Genetic Differences Between Wild Transplanted and Farmed Populations of Banggai Cardinalfish Pterapogon kauderni Based on Mitochondrial Control Region and SNP Polymorphism
Abstract
1. Introduction
2. Materials and Methods
2.1. Sources of Specimens
2.2. Genomic DNA Extraction
2.3. PCR and DNA Sequencing
2.4. CR Data Analysis
2.5. SLAF-Seq Analysis
2.6. SLAF-Seq Data Analysis
3. Results
3.1. Genetic Diversity and Differentiation Based on CR Polymorphism
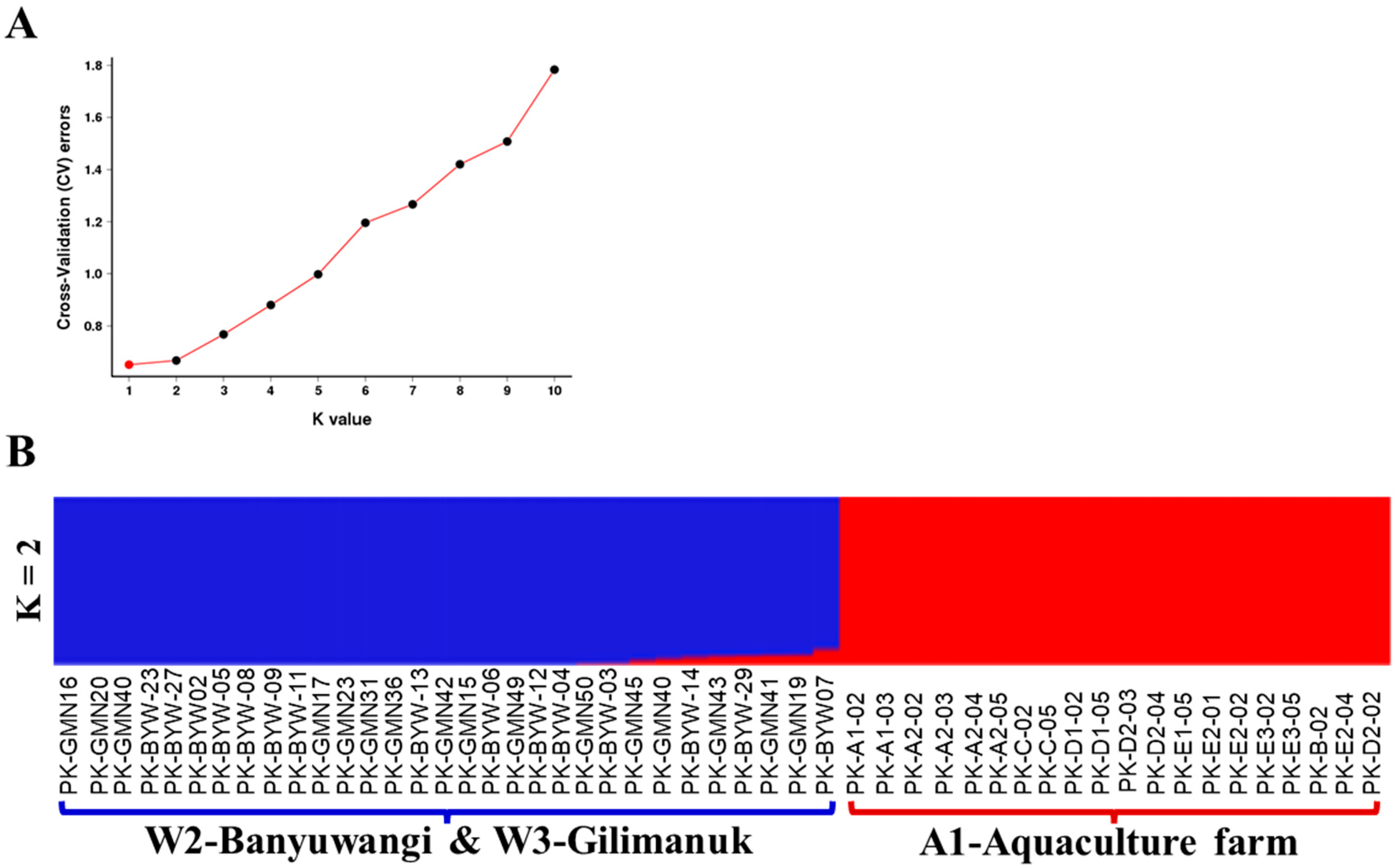
3.2. Genetic Diversity and Differentiation Based on SNPs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Putra, I.N.G.; Puspitha, N.L.P.R.; Suryaningtyas, E.W. Spread beyond the border: Small Scale genetic structure of the introduced Banggai cardinalfish (Pterapogon kauderni) population in the Bali Strait. Indonesian J. Mar. Sci. 2021, 26, 163–172. [Google Scholar] [CrossRef]
- Allen, G.R. Threatened fishes of the world: Pterapogon kauderni Koumans, 1933 (Apogonidae). Environ. Biol. Fishes 2000, 57, 142. [Google Scholar] [CrossRef]
- CITES. Convention on the International Trade in Endangered Species. COP 14 Prop. XX. Consideration of Proposals for Amendment of Appendices I and II. Proposal: Inclusion of the Banggai cardinalfish (Pterapogon kauderni, Koumans 1933) in Appendix II of CITES. In Proceedings of the Fourteenth Meeting of the Conference of the Parties, The Hague, The Netherlands, 3–15 June 2007. Available online: www.cites.org (accessed on 1 August 2025).
- Allen, G.R.; Donaldson, T.J. Pterapogon kauderni. The IUCN Red List of Threatened Species 2007: E.T63572A12692964., The IUCN Red List of Threatened Species 2007. Available online: https://doi.org/10.2305/IUCN.UK.2007.RLTS.T63572A12692964.en (accessed on 1 August 2025).
- Allen, G.; Erdmann, M. Reef Fishes of the East Indies; Tropical Reef Research: Perth, Australia, 2012; Volume I–III. [Google Scholar]
- Erdmann, M.V.; Vagelli, A. Banggai cardinalfish invade Lembeh Strait. Coral Reefs 2001, 20, 252–253. [Google Scholar] [CrossRef]
- Moore, A.; Ndobe, S. Discovery of an introduced Banggai Cardinalfish population in Palu Bay, Central Sulawesi, Indonesia. Coral Reefs 2007, 26, 569. [Google Scholar] [CrossRef]
- Moore, A.M.; Ndobe, S.; Jompa, J. A site based conservation approach to promote the recovery of Banggai cardinalfish (Pterapogon kauderni) endemic populations. Coast. Ocean J. 2017, 1, 63–72. [Google Scholar] [CrossRef]
- Lunn, K.E.; Moreau, M.A. Unmonitored trade in marine ornamental fishes: The case of Indonesia’s Banggai cardinalfish (Pterapogon kauderni). Coral Reefs 2004, 23, 344–351. [Google Scholar] [CrossRef]
- Bernardi, G.; Vagelli, A. Population structure in Banggai cardinalfish, Pterapogon kauderni, a coral reef species lacking a pelagic larval phase. Mar. Biol. 2004, 145, 803–810. [Google Scholar] [CrossRef]
- Hoffman, E.A.; Kolm, N.; Berglund, A.; Arguello, J.R.; Jones, A.G. Genetic structure in the coral-reef-associated Banggai cardinalfish, Pterapogon kauderni. Mol. Ecol. 2005, 14, 1367–1375. [Google Scholar] [CrossRef] [PubMed]
- Vagelli, A.; Burford, M.; Bernardi, G. Fine scale dispersal in Banggai Cardinalfish, Pterapogon kauderni, a coral reef species lacking a pelagic larval phase. Mar. Genom. 2009, 1, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Pouil, S.; Tlusty, M.; Rhyne, A.; Metian, M. Aquaculture of marine ornamental fish: Overview of the production trends and the role of academia in research progress. Rev. Aquac. 2020, 12, 1217–1230. [Google Scholar] [CrossRef]
- Lal, K.K.; Kumar, T.T.A. Marine Ornamental Aquaculture: Measure Towards Biodiversity Conservation and Livelihood Promotion. J. Indian Soc. Coastal Agric. Res. 2021, 39, 49–54. [Google Scholar] [CrossRef]
- Sun, X.; Liu, D.; Zhang, X.; Li, W.; Liu, H.; Hong, W.; Jiang, C.; Guan, N.; Ma, C.; Zeng, H.; et al. SLAF-seq: An efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 2016, 8, e58700. [Google Scholar] [CrossRef]
- Khamnamtong, B.; Chaimongkol, A.; Prasertlux, S.; Janpoom, S.; Chaimongkol, J.; Tang, S.; Ittarat, W.; Songsangjinda, P.; Sakamoto, T.; Sae-Lim, P.; et al. Population genomics and application for growth improvement of domesticated Asian seabass Lates calcarifer from Thailand. Diversity 2025, 17, 383. [Google Scholar] [CrossRef]
- Lee, W.J.; Conroy, J.; Howell, W.H.; Kocher, T.D. Structure and evolution of teleost mitochondrial control regions. J. Mol. Evol. 1995, 41, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. Clustal W: Improving the sensitivity of progressive multiple sequence weighting, position-specific gap penalties and weight metric choices. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Met. Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimation F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Price, A.; Patterson, N.; Plenge, R.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Abraham, G.; Inouye, M. Fast principal component analysis of large-scale genome-wide data. PLoS ONE 2014, 9, e93766. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Janes, J.K.; Miller, J.M.; Dupuis, J.R.; Malenfant, R.M.; Gorrell, J.C.; Cullingham, C.I.; Andrew, R.L. The K = 2 conundrum. Mol. Ecol. 2017, 26, 3594–3602. [Google Scholar] [CrossRef]
- Clark, R.D.; Pinsky, M.L. Global patterns of nuclear and mitochondrial genetic diversity in marine fishes. Ecol. Evol. 2024, 14, e11365. [Google Scholar] [CrossRef] [PubMed]
- Ndobe, S.; Moore, A.; Yasir, I.; Jompa, J. Banggai cardinalfish conservation: Priorities, opportunities, and risks. IOP Conf. Ser. Earth Environ. Sci. 2019, 253, 012033. [Google Scholar] [CrossRef]
- Arbi, U.Y.; Vimono, I.B.; Kusumawardhani, N.R.; Anshari, L. Population status and microhabitat preferences of endemic Banggai cardinalfish (Pterapogon kauderni) in the introduced habitat in Kendari Bay, Indonesia. IOP Conf. Ser. Earth Environ. Sci. 2022, 1119, 012015. [Google Scholar] [CrossRef]
- Madduppa, H.H.; Timm, J.; Kochzius, M. Reduced genetic diversity in the clown anemonefish Amphiprion ocellaris in exploited reefs of Spermonde Archipelago, Indonesia. Front. Mar. Sci. 2018, 5, 80. [Google Scholar] [CrossRef]
- Villanueva, B.; Fernández, A.; Peiró-Pastor, R.; Peñaloza, C.; Houston, R.D.; Sonesson, A.K.; Tsigenopoulos, C.S.; Bargelloni, L.; Gamsız, K.; Karahan, B.; et al. Population structure and genetic variability in wild and farmed Mediterranean populations of gilthead seabream and European seabass inferred from a 60K combined species SNP array. Aquac. Rep. 2022, 24, 101145. [Google Scholar] [CrossRef]
- Gkagkavouzis, K.; Papakostas, S.; Maroso, F.; Karaiskou, N.; Carr, A.; Nielsen, E.E.; Bargelloni, L.; Triantafyllidis, A. Investigating genetic diversity and genomic signatures of hatchery-induced evolution in gilthead seabream (Sparus aurata) populations. Diversity 2021, 13, 563. [Google Scholar] [CrossRef]
- Wenne, R.; Bernaś, R.; Kijewska, A.; Poćwierz-Kotus, A.; Strand, J.; Petereit, C.; Plauška, K.; Sics, I.; Árnyasi, M.; Kent, M.P. SNP genotyping reveals substructuring in weakly differentiated populations of Atlantic cod (Gadus morhua) from diverse environments in the Baltic Sea. Sci Rep. 2020, 10, 9738. [Google Scholar] [CrossRef]
- Cui, A.; Xu, Y.; Kikuchi, K.; Jiang, Y.; Wang, B.; Koyama, T.; Liu, X. Comparative analysis of genetic structure and diversity in five populations of yellowtail kingfish (Seriola aureovittata). J. Mar. Sci. Eng. 2023, 11, 1583. [Google Scholar] [CrossRef]
- Glover, K.A.; Dahle, G.; Jørstad, K.E. Genetic identification of farmed and wild Atlantic cod, Gadus morhua, in coastal Norway. ICES J. Mar. Sci. 2011, 68, 901–910. [Google Scholar] [CrossRef][Green Version]

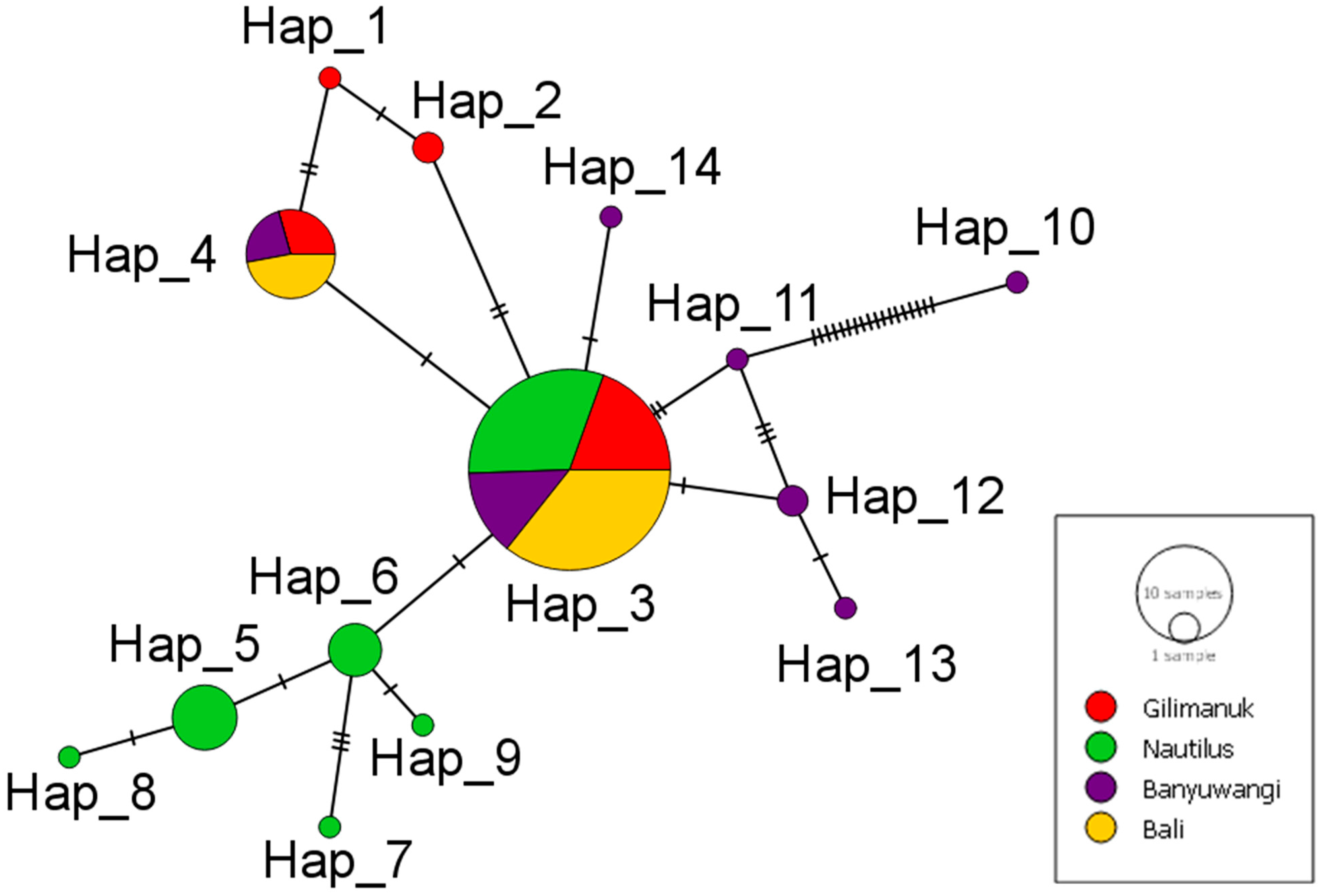
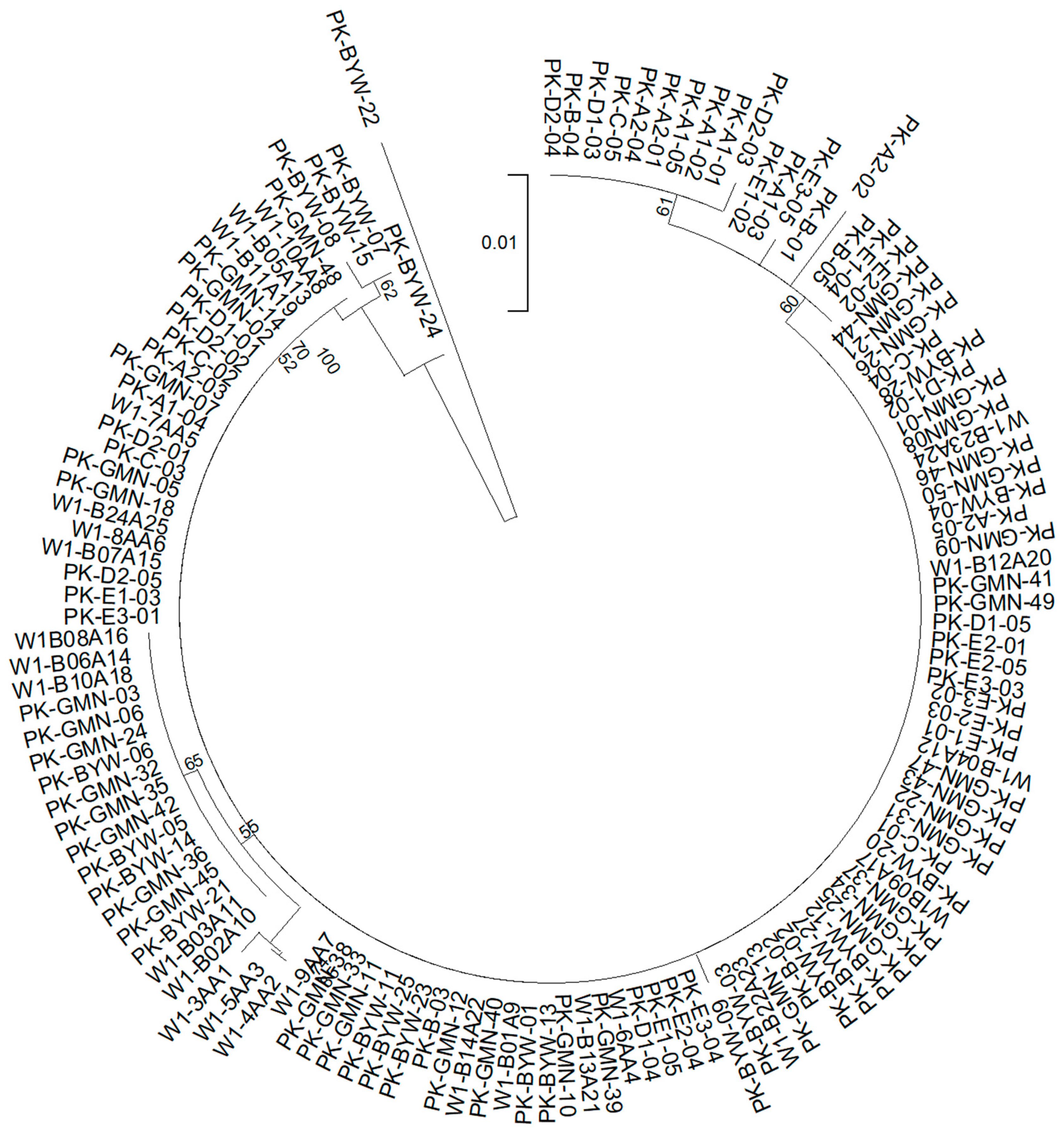
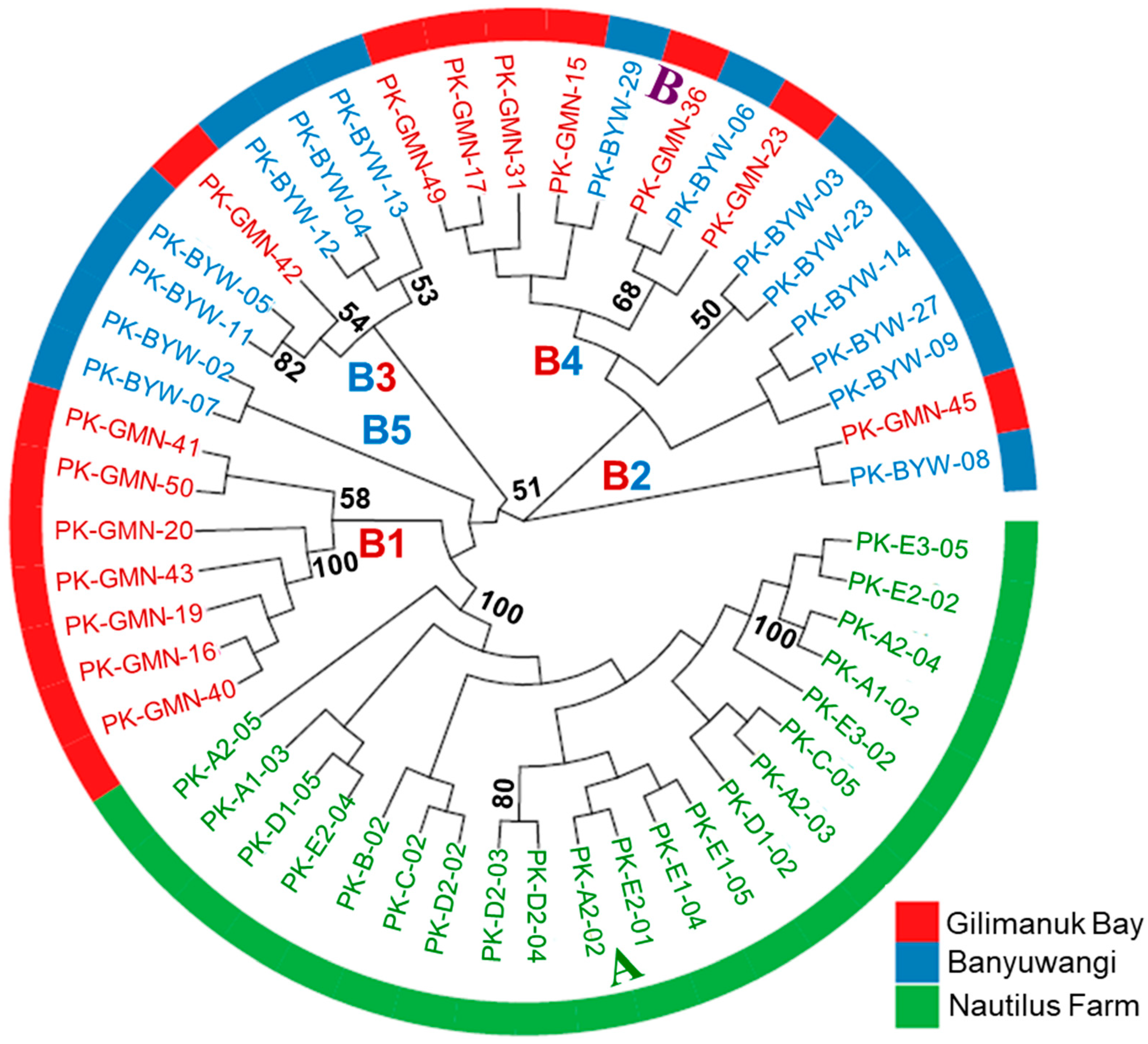
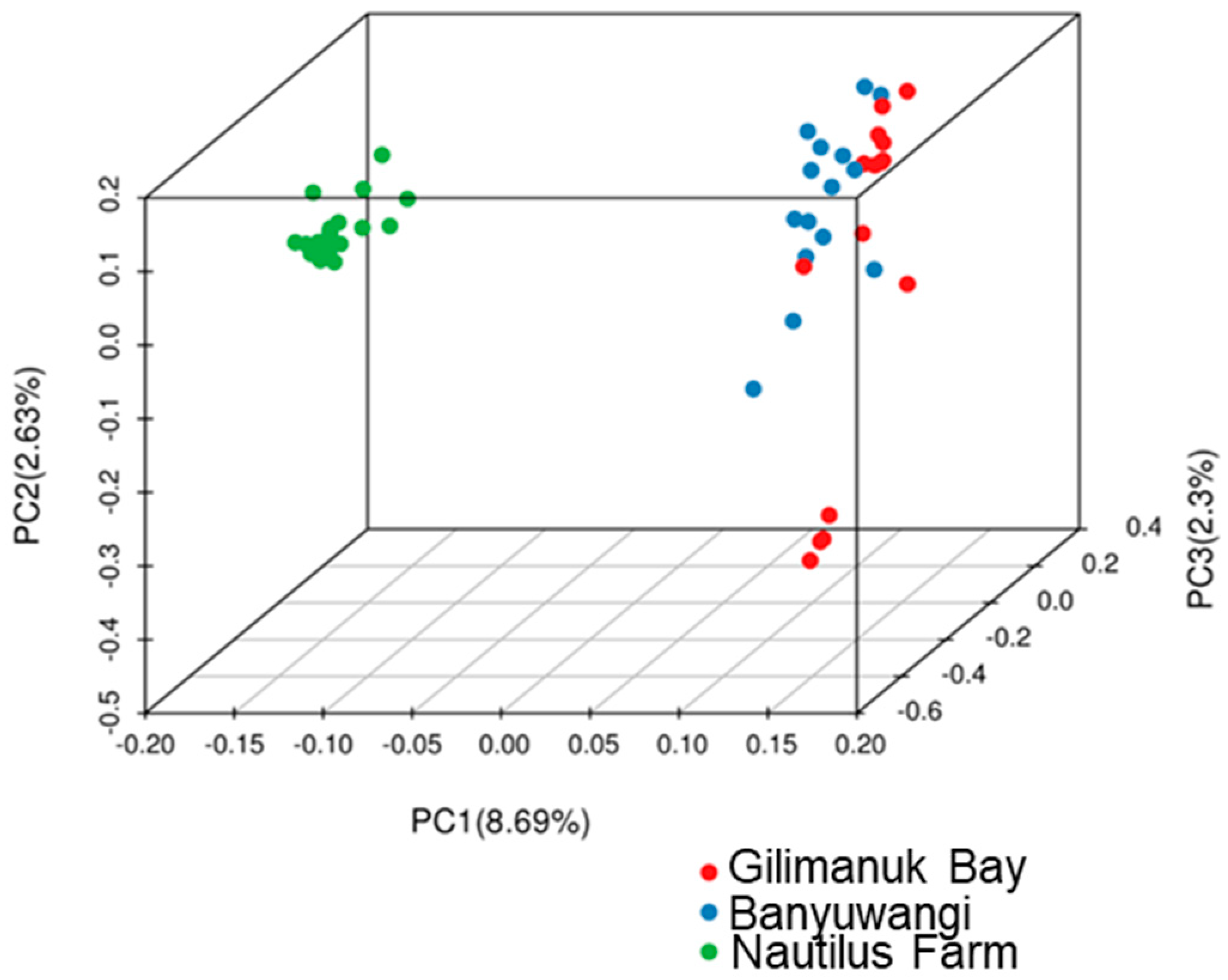
| Population | Sampling Year | Sample Size (N) | NH | Hd | π | k | References |
|---|---|---|---|---|---|---|---|
| Wild transplanted stocks | |||||||
| W1-Gilimanuk Bay | 2019 | 25 | 4 | 0.510 | 0.00186 | 0.820 | [1] |
| W2-Banyuwangi (2024) | 2024 | 22 | 7 | 0.688 | 0.00721 | 3.143 | This study |
| W3-Gilimanuk Bay (2024) | 2024 | 39 | 2 | 0.335 | 0.00074 | 0.335 | This study |
| Cultured stock | |||||||
| A1-Nautilus Park | 2024 | 45 | 6 | 0.602 | 0.00257 | 1.156 | This study |
| Total samples | 131 | 14 | 0.538 | 0.00283 | 1.227 | This study |
| W1-Gilimanuk [1] | A1-Aquaculture (2024) | W2-Banyuwangi (2024) | W3-Gilimanuk (2024) | |
|---|---|---|---|---|
| W1-Gilimanuk [1] | - | |||
| A1-Aquaculture (2024) | 0.00061 | - | ||
| W2-Banyuwangi (2024) | 0.00011 | 0.00059 | - | |
| W3-Gilimanuk (2024) | 0.00002 | 0.00053 | 0.00006 | - |
| W1-Gilimanuk [1] | A1-Aquaculture (2024) | W2-Banyuwangi (2024) | W3-Gilimanuk (2024) | |
|---|---|---|---|---|
| W1-Gilimanuk [1] | - | ND | ND | ND |
| A1-Aquaculture (2024) | 0.0240 ns | - | 0.1261 *** | 0.1234 *** |
| W2-Banyuwangi (2024) | 0.2147 *** | 0.2454 *** | - | 0.0035 *** |
| W3-Gilimanuk (2024) | 0.0292 ns | 0.0416 *** | 0.1474 *** | - |
| Parameter | A1-Nautilus Park | W2-Banyuwangi | W3-Gilimanuk Bay |
|---|---|---|---|
| Total reads | 342,000,000 | 289,000,000 | 320,000,000 |
| Average reads/sample | 16,269,337 | 19,236,880 | 21,318,605 |
| Q30 (%) | 94.86–96.27 | 95.31–96.29 | 95.29–96.28 |
| (95.91) | (95.89) | (95.93) | |
| GC (%) | 40.32–42.77 | 40.65–42.45 | 40.62–42.93 |
| (41.10) | (41.38) | (41.48) | |
| Average nucleotide sequenced/sample | 5,908,063 | 6,212,348 | 6,379,631 |
| Average depth | 9.74 | 10.15 | 10.38 |
| Total SLAF number | 12,672,870 | 9,108,019 | 9,168,429 |
| Average SLAF number | 603,470 | 607,201.3 | 611,228.6 |
| N | 21 | 15 | 15 |
| Parameters/Group | A1-Nautilus Park | W2-Banyuwangi | W3-Gilimanuk Bay |
|---|---|---|---|
| Average MAF | 0.217 | 0.222 | 0.219 |
| No. of polymorphic markers | 11137 | 11132 | 11147 |
| Observed no. of allele (Ao) | 1.000–2.000 (1.879) | 1.000–2.000 (1.862) | 1.000–2.000 (1.863) |
| Expected no. of allele (Ae) | 1.000–2.000 (1.417) | 1.000–2.000 (1.438) | 1.000–2.000 (1.434) |
| Observed heterozygosity (Ho) | 0.048–1.000 (0.262) | 0.067–1.000 (0.288) | 0.067–1.000 (0.282) |
| Expected heterozygosity (He) | 0.038–0.500 (0.296) | 0.007–0.500 (0.312) | 0.007–0.500 (0.310) |
| PIC | 0.045–0.375 (0.243) | 0.062–0.375 (0.255) | 0.062–0.375 (0.253) |
| Nei diversity index | 0.048–0.667 (0.307) | 0.067–0.667 (0.328) | 0.067–0.667 (0.325) |
| Shannon Wiener index | 0.113–0.693 (0.460) | 0.146–0.693(0.480) | 0.146–0.693(0.477) |
| Sources of Variation | df | Sum of Square | Variance Component | Percentage of Variation | F-Statistics | p-Values |
|---|---|---|---|---|---|---|
| Within individuals | 51 | 18254.00 | 357.92 | 101.21 | FIT = −0.0121 | 0.9863 |
| Among individuals Within populations | 49 | 12763.72 | −48.72 | −13.78 | FIS = −0.1576 | 1.0000 |
| Among populations | 1 | 2455.71 | 44.43 | 12.56 | FST = 0.1256 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prasertlux, S.; Janpoom, S.; Ratdee, O.; Tang, S.; Ittarat, W.; Klinbunga, S.; Khamnamtong, B. Genetic Differences Between Wild Transplanted and Farmed Populations of Banggai Cardinalfish Pterapogon kauderni Based on Mitochondrial Control Region and SNP Polymorphism. Diversity 2025, 17, 754. https://doi.org/10.3390/d17110754
Prasertlux S, Janpoom S, Ratdee O, Tang S, Ittarat W, Klinbunga S, Khamnamtong B. Genetic Differences Between Wild Transplanted and Farmed Populations of Banggai Cardinalfish Pterapogon kauderni Based on Mitochondrial Control Region and SNP Polymorphism. Diversity. 2025; 17(11):754. https://doi.org/10.3390/d17110754
Chicago/Turabian StylePrasertlux, Sirikan, Sirithorn Janpoom, Onchuda Ratdee, Sureerat Tang, Wanwipa Ittarat, Sirawut Klinbunga, and Bavornlak Khamnamtong. 2025. "Genetic Differences Between Wild Transplanted and Farmed Populations of Banggai Cardinalfish Pterapogon kauderni Based on Mitochondrial Control Region and SNP Polymorphism" Diversity 17, no. 11: 754. https://doi.org/10.3390/d17110754
APA StylePrasertlux, S., Janpoom, S., Ratdee, O., Tang, S., Ittarat, W., Klinbunga, S., & Khamnamtong, B. (2025). Genetic Differences Between Wild Transplanted and Farmed Populations of Banggai Cardinalfish Pterapogon kauderni Based on Mitochondrial Control Region and SNP Polymorphism. Diversity, 17(11), 754. https://doi.org/10.3390/d17110754





