Mitochondrial Phylogeography and Population History of the Balkan Short-Tailed Mouse (Mus macedonicus Petrov and Ružić, 1983) in Turkey and Surrounding Areas
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Sequencing
2.2. Genetic Diversity Estimates
2.3. Phylogenetic Analysis
2.4. Demographic Analysis
3. Results
3.1. Haplotype Distribution Within Turkey
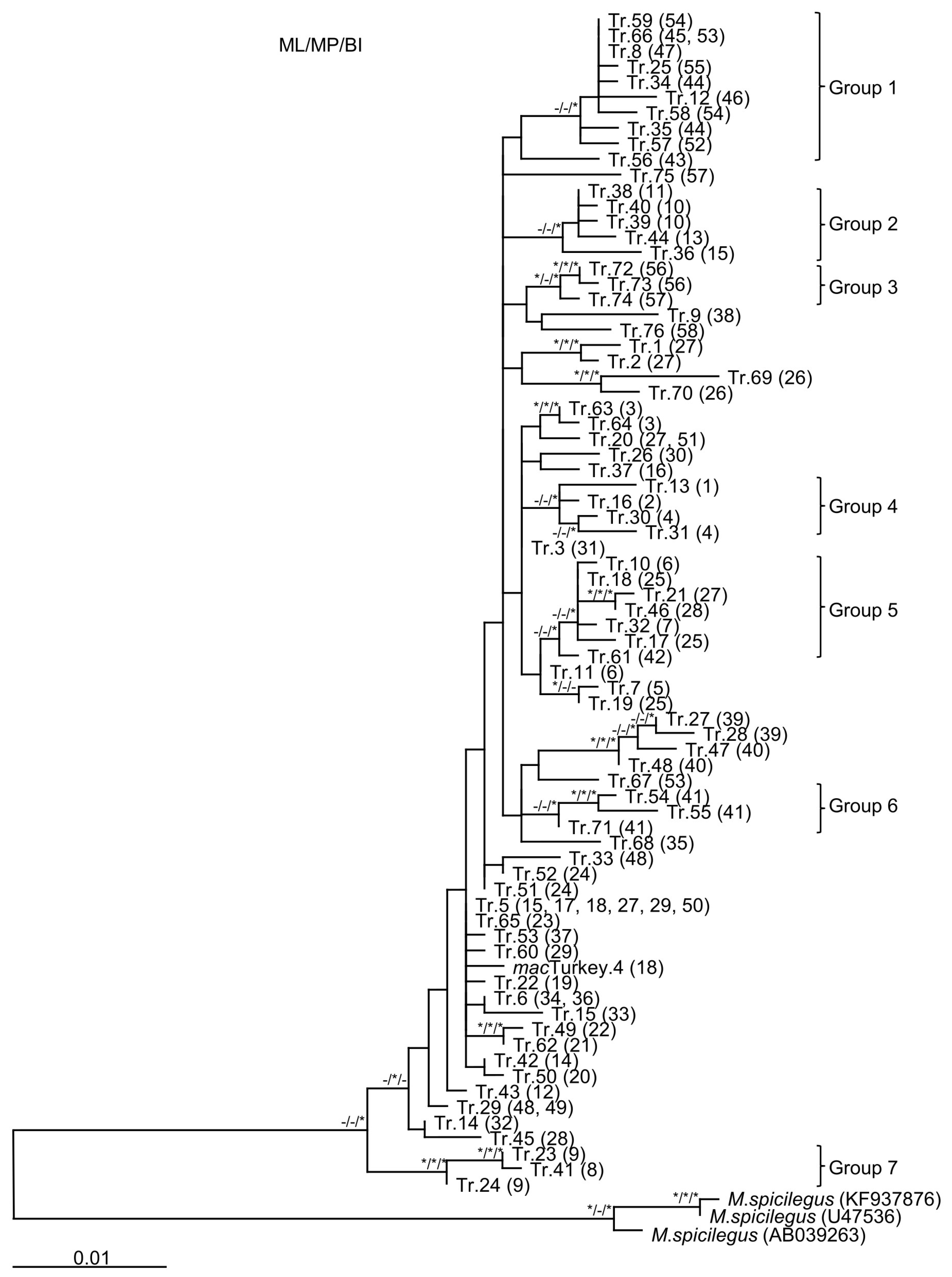
3.2. Phylogenetic Analysis of the Turkish Data
3.3. Demographic History: Genetic Diversity Within Turkey and Tests of Neutrality
3.4. Phylogenetic Analysis of D-Loop Across the Species Range
3.5. Demography of Mus macedonicus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BI | Bayesian inference |
| bp | base pairs |
| BSP | Bayesian Skyline Plot |
| DDBJ | DNA Data Bank of Japan |
| GTR | general time reversible |
| IUCN | International Union for the Conservation of Nature |
| kya | kilo-years ago |
| LGM | Last Glacial Maximum |
| MCMC | Markov chain Monte Carlo |
| MJ | Median-Joining |
| ML | Maximum Likelihood |
| MP | Maximum Parsimony |
| OMÜ | Ondokuz Mayis University |
| TBR | tree bisection and reconnection |
References
- Avise, J.C. Phylogeography. The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Hewitt, G.M. Post-glacial re-colonization of European biota. Biol. J. Linn. Soc. 1999, 68, 87–112. [Google Scholar] [CrossRef]
- Weiss, S.; Ferrand, N. Phylogeography of Southern European Refugia; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Bilgin, R.; Gürün, K.; Rebelo, H.; Puechmaille, S.J.; Maracı, Ö.; Presetnik, P.; Benda, P.; Hulva, P.; Ibáñez, C.; Hamidovic, D.; et al. Circum-Mediterranean phylogeography of a bat coupled with past environmental niche modeling: A new paradigm for the recolonization of Europe? Mol. Phylogenet. Evol. 2016, 99, 323–336. [Google Scholar] [CrossRef]
- Médail, F.; Diadema, K. Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J. Biogeogr. 2009, 36, 1333–1345. [Google Scholar] [CrossRef]
- Bilgin, R. Back to the suture: The distribution of intraspecific genetic diversity in and around Anatolia. Int. J. Mol. Sci. 2011, 12, 4080–4103. [Google Scholar] [CrossRef]
- Albayrak, T.; Tunçel, T.; Öğe, P.; Tietze, D.T.; Forcina, G. Anatolia: A hotspot of avian genetic diversity in the Western Palaearctic. Diversity 2024, 16, 339. [Google Scholar] [CrossRef]
- Seddon, J.M.; Santucci, F.; Reeve, N.; Hewitt, G.M. Caucasus Mountains divide postulated postglacial colonization routes in the white-breasted hedgehog, Erinaceus concolor. J. Evol. Biol. 2002, 15, 463–467. [Google Scholar] [CrossRef]
- Michaux, J.R.; Libois, R.; Paradis, E.; Filippucci, M.G. Phylogeographic history of the yellow-necked fieldmouse (Apodemus flavicollis) in Europe and in the Near and Middle East. Mol. Phylogenet. Evol. 2004, 32, 788–798. [Google Scholar] [CrossRef]
- Gündüz, İ.; Jaarola, M.; Tez, C.; Yeniyurt, C.; Polly, P.D.; Searle, J.B. Multigenic and morphometric differentiation of ground squirrels (Spermophilus, Scuiridae, Rodentia) in Turkey, with a description of a new species. Mol. Phylogenet. Evol. 2007, 43, 916–935. [Google Scholar] [CrossRef]
- Dubey, S.; Cosson, J.-F.; Vohralík, V.; Kryštufek, B.; Diker, E.; Vogel, P. Molecular evidence of Pleistocene bidirectional faunal exchange between Europe and the Near East: The case of the bicoloured shrew (Crocidura leucodon, Soricidae). J. Evol. Biol. 2007, 20, 1799–1808. [Google Scholar] [CrossRef]
- Demirbaş, Y.; Albayrak, İ.; Koca, A.Ö.; Stefanović, M.; Knauer, F.; Suchentrunk, F. Spatial genetics of brown hares (Lepus europaeus Pallas, 1778) from Turkey: Different gene pool architecture on either side of the Bosphorus? Mamm. Biol. 2019, 94, 77–85. [Google Scholar] [CrossRef]
- Arslan, Y.; Demïrtaş, S.; Herman, J.S.; Pustilnik, J.D.; Searle, J.B.; Gündüz, İ. The Anatolian glacial refugium and human-mediated colonization: A phylogeographical study of the stone marten (Martes foina) in Turkey. Biol. J. Linn. Soc. 2020, 129, 470–491. [Google Scholar] [CrossRef]
- Özmen, M.; Demirtaş, S.; Herman, J.S.; Gündüz, İ. Mitochondrial genetic variation in long-eared hedgehogs, Hemiechinus auritus, from the Anatolian Peninsula and Cyprus. Mammalia 2024, 88, 52–62. [Google Scholar] [CrossRef]
- Macholán, M. Mus macedonicus Petrov and Ružić, 1983. In The Atlas of European Mammals; Mitchell-Jones, A.J., Amori, G., Bogdanowicz, W., Kryštufek, B., Reijnders, P.J.H., Spitzenberger, F., Stubbe, M., Thissen, J.B.M., Vohralík, V., Zima, J., Eds.; Poyser: London, UK, 1999; pp. 284–285. [Google Scholar]
- Prager, E.M.; Tichy, H.; Sage, R.D. Mitochondrial DNA sequence variation in the eastern house mouse, Mus musculus: Comparison with other house mice and report of a 75-bp repeat. Genetics 1996, 143, 427–446. [Google Scholar] [CrossRef]
- Prager, E.M.; Orrego, C.; Sage, R.D. Genetic variation and phylogeography of central Asian and other house mice, including a major new mitochondrial lineage in Yemen. Genetics 1998, 150, 835–861. [Google Scholar] [CrossRef]
- Gündüz, İ.; Tez, C.; Malikov, V.; Vaziri, A.; Polyakov, A.V.; Searle, J.B. Mitochondrial DNA and chromosomal studies of wild mice (Mus) from Turkey and Iran. Heredity 2000, 84, 458–467. [Google Scholar] [CrossRef]
- Orth, A.; Auffray, J.-C.; Bonhomme, F. Two deeply divergent mitochondrial lineages in the wild mouse Mus macedonicus reveal multiple glacial refuges south of Caucasus. Heredity 2002, 89, 353–357. [Google Scholar] [CrossRef]
- Macholán, M.; Vyskocilová, M.; Bonhomme, F.; Krystufek, B.; Orth, A.; Vohralík, V. Genetic variation and phylogeography of free-living mouse species (genus Mus) in the Balkans and the Middle East. Mol. Ecol. 2007, 16, 4774–4788. [Google Scholar] [CrossRef]
- Rajabi-Maham, H.; Azizi, V. Phylogeographical study of short-tailed mouse species (Mus macedonicus Petrov & Ruzic, 1983) in North West of Iran. J. Anim. Res. 2013, 26, 289–297. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Piálek, J.; Ďureje, Ľ.; Hiadlovská, Z.; Kreisinger, J.; Aghová, T.; Bryjová, A.; Čížková, D.; Goüy de Bellocq, J.; Hejlová, H.; Janotová, K.; et al. Phenogenomic resources immortalized in a panel of wild-derived strains of five species of house mice. Sci. Rep. 2025, 15, 12060. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods); Version 4; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2003, 17, 754–755. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Crandall, K.A. Intraspecific gene genealogies: Trees grafting into networks. Trends Ecol. Evol. 2001, 16, 37–45. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X. Statistical tests of neutrality of mutations against population growth hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Strobeck, C. Average number of nucleotide differences in a sample from a single subpopulation: A test for population subdivision. Genetics 1987, 117, 149–153. [Google Scholar] [CrossRef]
- Ramos-Onsins, S.E.; Rozas, J. Statistical properties of new neutrality test against population growth. Mol. Biol. Evol. 2002, 19, 2092–2100. [Google Scholar] [CrossRef]
- Rogers, A.R.; Harpending, H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar] [CrossRef]
- Rogers, A.R. Genetic evidence for a Pleistocene population explosion. Evolution 1995, 49, 608–615. [Google Scholar] [CrossRef]
- Harpending, R.C. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum. Biol. 1994, 66, 591–600. [Google Scholar]
- Schneider, S.; Excoffier, L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: Application to human mitochondrial DNA. Genetics 1999, 152, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef] [PubMed]
- García-Rodríguez, O.; Andreou, D.; Herman, J.S.; Mitsainas, G.P.; Searle, J.B.; Bonhomme, F.; Hadjisterkotis, E.; Schutkowski, H.; Stafford, R.; Stewart, J.R.; et al. Cyprus as an ancient hub for house mice and humans. J. Biogeogr. 2018, 45, 2618–2630. [Google Scholar] [CrossRef]
- Rajabi-Maham, H.; Orth, A.; Bonhomme, F. Phylogeography and postglacial expansion of Mus musculus domesticus inferred from mitochondrial DNA coalescent, from Iran to Europe. Mol. Ecol. 2008, 17, 627–641. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A.; Shapiro, B.; Pybus, O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005, 22, 1185–1192. [Google Scholar] [CrossRef]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Auffray, J.-C.; Vanlerberghe, F.; Britton-Davidian, J. The house mouse progression in Eurasia: A palaeontological and archaeozoological approach. Biol. J. Linn. Soc. 1990, 41, 13–25. [Google Scholar] [CrossRef]
- Demirtaş, S.; Gündüz, N.E.; Bilton, D.T. Ecological niche modeling of the Macedonian mouse, Mus macedonicus (Mammalia, Rodentia), under climate change conditions. Israel J. Ecol. Evol. 2022, 69, 28–36. [Google Scholar] [CrossRef]
- Turkozan, O.; Karacaoğlu, Ç.; Parham, J.F. Reconstructions of the past distribution of Testudo graeca mitochondrial lineages in the Middle East and Transcaucasia support multiple refugia since the Last Glacial Maximum. Herpetol. J. 2021, 31, 10–17. [Google Scholar] [CrossRef]
- Şenkul, Ç.; Doğan, U. Vegetation and climate of Anatolia and adjacent regions during the Last Glacial period. Quat. Int. 2013, 302, 110–122. [Google Scholar] [CrossRef]
- Marciniak, A.; Czerniak, L. Social transformations in the Late Neolithic and the Early Chalcolithic periods in central Anatolia. Anatolian Stud. 2007, 57, 115–130. [Google Scholar] [CrossRef]
- Rowan, Y.M.; Golden, J. The Chalcolithic period of the Southern Levant: A synthetic review. J. World Prehist. 2009, 22, 1–92. [Google Scholar] [CrossRef]
- Kotlík, P.; Marková, S.; Horníková, M.; Escalante, M.A.; Searle, J.B. The bank vole (Clethrionomys glareolus) as a model system for adaptive phylogeography in the European theater. Front. Ecol. Evol. 2022, 10, 866605. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gündüz, İ.; Özçam, P.; Demirtaş, S.; Herman, J.S.; Searle, J.B. Mitochondrial Phylogeography and Population History of the Balkan Short-Tailed Mouse (Mus macedonicus Petrov and Ružić, 1983) in Turkey and Surrounding Areas. Diversity 2025, 17, 740. https://doi.org/10.3390/d17110740
Gündüz İ, Özçam P, Demirtaş S, Herman JS, Searle JB. Mitochondrial Phylogeography and Population History of the Balkan Short-Tailed Mouse (Mus macedonicus Petrov and Ružić, 1983) in Turkey and Surrounding Areas. Diversity. 2025; 17(11):740. https://doi.org/10.3390/d17110740
Chicago/Turabian StyleGündüz, İslam, Pınar Özçam, Sadık Demirtaş, Jeremy S. Herman, and Jeremy B. Searle. 2025. "Mitochondrial Phylogeography and Population History of the Balkan Short-Tailed Mouse (Mus macedonicus Petrov and Ružić, 1983) in Turkey and Surrounding Areas" Diversity 17, no. 11: 740. https://doi.org/10.3390/d17110740
APA StyleGündüz, İ., Özçam, P., Demirtaş, S., Herman, J. S., & Searle, J. B. (2025). Mitochondrial Phylogeography and Population History of the Balkan Short-Tailed Mouse (Mus macedonicus Petrov and Ružić, 1983) in Turkey and Surrounding Areas. Diversity, 17(11), 740. https://doi.org/10.3390/d17110740






