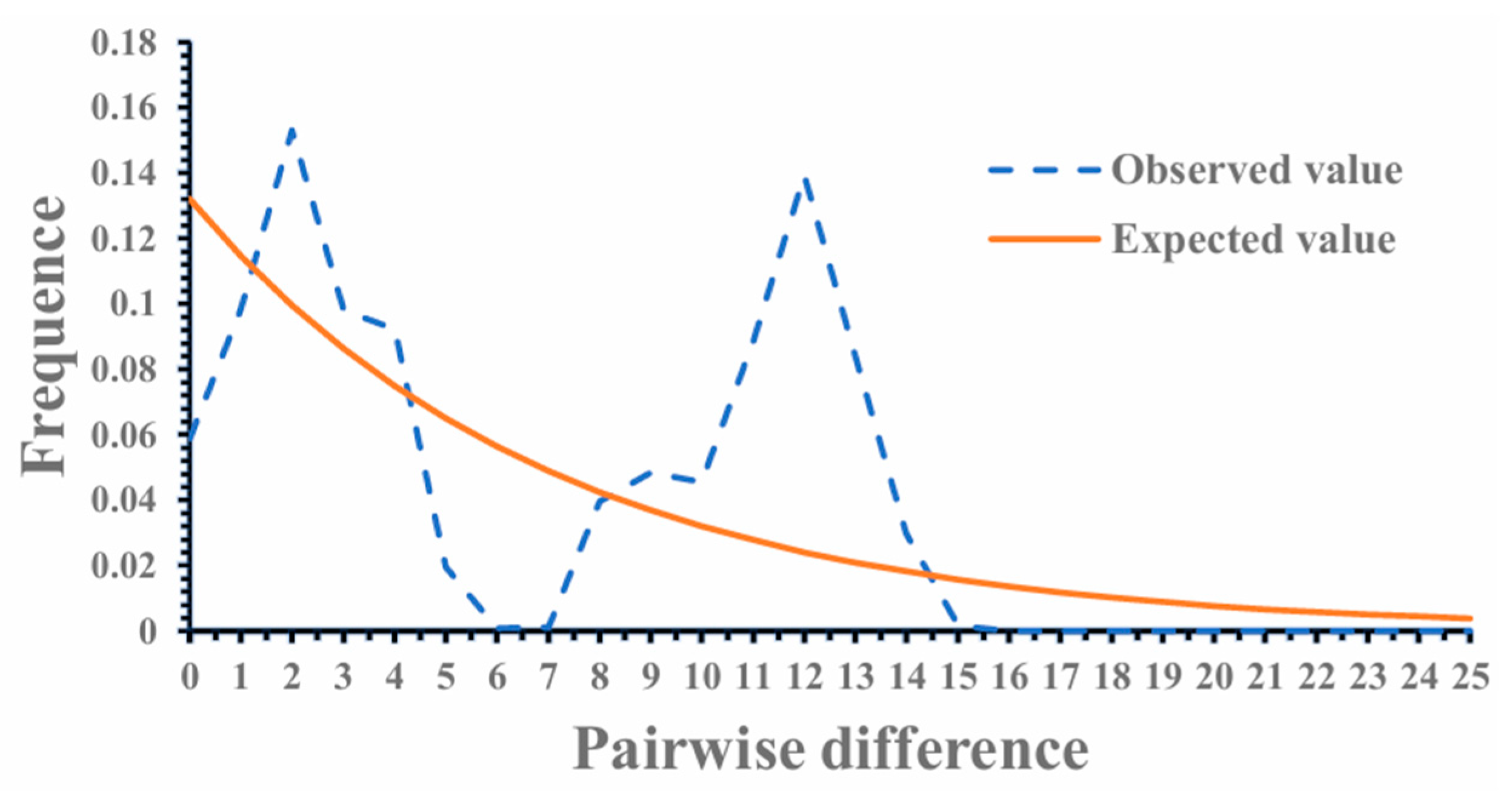
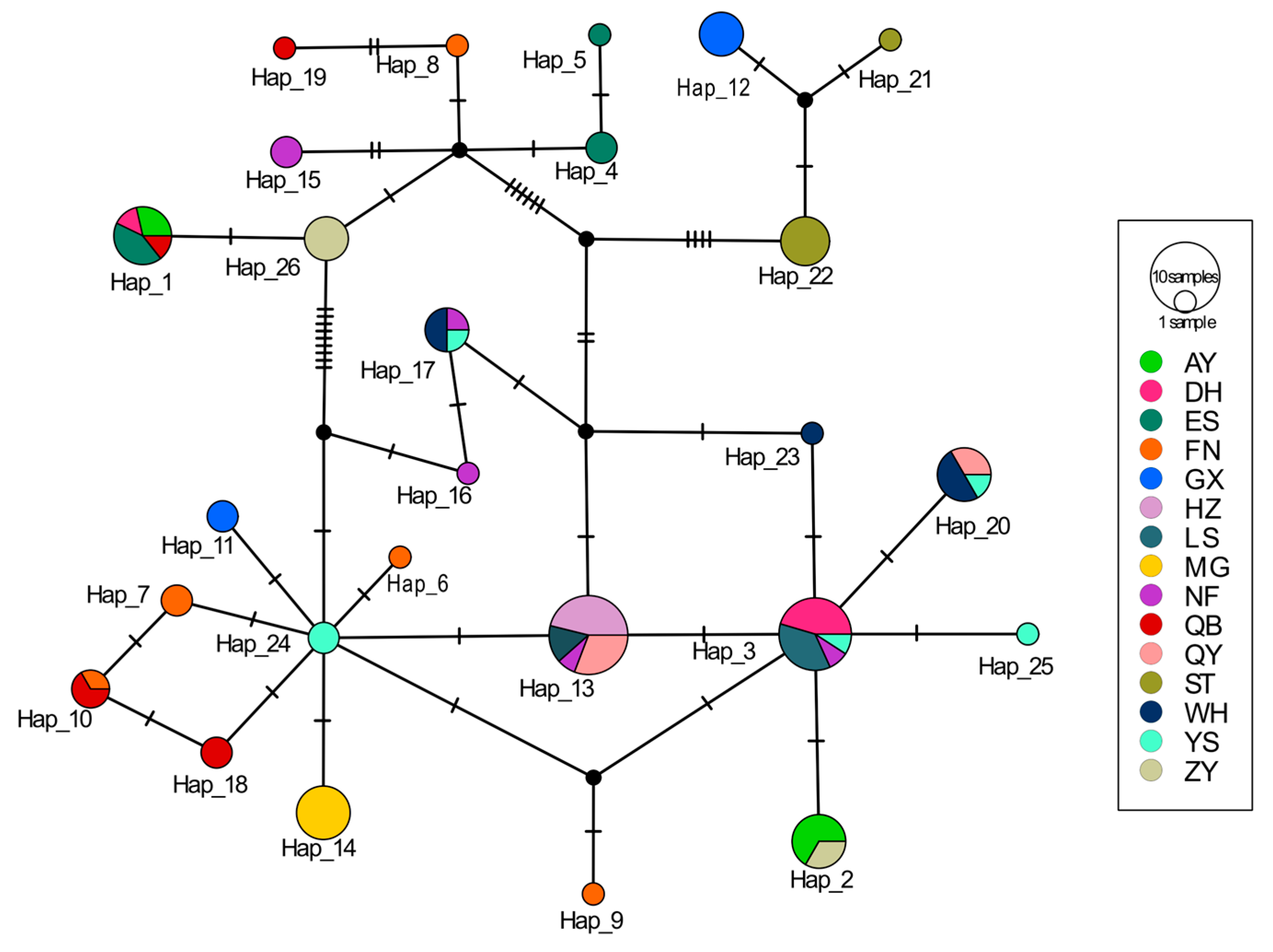
| Population | Haplotypes (Hap) | Segregating Site (S) | Haplotype Diversity (Hd) | Nucleotide Diversity (Pi) | Average Number of Difference (K) |
|---|---|---|---|---|---|
| AY | H1(2), H2(4) | 13 | 0.533 ± 0.172 | 0.01127 ± 0.00364 | 6.93333 |
| DH | H1(1), H3(5) | 12 | 0.333 ± 0.215 | 0.00650 ± 0.00420 | 4.00000 |
| ES | H1(3), H4(2), H5(1) | 4 | 0.733 ± 0.155 | 0.00347 ± 0.00074 | 2.13333 |
| FN | H6(1), H7(2), H8(1), H9(1), H10(1) | 15 | 0.933 ± 0.122 | 0.00932 ± 0.00383 | 5.73333 |
| GX | H11(2), H12(4) | 11 | 0.533 ± 0.172 | 0.00954 ± 0.00308 | 5.86667 |
| HZ | H13(6) | 0 | 0 | 0 | 0 |
| LS | H3(4), H13(2) | 1 | 0.533 ± 0.172 | 0.00087 ± 0.00028 | 0.53333 |
| MG | H14(6) | 0 | 0 | 0 | 0 |
| NF | H3(1), H13(1), H15(2), H16(1), H17(1) | 13 | 0.933 ± 0.122 | 0.01171 ± 0.00270 | 7.20000 |
| QB | H1(1), H10(2), H18(2), H19(1) | 16 | 0.867 ± 0.129 | 0.01225 ± 0.00374 | 7.53333 |
| QY | H13(4), H20(2) | 2 | 0.533 ± 0.172 | 0.00173 ± 0.00056 | 1.06667 |
| ST | H21(1), H22(5) | 2 | 0.333 ± 0.215 | 0.00108 ± 0.00070 | 0.66667 |
| WH | H17(2), H20(3), H23(1) | 4 | 0.733 ± 0.155 | 0.00369 ± 0.00080 | 2.26667 |
| YS | H3(1), H17(1), H20(1), H24(2), H25(1) | 6 | 0.933 ± 0.122 | 0.00401 ± 0.00075 | 2.46667 |
| ZY | H2(2), H26(4) | 12 | 0.533 ± 0.172 | 0.01041 ± 0.00336 | 6.40000 |
| Total | Hap-1~Hap-26 | 28 | 0.941 ± 0.010 | 0.01068 ± 0.00079 | 6.57104 |
| Population | Tajima’s D | p Value | Fu’s Fs | p Value |
|---|---|---|---|---|
| AY | 1.33148 | p > 0.10 | 6.639 | / |
| DH | −1.45326 | 0.10 > p > 0.05 | 4.828 | / |
| ES | 1.18059 | p > 0.10 | 1.141 | / |
| FN | −1.12437 | p > 0.10 | −0.114 | / |
| GX | 1.31709 | p > 0.10 | 6.057 | / |
| HZ | - | - | 0 | 0 |
| LS | 0.85057 | p > 0.10 | 0.625 | / |
| MG | - | - | 0 | 0 |
| NF | 1.07021 | p > 0.10 | 0.271 | / |
| QB | 0.46426 | p > 0.10 | 1.986 | / |
| QY | 1.03194 | p > 0.10 | 1.723 | / |
| ST | −1.13197 | p > 0.10 | 0.952 | / |
| WH | 1.59319 | p > 0.10 | 1.256 | / |
| YS | −0.35084 | p > 0.10 | −1.672 | / |
| ZY | 1.32483 | p > 0.10 | 6.357 | / |
| Total | 0.12833 | p > 0.10 | −4.001 | / |
| Source of Variation | Df | Sum of Squares | Variance Components | Percentage of Variation | Fixation Indices | p-Value |
|---|---|---|---|---|---|---|
| Among populations | 14 | 160.411 | 1.61632 Va | 47.87% | ||
| Within populations | 75 | 132.000 | 1.76000 Vb | 52.13% | ||
| Total | 89 | 292.411 | 3.37632 | 100% | FST:0.47872 | 0.00000 |
| Source of Variation | Df | Sum of Squares | Variance Components | Percentage of Variation | Fixation Indices | p-Value |
|---|---|---|---|---|---|---|
| Among groups | 1 | 33.094 | 0.58252 Va | 15.82% | FCT:0.15823 | 0.00000 |
| Among populations Within groups | 13 | 127.317 | 1.33893 Vb | 36.37% | FSC:0.43206 | 0.00000 |
| Within populations | 75 | 132.000 | 1.76000 Vc | 47.81% | ||
| Total | 89 | 292.411 | 3.68145 | 100% | FST:0.52193 | 0.00978 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, J.; Zhang, J.; Liu, J.; Wang, Z.; Guo, Y.; Yang, J.; Wang, Z. Establishment of a COI Haplotype Baseline and Genetic Diversity Evaluation of Vespa soror (Hymenoptera: Vespidae) in Southern China Based on Mitochondrial Gene Sequences. Diversity 2025, 17, 756. https://doi.org/10.3390/d17110756
Cui J, Zhang J, Liu J, Wang Z, Guo Y, Yang J, Wang Z. Establishment of a COI Haplotype Baseline and Genetic Diversity Evaluation of Vespa soror (Hymenoptera: Vespidae) in Southern China Based on Mitochondrial Gene Sequences. Diversity. 2025; 17(11):756. https://doi.org/10.3390/d17110756
Chicago/Turabian StyleCui, Junming, Jinlu Zhang, Jun Liu, Zian Wang, Yanhe Guo, Jun Yang, and Zhenji Wang. 2025. "Establishment of a COI Haplotype Baseline and Genetic Diversity Evaluation of Vespa soror (Hymenoptera: Vespidae) in Southern China Based on Mitochondrial Gene Sequences" Diversity 17, no. 11: 756. https://doi.org/10.3390/d17110756
APA StyleCui, J., Zhang, J., Liu, J., Wang, Z., Guo, Y., Yang, J., & Wang, Z. (2025). Establishment of a COI Haplotype Baseline and Genetic Diversity Evaluation of Vespa soror (Hymenoptera: Vespidae) in Southern China Based on Mitochondrial Gene Sequences. Diversity, 17(11), 756. https://doi.org/10.3390/d17110756






