Rhizosphere Bacterial Diversity and Community Structure of Kobresia humilis in the Alpine Meadow of Eastern Qinghai–Tibetan Plateau and Its Response to Environmental Variables
Abstract
1. Introduction
2. Materials and Methods
2.1. Root and Rhizosphere Soil Samples
2.2. Soil Physicochemical Properties
2.3. DNA Extraction and Polymerase Chain Reaction (PCR) Amplification
2.4. Illumina MiSeq Sequencing and Sequence Analysis
2.5. Statistical Analysis
3. Results
3.1. Bacterial Community Sequencing Results and OTU Classification
3.2. Bacterial Diversity and Community Composition of K. humilis in Alpine Meadows on the QTP
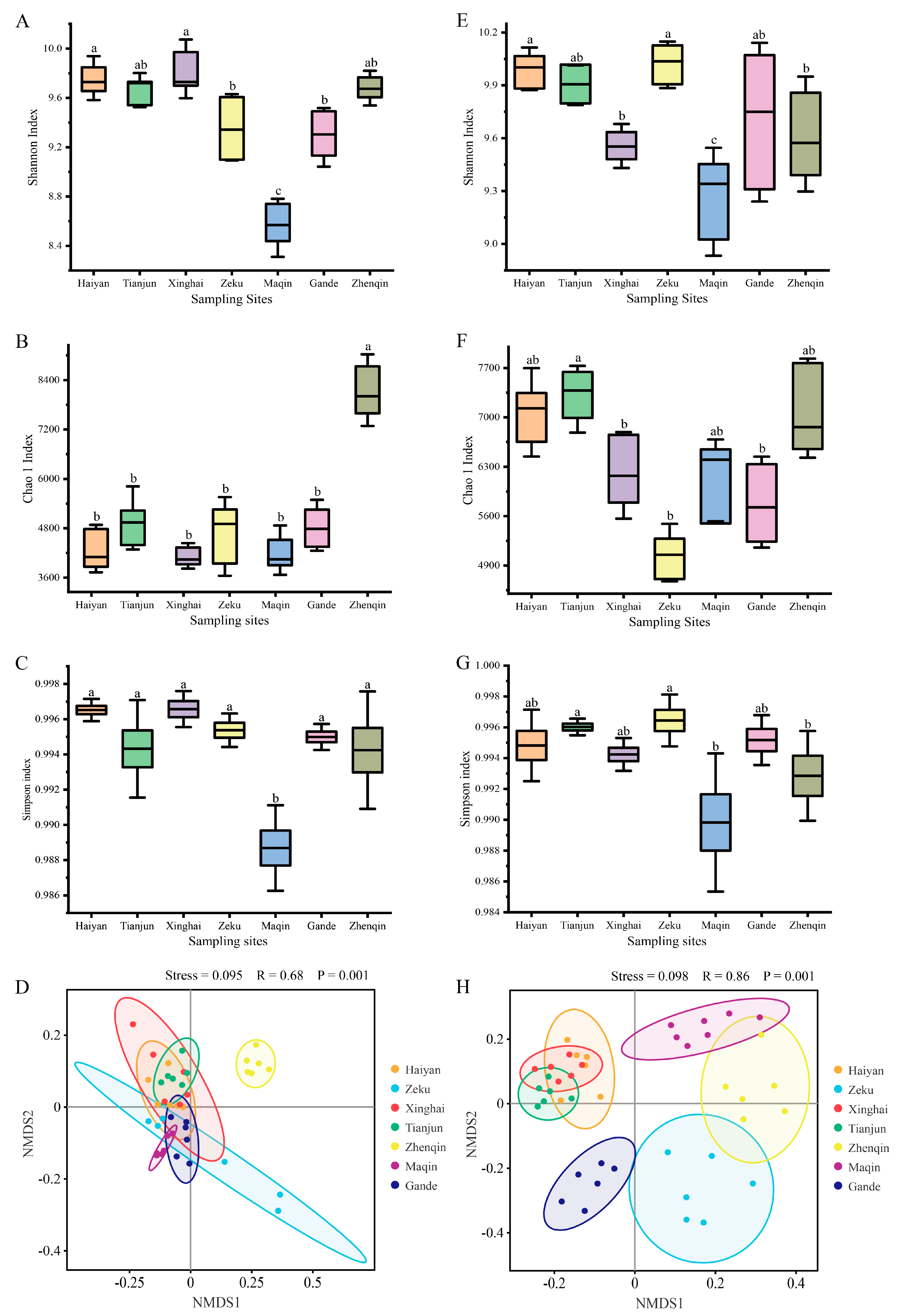
3.2.1. Alpha- and Beta-Diversity of K. humilis Communities
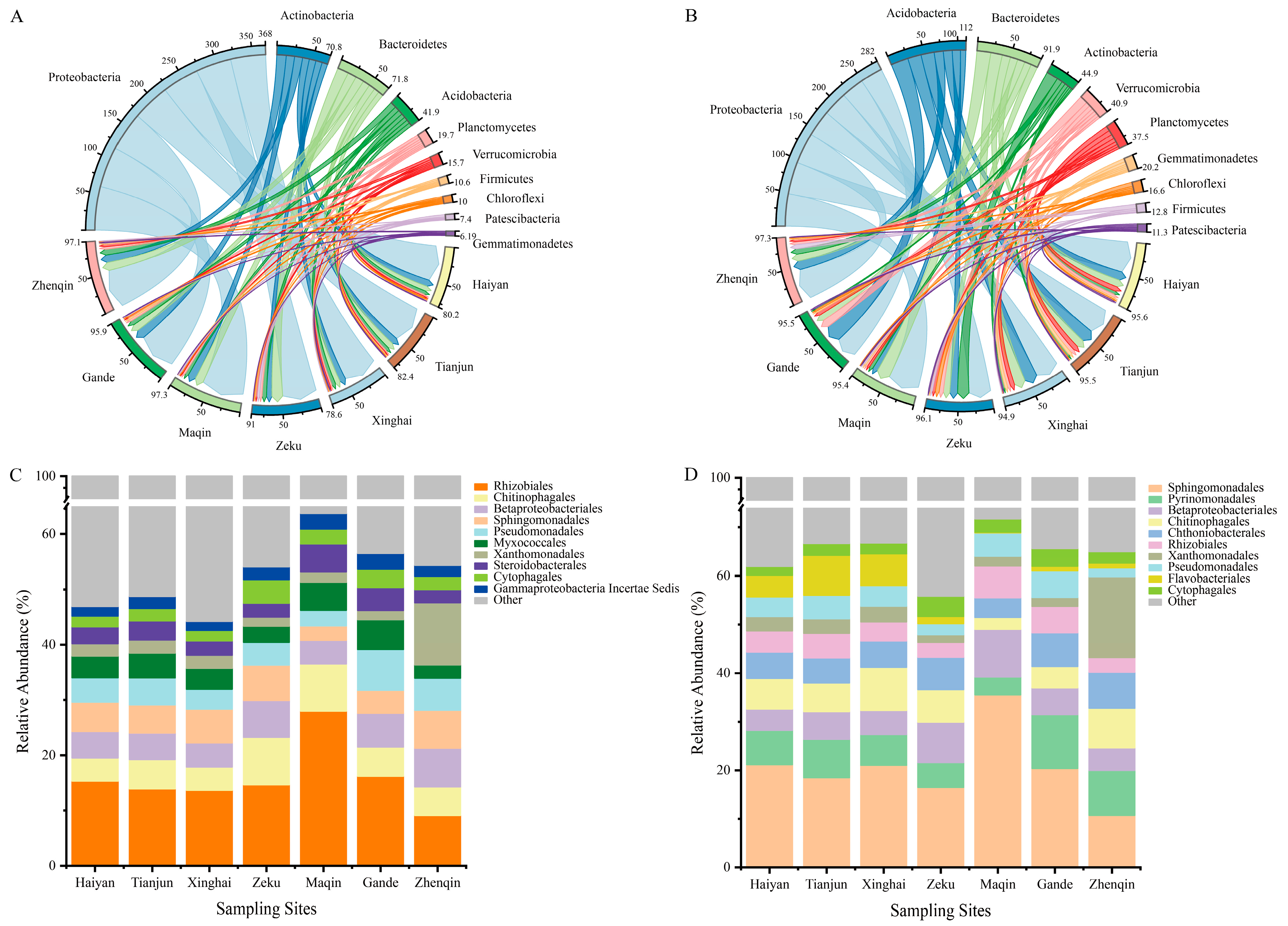
3.2.2. Changes in the Bacterial Community Composition of K. humilis
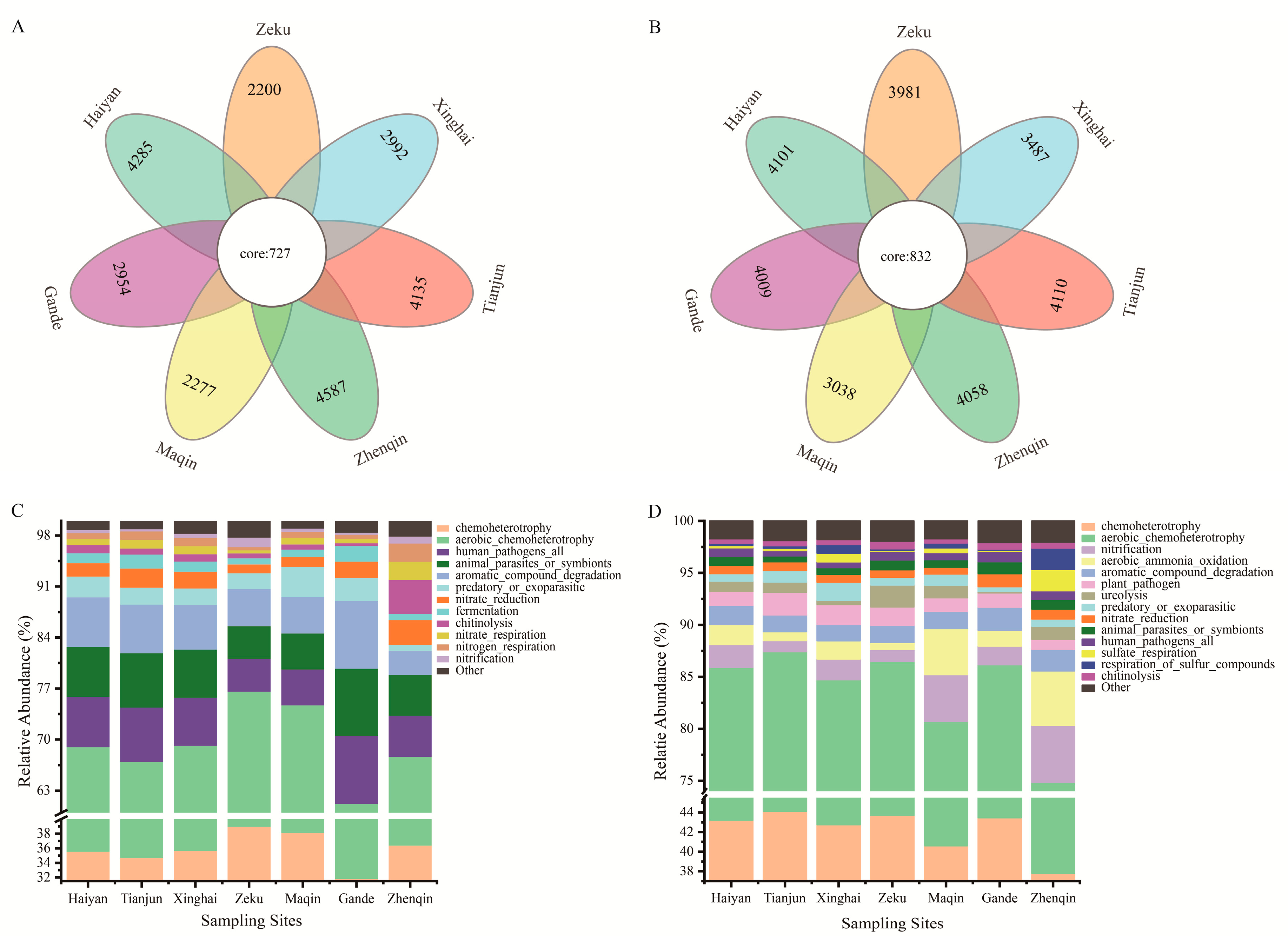
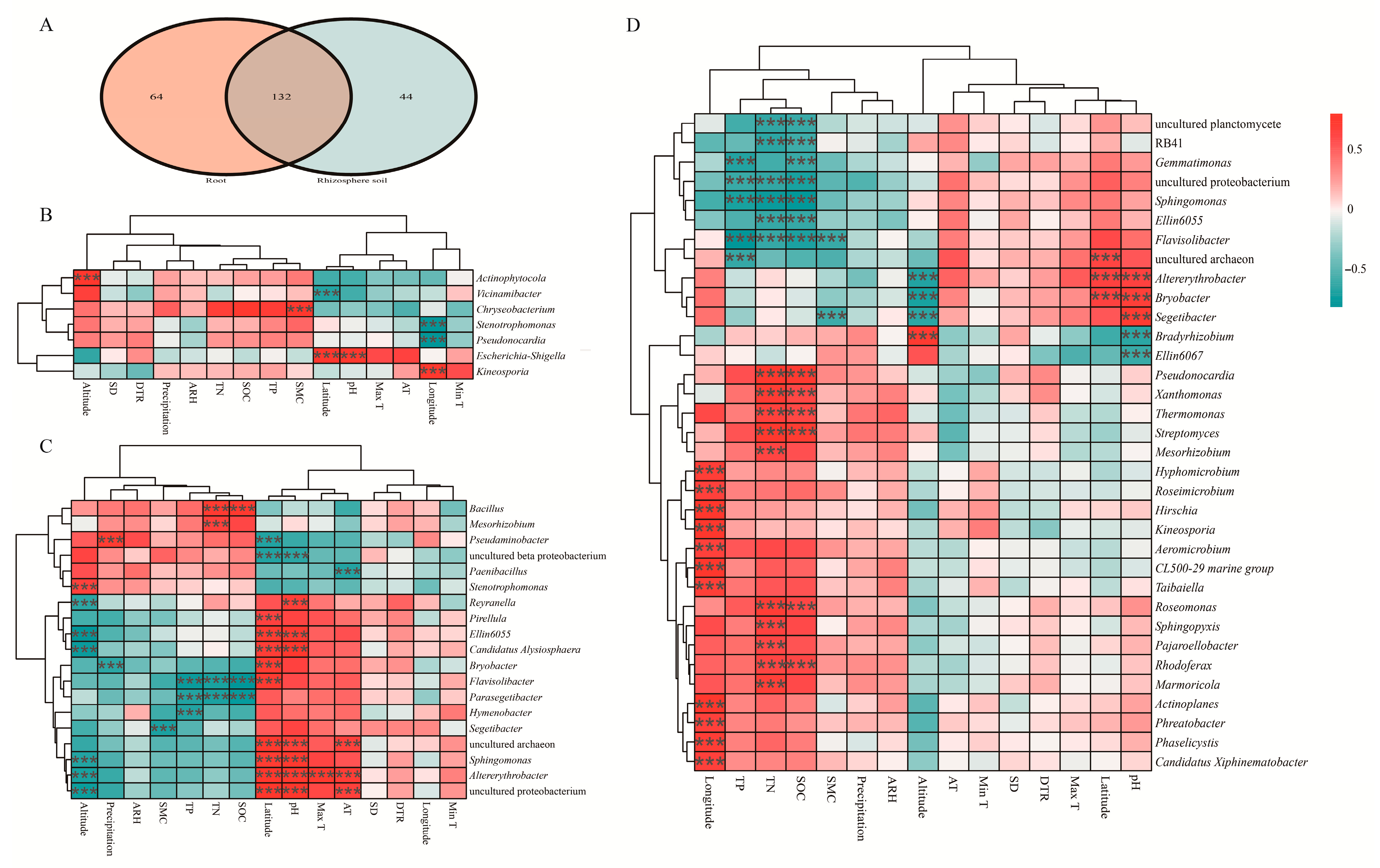
3.2.3. Core Bacterial Community Composition and Functional Prediction
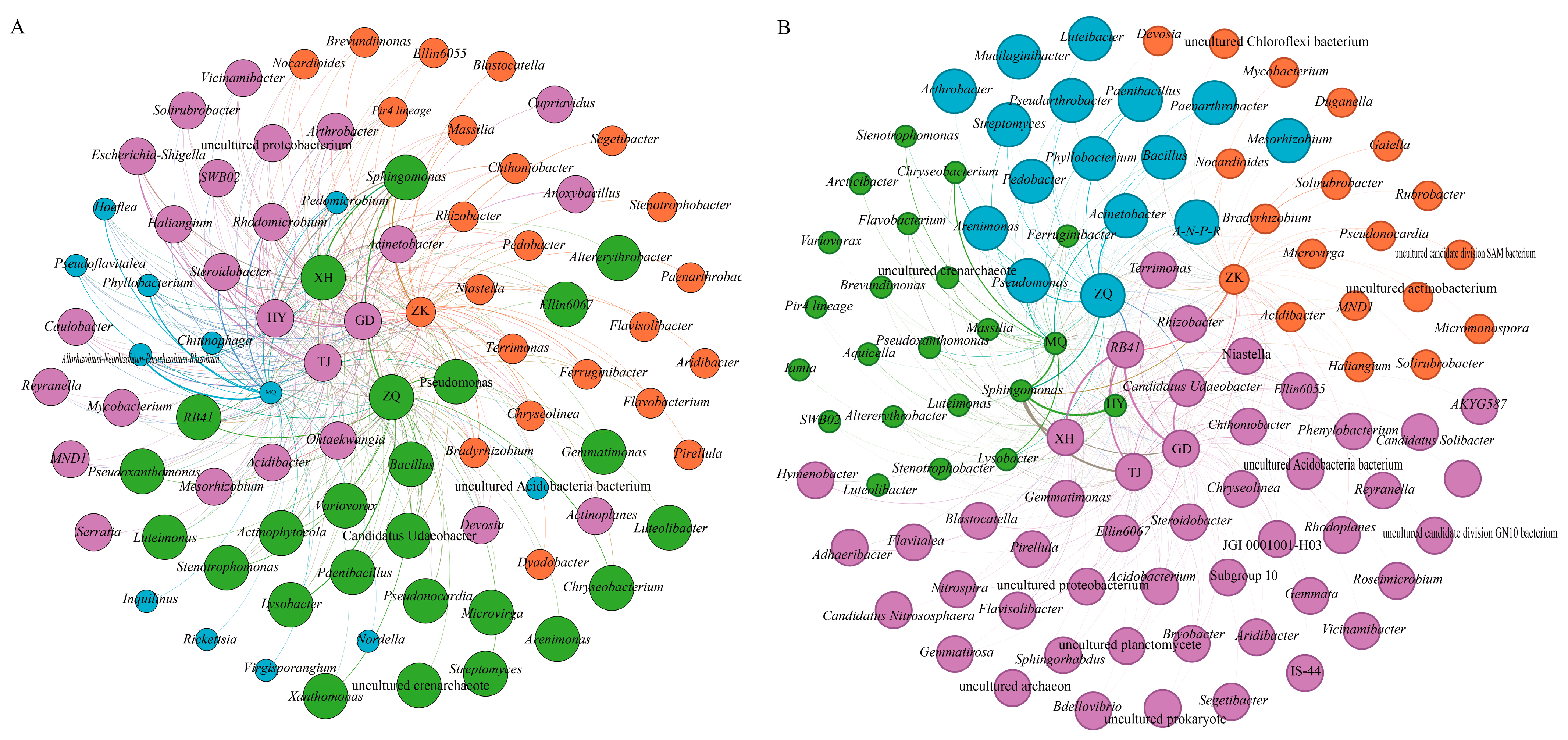
3.2.4. Co-Occurrence Network of Bacterial Communities
3.3. Correlation Between Bacteria Microbiota and Environmental Variables
3.3.1. Differences in the Soil Physicochemical Properties of Seven Alpine Meadow
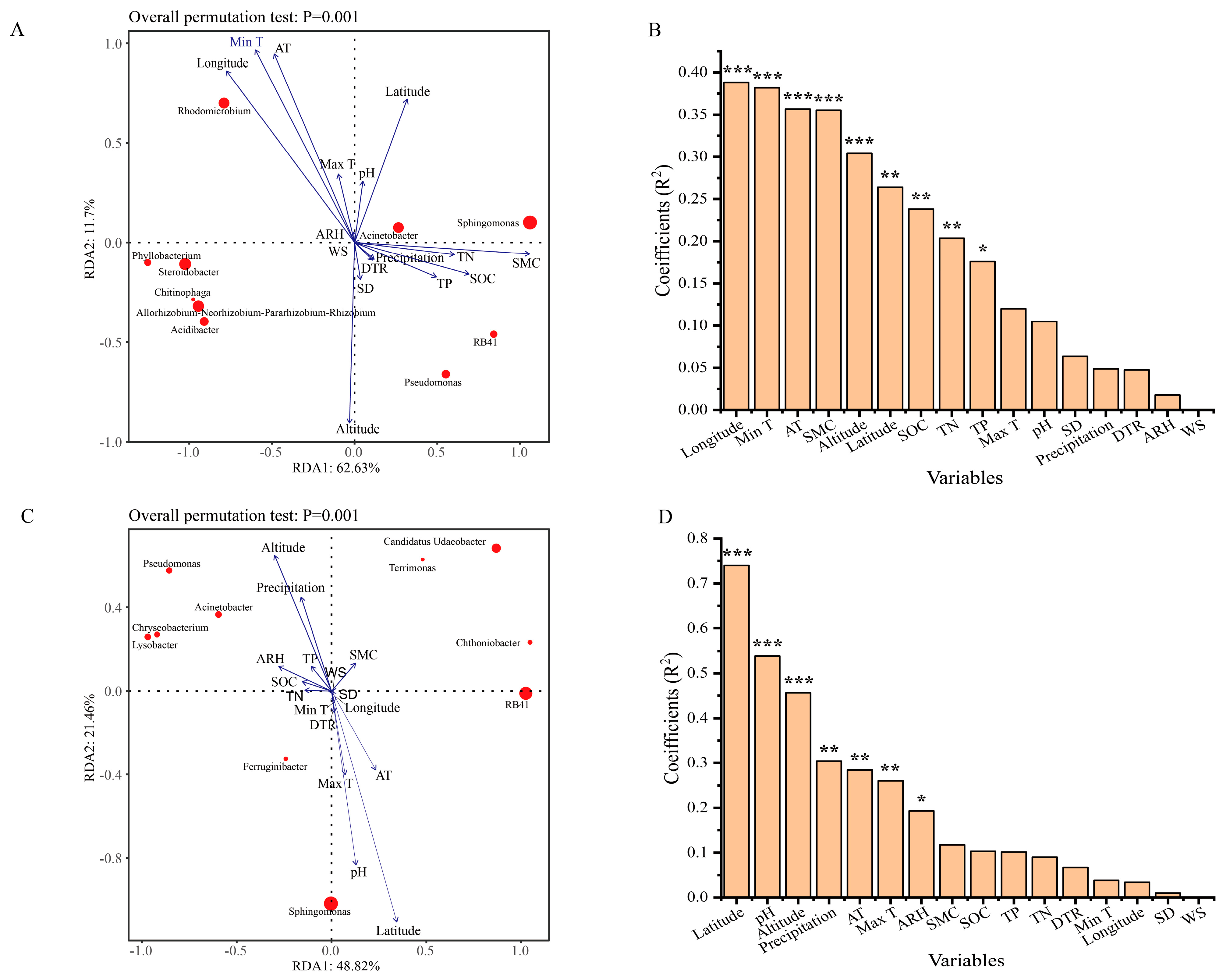
3.3.2. Redundancy Analysis of the Abundant Bacterial Genera and Environmental Variables
3.3.3. Correlation Analysis of the Core Bacteria Microbiota and Environmental Variables
4. Discussion
4.1. Abundant Potential Bacterial Resources Were Conserved in the Rhizosphere of K. humilis on the QTP
4.2. Geographic Differentiation of Bacterial Community Associated with K. humilis on the QTP
4.3. Core Bacteria Associated with K. humilis Kept Stable in Variable Environments on the QTP
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.F.; Lv, W.W.; Xue, K.; Wang, S.P.; Zhang, L.R.; Hu, R.H.; Zeng, H.; Xu, X.L.; Li, Y.M.; Jiang, L.L.; et al. Grassland changes and adaptive management on the Qinghai–Tibetan Plateau. Nat. Rev. Earth Environ. 2022, 3, 668–683. [Google Scholar] [CrossRef]
- Hu, J.P.; Zhang, M.X.; Lü, Z.L.; He, Y.Y.; Yang, X.X.; Khan, A.; Xiong, Y.C.; Fang, X.L.; Dong, Q.M.; Zhang, J.L. Grazing practices affect phyllosphere and rhizosphere bacterial communities of Kobresia humilis by altering their network stability. Sci. Total Environ. 2023, 900, 165814. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.M.; Wei, K.T.; Ma, K.K.; Xu, C.L.; Yu, X.J. Rest grazing from the critical period of soil thawing promotes the propagation of Kobresia humilis in alpine meadow. Ecol. Eng. 2022, 179, 106634. [Google Scholar] [CrossRef]
- Li, J.X.; Wu, L.L.; Zhou, Y.Z.; Xie, Y.L.; Lu, F.W.; Chang, F.F.; Yang, X.; Han, X.Z.; Cheng, M.X. Kobresia humilis via root-released flavonoids recruit Bacillus for promoted growth. Microbiol. Res. 2024, 287, 127866. [Google Scholar] [CrossRef]
- Wang, S.R.; Jia, C.C.; Zhao, D.Y.; Zeng, J.; Xing, P.; Liu, Y.Q.; Wu, Q.L. Disentangling the assembly mechanisms of bacterial communities in a transition zone between the alpine steppe and alpine meadow ecosystems on the Tibetan Plateau. Sci. Total Environ. 2022, 847, 157446. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.T.; Liu, G.B.; Zhang, C.; Wang, G.L. Bacterial richness is negatively related to potential soil multifunctionality in a degraded alpine meadow. Ecol. Indic. 2021, 121, 106996. [Google Scholar] [CrossRef]
- Li, E.Q.; de Jonge, R.; Liu, C.; Jiang, H.N.; Friman, V.-P.; Pieterse, C.M.; Bakker, P.A.; Jousset, A. Rapid evolution of bacterial mutualism in the plant rhizosphere. Nat. Commun. 2021, 12, 3829. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.J.; Long, C.L.; Lam, E. Roles of plant-associated microbiota in traditional herbal medicine. Trends Plant Sci. 2018, 23, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.M.; Luo, W.; Liu, S.; Chen, W.M.; Chen, C.; Jiao, S.; Wei, G.H. Soil potassium is correlated with root secondary metabolites and root-associated core bacteria in licorice of different ages. Plant Soil 2020, 456, 61–79. [Google Scholar] [CrossRef]
- He, M.J.; Sun, W.M.; Cui, S.L.; Mu, G.J.; Liu, L.F.; Guo, W. Analysis of microbial diversity and community structure of peanut pod and its surrounding soil in peanut rot epidemic area. Curr. Microbiol. 2021, 78, 2173–2182. [Google Scholar] [CrossRef]
- Rivas, G.A.; Semorile, L.; Delfederico, L. Microbial diversity of the soil, rhizosphere and wine from an emerging wine-producing region of Argentina. LWT 2022, 153, 112429. [Google Scholar] [CrossRef]
- Wang, C.Y.; Wang, J.B.; Zhang, F.W.; Yang, Y.S.; Luo, F.L.; Li, Y.N.; Li, J.X. Stability response of alpine meadow communities to temperature and precipitation changes on the Northern Tibetan Plateau. Ecol. Evol. 2022, 12, e8592. [Google Scholar] [CrossRef]
- Ling, N.; Wang, T.T.; Kuzyakov, Y. Rhizosphere bacteriome structure and functions. Nat. Commun. 2022, 13, 836. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Tan, T.Y.; Jiang, S.J. Divergent responses of plant and soil microbial community to short-term nutrient addition in alpine grassland on the Qinghai-Tibetan Plateau. Front. Ecol. Evol. 2022, 10, 1056111. [Google Scholar] [CrossRef]
- Yuan, X.; Chen, Y.; Qin, W.K.; Xu, T.L.; Mao, Y.H.; Wang, Q.; Chen, K.L.; Zhu, B. Plant and microbial regulations of soil carbon dynamics under warming in two alpine swamp meadow ecosystems on the Tibetan Plateau. Sci. Total Environ. 2021, 790, 148072. [Google Scholar] [CrossRef]
- Guo, J.; Xie, Z.L.; Meng, Q.; Xu, H.Y.; Peng, Q.Q.; Wang, B.; Dong, D.Y.; Yang, J.B.; Jia, S.B. Distribution of rhizosphere fungi of Kobresia humilis on the Qinghai-Tibet Plateau. PeerJ 2024, 12, e16620. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.; Fernandes, T.; Martins, F.; Pereira, J.A.; Tavares, R.M.; Santos, P.M.; Baptista, P.; Lino-Neto, T. Illuminating Olea europaea L. endophyte fungal community. Microbiol. Res. 2021, 245, 126693. [Google Scholar] [CrossRef] [PubMed]
- Cota-Sánchez, J.H.; Remarchuk, K.; Ubayasena, K. Ready-to-use DNA extracted with a CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant Mol. Biol. Rep. 2006, 24, 161–167. [Google Scholar] [CrossRef]
- Hussain, M.; Xuan, P.X.; Xin, Y.; Ma, H.K.; Zhou, Y.H.; Wen, S.H.; Wan, T.Y.; Hu, J.Y.; Li, Y.Z.; Kang, S.; et al. Redundancy in microbiota-mediated suppression of the soybean cyst nematode. Microbiome 2024, 12, 125. [Google Scholar] [CrossRef]
- Zancarini, A.; Le Signor, C.; Terrat, S.; Aubert, J.; Salon, C.; Munier-Jolain, N.; Mougel, C. Medicago truncatula genotype drives the plant nutritional strategy and its associated rhizosphere bacterial communities. New Phytol. 2025, 245, 767–784. [Google Scholar] [CrossRef]
- Sansupa, C.; Wahdan, S.F.M.; Hossen, S.; Disayathanoowat, T.; Wubet, T.; Purahong, W. Can we use functional annotation of prokaryotic taxa (FAPROTAX) to assign the ecological functions of soil bacteria? Appl. Sci. 2021, 11, 688. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, D.G.; Jiang, Z.H.; Sun, P.; Xiao, H.L.; Wu, Y.X.; Chen, J.G. Changes in the soil microbial communities of alpine steppe at Qinghai-Tibetan Plateau under different degradation levels. Sci. Total Environ. 2019, 651, 2281–2291. [Google Scholar] [CrossRef]
- Zhao, Q.; Xie, F.X.; Zhang, F.F.; Zhou, K.; Sun, H.B.; Zhao, Y.J.; Yang, Q. Analysis of bacterial community functional diversity in late-stage shrimp (Litopenaeus vannamei) ponds using Biolog EcoPlates and PICRUSt2. Aquaculture 2022, 546, 737288. [Google Scholar] [CrossRef]
- Cong, J.; Cong, W.; Lu, H.; Zhang, Y.G. Distinct elevational patterns and their linkages of soil bacteria and plant community in an alpine meadow of the Qinghai–Tibetan plateau. Microorganisms 2022, 10, 1049. [Google Scholar] [CrossRef]
- An, Z.G.; Guo, F.X.; Chen, Y.; Bai, G.; Chen, Z.J. Rhizosphere bacterial and fungal communities during the growth of Angelica sinensis seedlings cultivated in an Alpine uncultivated meadow soil. PeerJ 2020, 8, e8541. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, M.; Hagedorn, F.; Wipf, S.; Donhauser, J.; Vittoz, P.; Rixen, C.; Frossard, A.; Theurillat, J.-P.; Frey, B. The Soil microbiome of GLORIA mountain summits in the Swiss Alps. Front. Microbiol. 2019, 10, 1080. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.J.; Lü, Y.H.; Hu, J.; Yin, L.C. Geodiversity underpins biodiversity but the relations can be complex: Implications from two biodiversity proxies. Glob. Ecol. Conserv. 2021, 31, e01830. [Google Scholar] [CrossRef]
- Zhao, Y.N.; Mou, X.M.; Wei, M.; Li, X.G. Effect of vegetation mosaic on spatial heterogeneity of soil organic carbon mineralization and nitrification in an alpine meadow. Appl. Soil Ecol. 2021, 165, 104007. [Google Scholar] [CrossRef]
- Zhao, B.H.; Jiao, C.C.; Wang, S.R.; Zhao, D.Y.; Jiang, C.L.; Zeng, J.; Wu, Q.L. Contrasting assembly mechanisms explain the biogeographic patterns of benthic bacterial and fungal communities on the Tibetan Plateau. Environ. Res. 2022, 214, 113836. [Google Scholar] [CrossRef]
- Pervaiz, Z.H.; Contreras, J.; Hupp, B.M.; Lindenberger, J.H.; Chen, D.M.; Zhang, Q.M.; Wang, C.H.; Twigg, P.; Saleem, M. Root microbiome changes with root branching order and root chemistry in peach rhizosphere soil. Rhizosphere 2020, 16, 100249. [Google Scholar] [CrossRef]
- Hussain, M.; Heuer, H. Rice diterpenoids rewire microbiome to fight plant-parasitic nematodes. Trends Parasitol. 2025, 41, 421–423. [Google Scholar] [CrossRef]
- Wang, B.X.; Sugiyama, S. Phylogenetic signal of host plants in the bacterial and fungal root microbiomes of cultivated angiosperms. Plant J. 2020, 104, 522–531. [Google Scholar] [CrossRef]
- Ren, C.J.; Zhang, X.Y.; Zhang, S.H.; Wang, J.Y.; Xu, M.P.; Guo, Y.X.; Wang, J.; Han, X.H.; Zhao, F.Z.; Yang, G.H.; et al. Altered microbial CAZyme families indicated dead biomass decomposition following afforestation. Soil Biol. Biochem. 2021, 160, 108362. [Google Scholar] [CrossRef]
- Kertesz, M.A.; Kawasaki, A.; Stolz, A. Aerobic hydrocarbon-degrading Alphaproteobacteria: Sphingomonadales. In Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes; McGenity, T., Ed.; Handbook of Hydrocarbon and Lipid Microbiology; Springer: Cham, Switzerland, 2018; pp. 105–124. [Google Scholar] [CrossRef]
- Balázs, H.E.; Schmid, C.A.O.; Cruzeiro, C.; Podar, D.; Szatmari, P.M.; Buegger, F.; Hufnagel, G.; Radl, V.; Schröder, P. Post-reclamation microbial diversity and functions in hexachlorocyclohexane (HCH) contaminated soil in relation to spontaneous HCH tolerant vegetation. Sci. Total Environ. 2021, 767, 144653. [Google Scholar] [CrossRef]
- Ge, W.; Zhang, Z.Y.; Dong, C.B.; Han, Y.F.; Deshmukh, S.K.; Liang, Z.Q. Bacterial community analysis and potential functions of core taxa in different parts of the fungus Cantharellus cibarius. Pol. J. Microbiol. 2021, 70, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.M.; Tang, D.X.; Li, Q.Q.; Wang, Y.B.; Xu, Z.H.; Li, W.J.; Yu, H. Complex microbial communities inhabiting natural Cordyceps militaris and the habitat soil and their predicted functions. Anton. Leeuw. Int. J. G. 2021, 114, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wei, P.F.; Banda, J.F.; Ma, L.Q.; Mao, W.A.; Li, H.Y.; Hao, C.B.; Dong, H.L. Succession of microbial communities in waste soils of an iron mine in eastern China. Microorganisms 2021, 9, 2463. [Google Scholar] [CrossRef]
- Wang, Y.C.; Lu, G.X.; Yu, H.; Du, X.F.; He, Q.; Yao, S.T.; Zhao, L.R.; Huang, C.X.; Wen, X.C.; Deng, Y. Meadow degradation increases spatial turnover rates of the fungal community through both niche selection and dispersal limitation. Sci. Total Environ. 2021, 798, 149362. [Google Scholar] [CrossRef]
- Miehe, G.; Schleuss, P.-M.; Seeber, E.; Babel, W.; Biermann, T.; Braendle, M.; Chen, F.; Coners, H.; Foken, T.; Gerken, T.; et al. The Kobresia pygmaea ecosystem of the Tibetan highlands-Origin, functioning and degradation of the world’s largest pastoral alpine ecosystem: Kobresia pastures of Tibet. Sci. Total Environ. 2019, 648, 754–771. [Google Scholar] [CrossRef]
- Wang, X.Y.; Li, W.; Xiao, Y.T.; Cheng, A.Q.; Shen, T.M.; Zhu, M.; Yu, L.J. Abundance and diversity of carbon-fixing bacterial communities in karst wetland soil ecosystems. Catena 2021, 204, 105418. [Google Scholar] [CrossRef]
- Zhang, W.H.; Wan, W.J.; Liu, X.N.; Yang, Y.Y.; Liu, M.X. Stronger geographic limitations shape a rapid turnover and potentially highly connected network of core bacteria on microplastics. Microb. Ecol. 2023, 85, 1179–1189. [Google Scholar] [CrossRef]
- Liang, S.C.; Deng, J.J.; Jiang, Y.; Wu, S.J.; Zhou, Y.B.; Zhu, W.X. Functional distribution of bacterial community under different land use patterns based on FaProTax function prediction. Pol. J. Environ. Stud. 2020, 29, 1245–1261. [Google Scholar] [CrossRef]
- Yu, X.M.; Yan, M.; Cui, Y.L.; Liu, Z.Y.; Liu, H.; Zhou, J.; Liu, J.H.; Zeng, L.; Chen, Q.; Gu, Y.F.; et al. Effects of co-application of cadmium-immobilizing bacteria and organic fertilizers on Houttuynia cordata and microbial communities in a cadmium-contaminated field. Front. Microbiol. 2022, 12, 809834. [Google Scholar] [CrossRef]
- Omoregie, A.I.; Palombo, E.A.; Nissom, P.M. Bioprecipitation of calcium carbonate mediated by ureolysis: A review. Environ. Eng. Res. 2021, 26, 200379. [Google Scholar] [CrossRef]
- Yang, Z.C.; Peng, C.Y.; Cao, H.M.; Song, J.J.; Gong, B.; Li, L.; Wang, L.; He, Y.; Liang, M.; Lin, J.C.; et al. Microbial functional assemblages predicted by the FAPROTAX analysis are impacted by physicochemical properties, but C, N and S cycling genes are not in mangrove soil in the Beibu Gulf, China. Ecol. Indic. 2022, 139, 108887. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Trego, A.C.; McAteer, P.G.; Nzeteu, C.; Mahony, T.; Abram, F.; Ijaz, U.Z.; O’Flaherty, V. Combined stochastic and deterministic processes drive community assembly of anaerobic microbiomes during granule flotation. Front. Microbiol. 2021, 12, 666584. [Google Scholar] [CrossRef]
- Yu, Y.Y.; Cui, M.; Xiao, Y.T.; Chang, M.Y.; Wang, C.; Zhao, L.; Cao, Y.H.; Miao, Y.; Chen, Z.J.; Han, S.J.; et al. Quantitative estimation of stochastic and deterministic processes for soil prokaryotic community assembly in the Yellow River floodplain. Eur. J. Soil Sci. 2021, 72, 1462–1477. [Google Scholar] [CrossRef]
- Mawarda, P.C.; Lakke, S.L.; van Elsas, J.D.; Salles, J.F. Temporal dynamics of the soil bacterial community following Bacillus invasion. iScience 2022, 25, 104185. [Google Scholar] [CrossRef]
- Richter-Heitmann, T.; Hofner, B.; Krah, F.S.; Sikorski, J.; Wust, P.K.; Bunk, B.; Huang, S.; Regan, K.M.; Berner, D.; Boeddinghaus, R.S.; et al. Stochastic dispersal rather than deterministic selection explains the spatio-temporal distribution of soil bacteria in a temperate grassland. Front. Microbiol. 2021, 11, 1391. [Google Scholar] [CrossRef]
- Wang, L.; Lu, W.L.; Song, Y.; Liu, S.J.; Fu, Y.V. Using machine learning to identify environmental factors that collectively determine microbial community structure of activated sludge. Environ. Res. 2024, 260, 119635. [Google Scholar] [CrossRef]
- Guo, Y.Q.; Luo, H.; Wang, L.; Xu, M.Y.; Wan, Y.S.; Chou, M.X.; Shi, P.; Wei, G.H. Multifunctionality and microbial communities in agricultural soils regulate the dynamics of a soil-borne pathogen. Plant Soil 2021, 461, 309–322. [Google Scholar] [CrossRef]
- Zhou, Y.N.; Meng, F.F.; Ochieng, B.; Xu, J.N.; Zhang, L.; Kimirei, I.S.; Feng, M.H.; Zhu, L.F.; Wang, J.J. Climate and environmental variables drive stream biofilm bacterial and fungal diversity on tropical mountainsides. Microb. Ecol. 2024, 87, 28. [Google Scholar] [CrossRef] [PubMed]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Stone, B.W.; Li, J.H.; Koch, B.J.; Blazewicz, S.J.; Dijkstra, P.; Hayer, M.; Hofmockel, K.S.; Liu, X.J.; Mau, R.L.; Morrissey, E.M.; et al. Nutrients cause consolidation of soil carbon flux to small proportion of bacterial community. Nat. Commun. 2021, 12, 3381. [Google Scholar] [CrossRef]
- Liang, D.; Xiao, C.Y.X.; Song, F.P.; Li, H.T.; Liu, R.M.; Gao, J.G. Complete genome sequence and function gene identify of prometryne-degrading strain Pseudomonas sp. DY-1. Microorganisms 2021, 9, 1261. [Google Scholar] [CrossRef]
- Peng, Z.; Xiao, H.; He, X.; Xu, C.L.; Pan, T.T.; Yu, X.J. Different levels of rainfall and trampling change the reproductive strategy of Kobresia humilis in the Qinghai-Tibet Plateau. Rangeland J. 2020, 42, 143–152. [Google Scholar] [CrossRef]
- Lin, L.; Li, Y.K.; Xu, X.L.; Zhang, F.W.; Du, Y.G.; Liu, S.L.; Guo, X.W.; Cao, G.M. Predicting parameters of degradation succession processes of Tibetan Kobresia grasslands. Solid Earth 2015, 6, 1237–1246. [Google Scholar] [CrossRef]

| Sampling Sites | Soil Moisture Content (%) | Soil pH | TN (g kg−1) | TP (g kg−1) | Organic Carbon (g kg−1) |
|---|---|---|---|---|---|
| Haiyan | 18.09 ± 0.07 b | 7.87 ± 0.05 b | 10.43 ± 1.20 a | 2.43 ± 0.16 a | 125.03 ± 9.28 a |
| Tianjun | 6.59 ± 0.24 d | 8.46 ± 0.02 a | 4.82 ± 0.045 c | 1.82 ± 0.05 c | 51.83 ± 0.94 c |
| Xinghai | 13.33 ± 0.57 c | 7.87 ± 0.05 b | 4.41 ± 0.12 c | 1.61 ± 0.03 d | 45.32 ± 1.03 c |
| Zeku | 13.41 ± 1.03 c | 7.51 ± 0.01 c | 6.07 ± 0.35 b | 1.89 ± 0.07 c | 75.73 ± 5.82 b |
| Maqin | 6.71 ± 0.29 d | 7.48 ± 0.14 c | 4.89 ± 0.47 c | 1.81 ± 0.19 c | 54.51 ± 2.10 c |
| Gande | 18.80 ± 0.44 b | 7.03 ± 0.09 e | 4.91 ± 0.17 c | 2.05 ± 0.05 b | 57.46 ± 4.76 c |
| Zhenqin | 21.16 ± 0.41 a | 7.21 ± 0.07 d | 9.90 ± 0.43 a | 2.38 ± 0.10 a | 129.57 ± 17.02 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Q.; Guo, J.; Yang, Z.; Hou, X.; Yang, Z.; Zhu, Z. Rhizosphere Bacterial Diversity and Community Structure of Kobresia humilis in the Alpine Meadow of Eastern Qinghai–Tibetan Plateau and Its Response to Environmental Variables. Diversity 2025, 17, 723. https://doi.org/10.3390/d17100723
Peng Q, Guo J, Yang Z, Hou X, Yang Z, Zhu Z. Rhizosphere Bacterial Diversity and Community Structure of Kobresia humilis in the Alpine Meadow of Eastern Qinghai–Tibetan Plateau and Its Response to Environmental Variables. Diversity. 2025; 17(10):723. https://doi.org/10.3390/d17100723
Chicago/Turabian StylePeng, Qingqing, Jing Guo, Zengzeng Yang, Xianbin Hou, Zhengzhou Yang, and Zhengjie Zhu. 2025. "Rhizosphere Bacterial Diversity and Community Structure of Kobresia humilis in the Alpine Meadow of Eastern Qinghai–Tibetan Plateau and Its Response to Environmental Variables" Diversity 17, no. 10: 723. https://doi.org/10.3390/d17100723
APA StylePeng, Q., Guo, J., Yang, Z., Hou, X., Yang, Z., & Zhu, Z. (2025). Rhizosphere Bacterial Diversity and Community Structure of Kobresia humilis in the Alpine Meadow of Eastern Qinghai–Tibetan Plateau and Its Response to Environmental Variables. Diversity, 17(10), 723. https://doi.org/10.3390/d17100723






