Genetic Monitoring of the Endangered Acipenser dabryanus Using a High-Resolution MNP System
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Restriction-Site Associated DNA (RAD) Sequencing
2.3. MNP Marker Screening and Primer Design in A. dabryanus
2.4. MNP System Workflow: From Library Construction to High-Throughput Sequencing for A. dabryanus
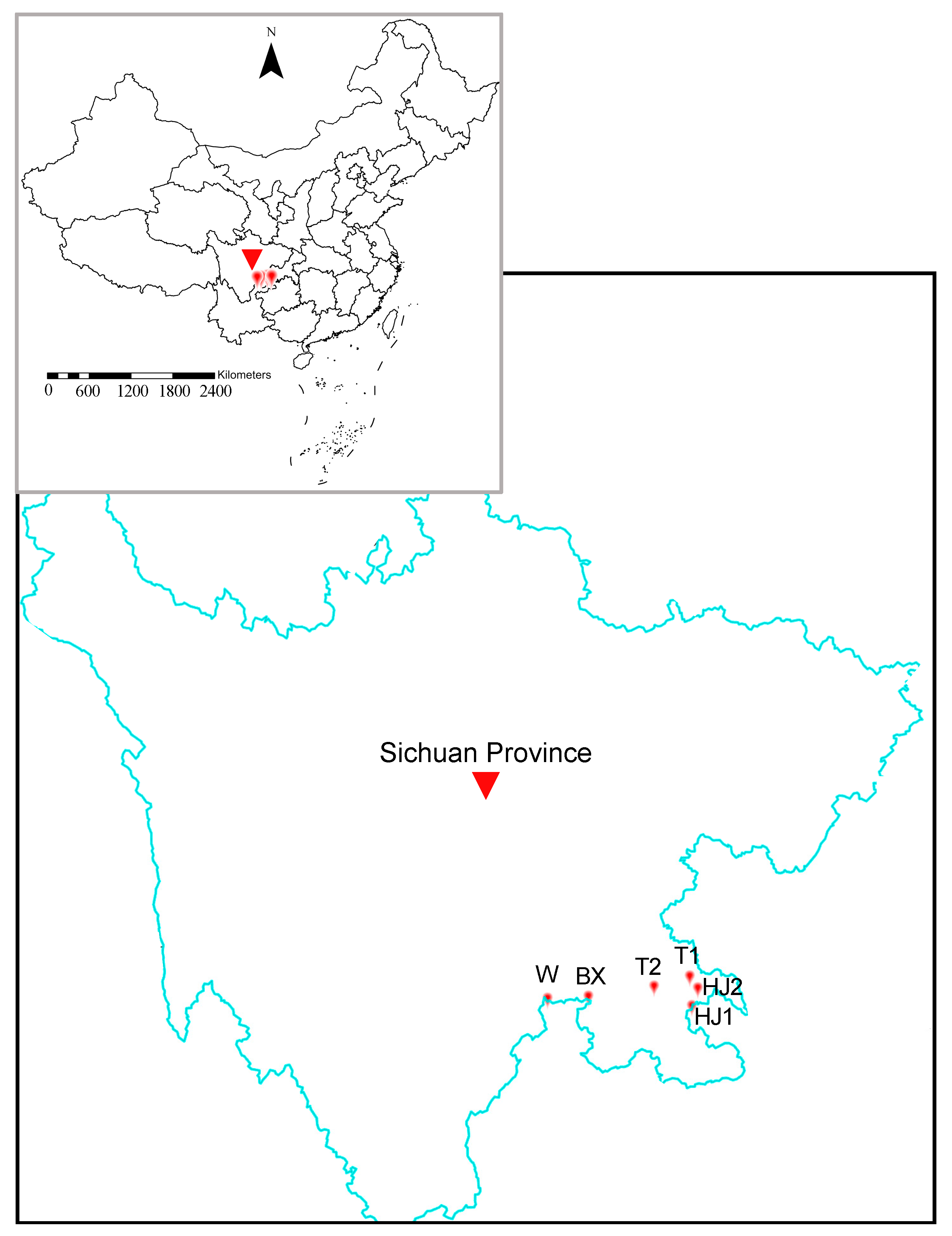
2.5. Validation and Application of the MNP System to Individual and Water Samples
2.6. Determination of Key Parameters of the MNP System
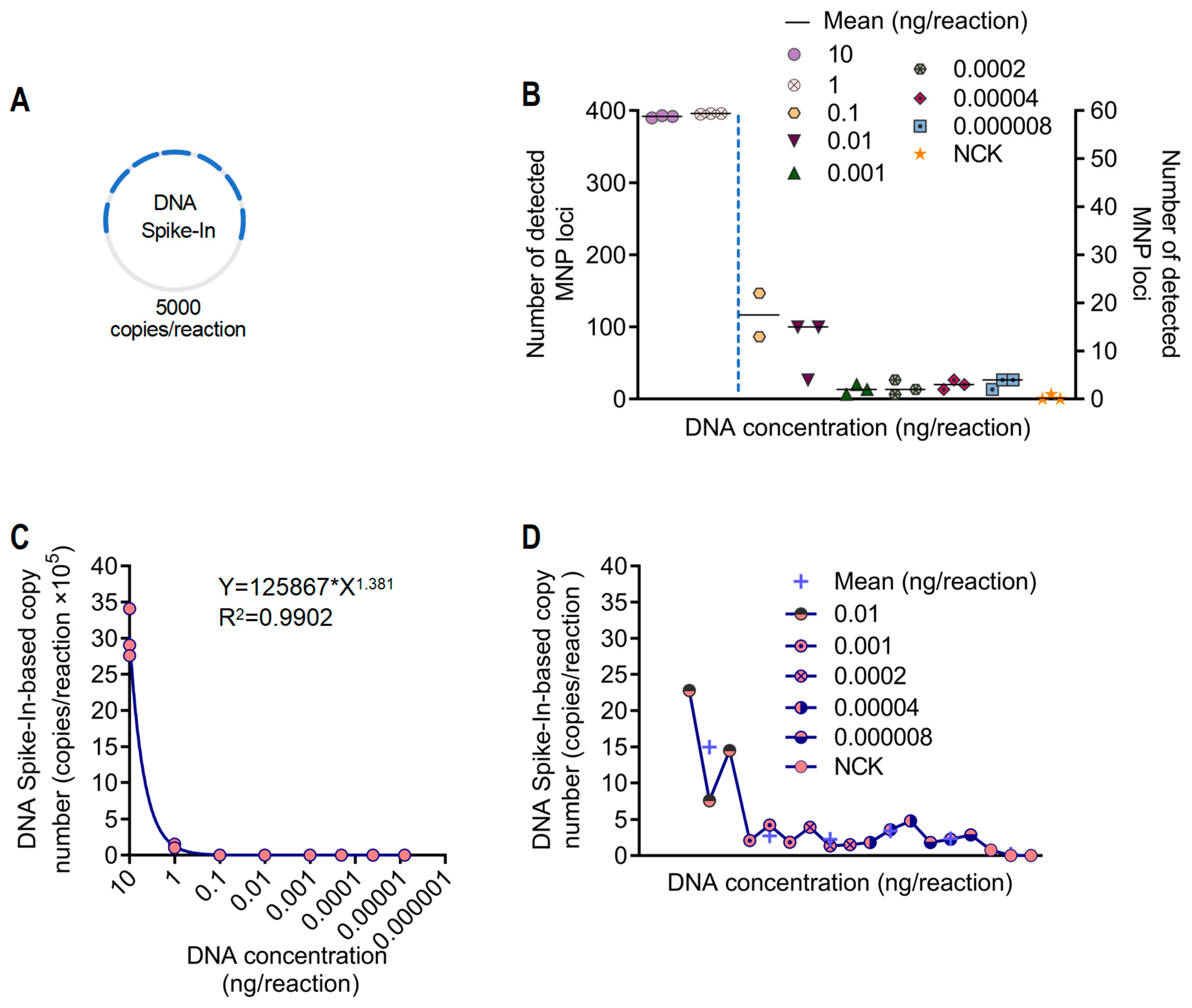
2.7. Testing Quantification and Sensitivity Assessment Based on Spike-In Normalization
3. Results
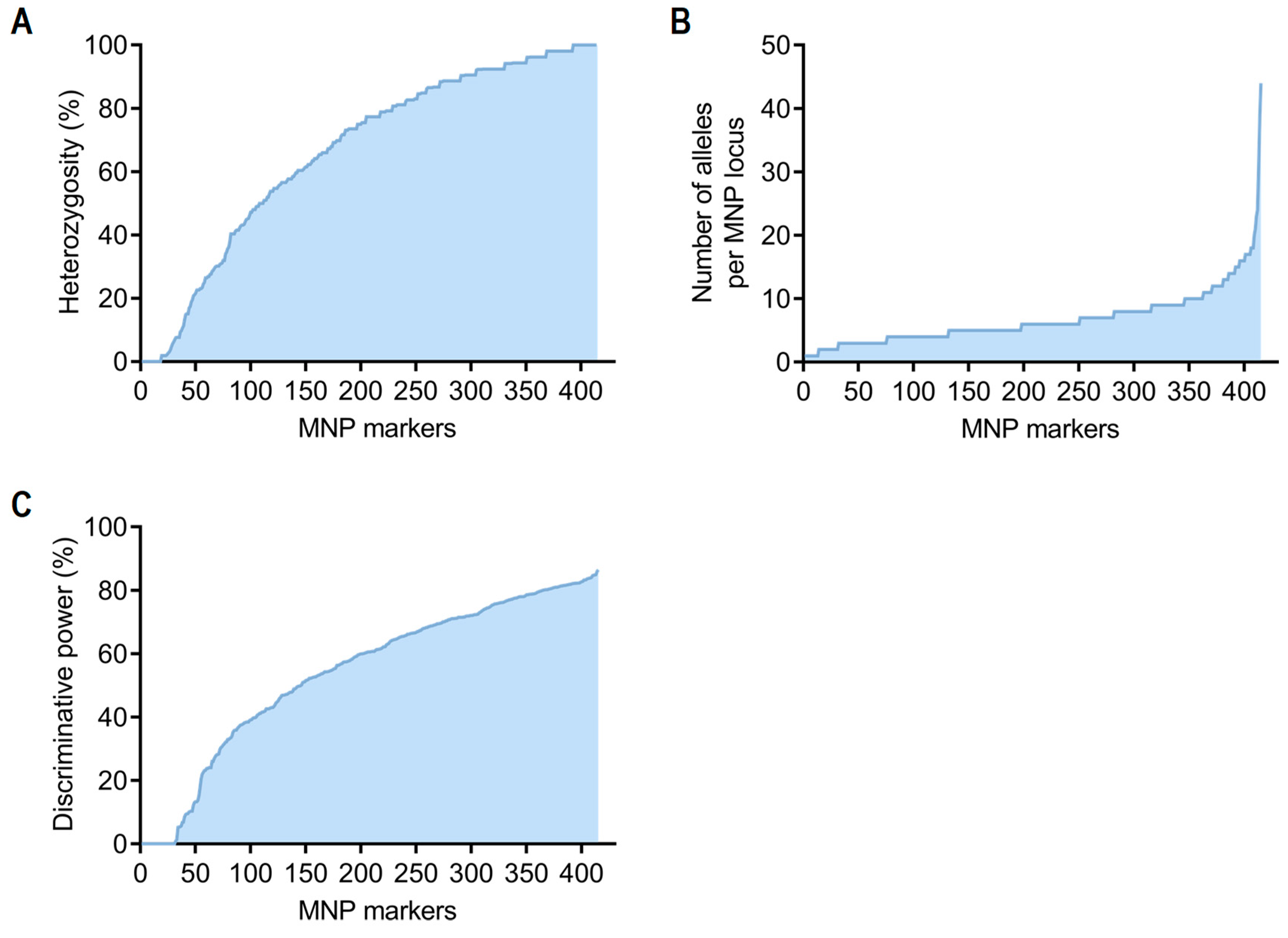
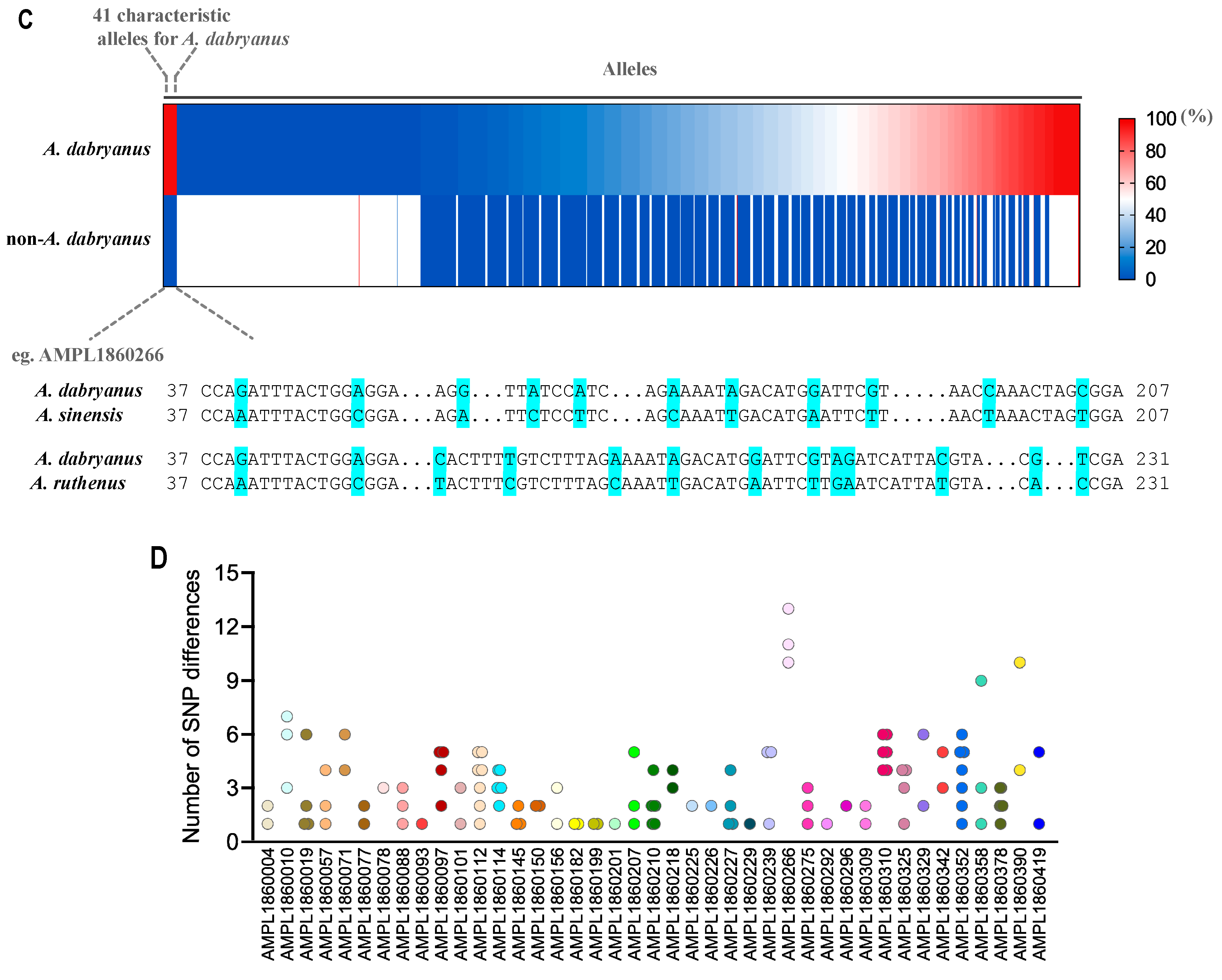
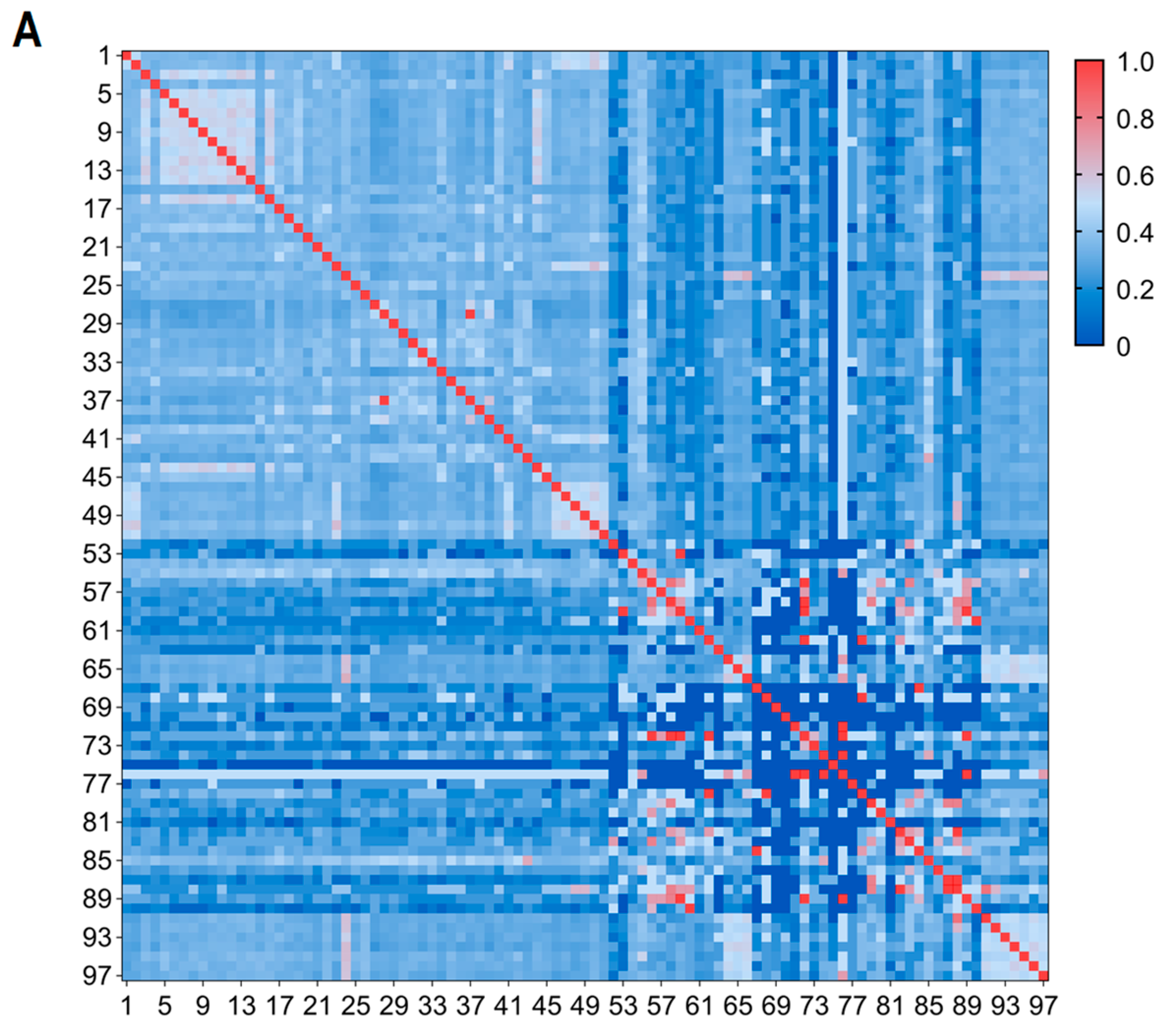
3.1. Evaluation of MNP Markers
3.2. Species Identification of A. dabryanus
3.3. Abundance Quantification and Sensitivity Analysis of A. dabryanus Individuals Based on DNA Spike-In
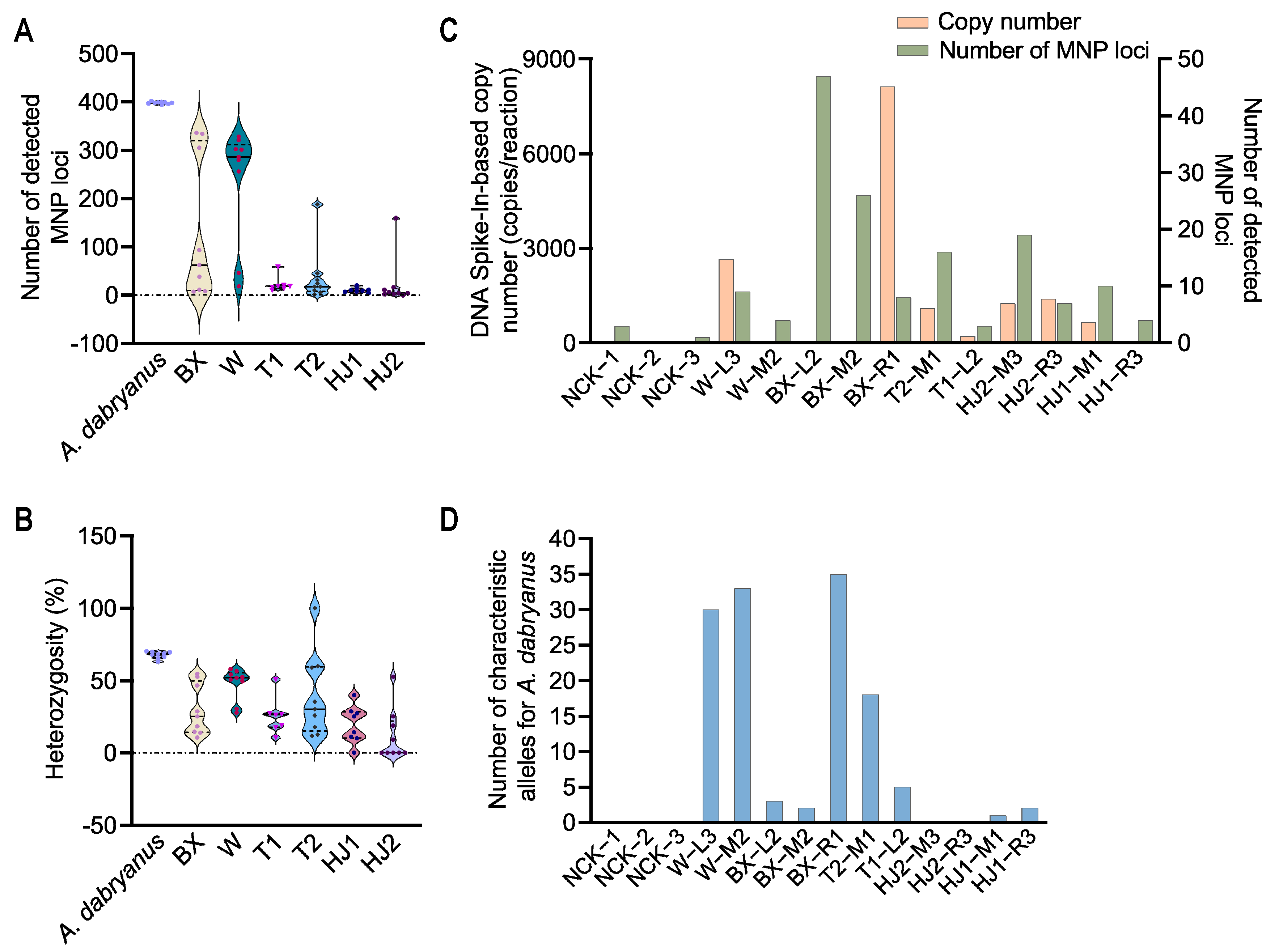
3.4. Application of the MNP System to Yangtze River Water Samples
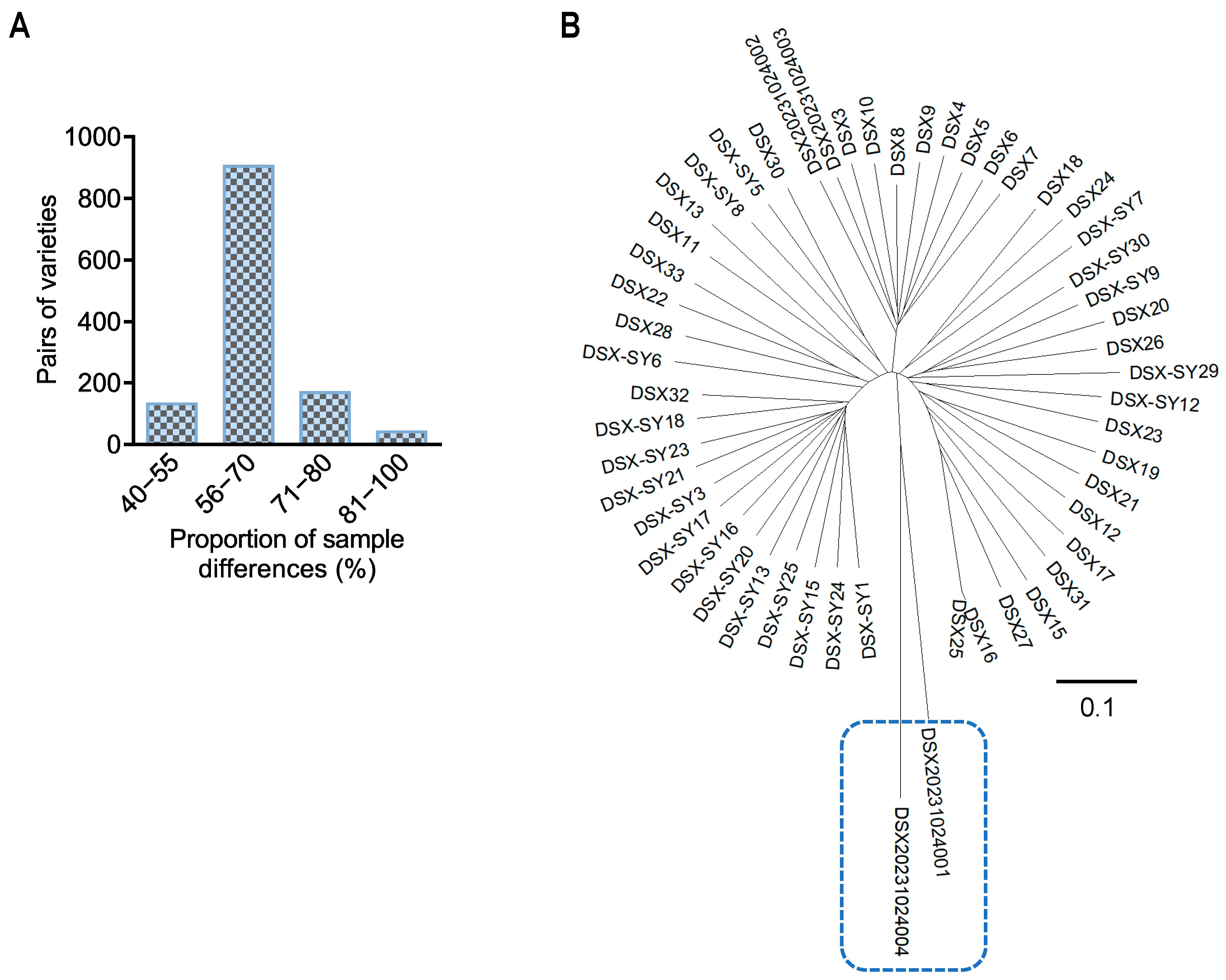
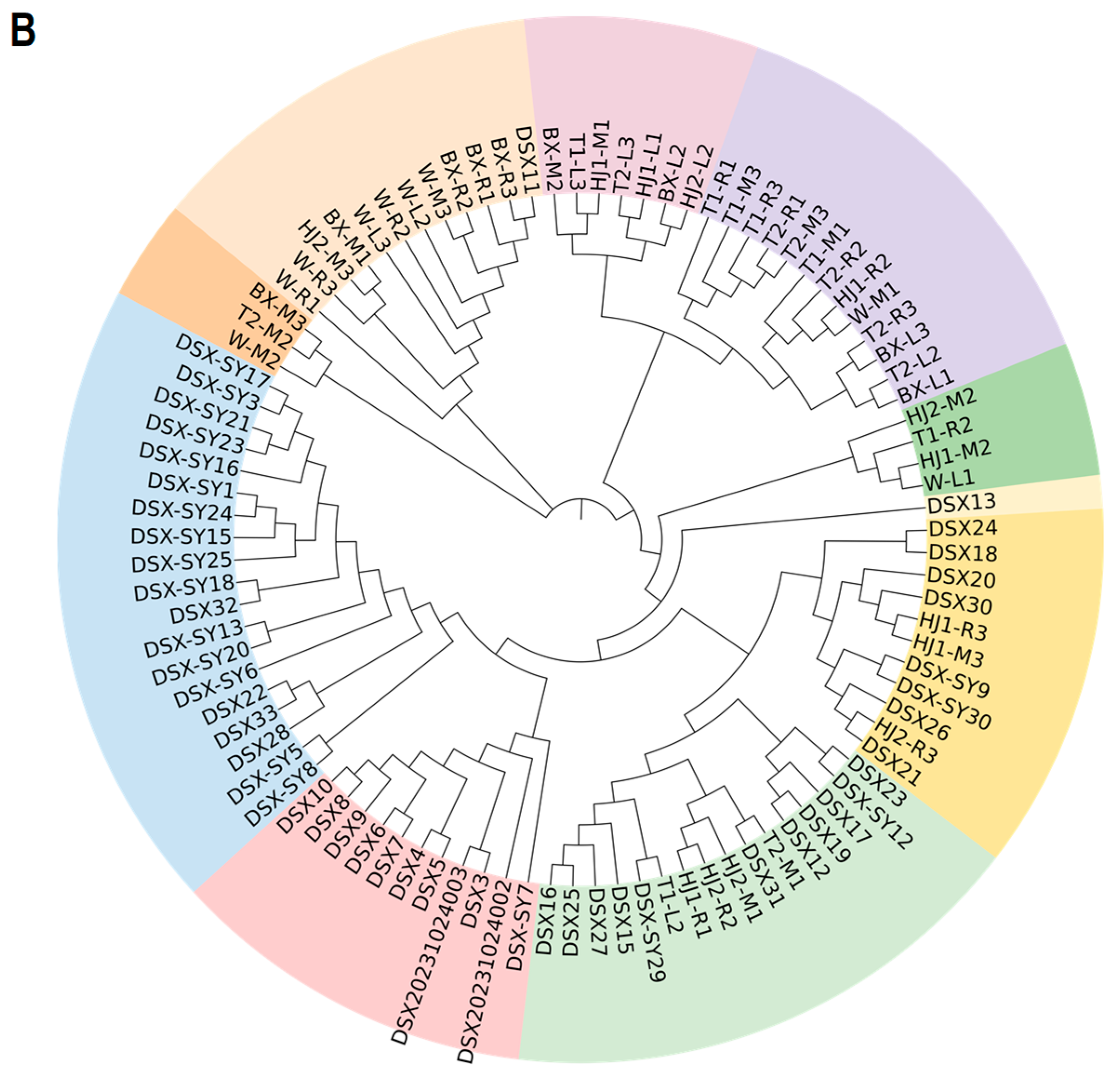
3.5. Correlation and Phylogenetic Analysis of A. dabryanus Individuals and Environmental Water Samples
4. Discussion
4.1. Differential Performance of the MNP System in Individual and Environmental Samples
4.2. Advantages of MNP System in eDNA Applications
4.3. Limitations of MNP Markers
4.4. Application of MNP System in Endangered Species Conservation: A Case Study of A. dabryanus
- Genetic Fingerprint Database: A genetic database will be created with allele genotypes from both wild and released individuals, using specific allelic markers to distinguish between them at the genetic level.
- Monitoring Released Individuals: DNA from water samples will be analyzed to detect genetic signals from released individuals, allowing tracking of their distribution across time and space.
- Allelic Gene Penetration Assessment: The specific allelic gene penetration rate (R1) will be calculated to assess the genetic contribution of released fish to the target gene pool. The formula for R1 is: R1 = SAt/SA, Where SAt is the number of specific alleles in released fish, and SA is the total number of alleles from both released and wild fish.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.; Kang, M.; Shen, L.; Wu, J.; Li, J.; Du, H.; Wang, C.; Yang, H.; Zhou, Q.; Liu, Z.; et al. Rapid change in Yangtze fisheries and its implications for global freshwater ecosystem management. Fish Fish. 2020, 21, 601–620. [Google Scholar] [CrossRef]
- Zhang, H.; Wei, Q.W.; Du, H.; Li, L.X. Present status and risk for extinction of the Dabry’s sturgeon (Acipenser dabryanus) in the Yangtze River watershed: A concern for intensified rehabilitation needs. J. Appl. Ichthyol. 2011, 27, 181–185. [Google Scholar] [CrossRef]
- WWF. Saving Sturgeons a Global Report on Their Status and Suggested Conservation Strategy; WWF International: Morges, Switzerland, 2020. [Google Scholar]
- Huang, Z.; Wang, L. Yangtze Dams Increasingly Threaten the Survival of the Chinese Sturgeon. Curr. Biol. 2018, 28, 3640–3647.e18. [Google Scholar] [CrossRef]
- Xie, P. Three-Gorges Dam: Risk to ancient fish. Science 2003, 302, 1149–1151. [Google Scholar] [CrossRef]
- Coble, A.A.; Flinders, C.A.; Homyack, J.A.; Penaluna, B.E.; Cronn, R.C.; Weitemier, K. eDNA as a tool for identifying freshwater species in sustainable forestry: A critical review and potential future applications. Sci. Total Environ. 2019, 649, 1157–1170. [Google Scholar] [CrossRef]
- Harrison, J.B.; Sunday, J.M.; Rogers, S.M. Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proc. Biol. Sci. 2019, 286, 20191409. [Google Scholar] [CrossRef]
- Pont, D.; Rocle, M.; Valentini, A.; Civade, R.; Jean, P.; Maire, A.; Roset, N.; Schabuss, M.; Zornig, H.; Dejean, T. Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation. Sci. Rep. 2018, 8, 10361. [Google Scholar] [CrossRef]
- Levy-Booth, D.J.; Campbell, R.G.; Gulden, R.H.; Hart, M.M.; Powell, J.R.; Klironomos, J.N.; Pauls, K.P.; Swanton, C.J.; Trevors, J.T.; Dunfield, K.E. Cycling of extracellular DNA in the soil environment. Soil. Biol. Biochem. 2007, 39, 2977–2991. [Google Scholar] [CrossRef]
- Pietramellara, G.; Ascher, J.; Borgogni, F.; Ceccherini, M.T.; Guerri, G.; Nannipieri, P. Extracellular DNA in soil and sediment: Fate and ecological relevance. Biol. Fert. Soils 2009, 45, 219–235. [Google Scholar] [CrossRef]
- Nevers, M.B.; Byappanahalli, M.N.; Morris, C.C.; Shively, D.; Przybyla-Kelly, K.; Spoljaric, A.M.; Dickey, J.; Roseman, E.F. Environmental DNA (eDNA): A tool for quantifying the abundant but elusive round goby (Neogobius melanostomus). PLoS ONE 2018, 13, e0191720. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, C.S.; Deconinck, D.; Eljasik, P.; Sobczak, M.; Derycke, S.; Panicz, R.; Kane, N.; Mazloomrezaei, M.; Devlin, R.H.; Faria, M.A. A fast HRMA tool to authenticate eight salmonid species in commercial food products. Food Chem. Toxicol. 2021, 156, 112440. [Google Scholar] [CrossRef]
- Giusti, A.; Tinacci, L.; Sotelo, C.G.; Marchetti, M.; Guidi, A.; Zheng, W.; Armani, A. Seafood Identification in Multispecies Products: Assessment of 16S rRNA, Cytb, and COI Universal Primers’ Efficiency as a Preliminary Analytical Step for Setting up Metabarcoding Next-Generation Sequencing Techniques. J. Agric. Food Chem. 2017, 65, 2902–2912. [Google Scholar] [CrossRef]
- Milan, D.T.; Mendes, I.S.; Damasceno, J.S.; Teixeira, D.F.; Sales, N.G.; Carvalho, D.C. New 12S metabarcoding primers for enhanced Neotropical freshwater fish biodiversity assessment. Sci. Rep. 2020, 10, 17966. [Google Scholar] [CrossRef]
- Guichoux, E.; Lagache, L.; Wagner, S.; Chaumeil, P.; Leger, P.; Lepais, O.; Lepoittevin, C.; Malausa, T.; Revardel, E.; Salin, F.; et al. Current trends in microsatellite genotyping. Mol. Ecol. Resour. 2011, 11, 591–611. [Google Scholar] [CrossRef]
- Mastrochirico-Filho, V.A.; Del Pazo, F.; Hata, M.E.; Villanova, G.V.; Foresti, F.; Vera, M.; Martinez, P.; Porto-Foresti, F.; Hashimoto, D.T. Assessing Genetic Diversity for a Pre-Breeding Program in Piaractus mesopotamicus by SNPs and SSRs. Genes 2019, 10, 668. [Google Scholar] [CrossRef] [PubMed]
- Bylemans, J.; Furlan, E.M.; Gleeson, D.M.; Hardy, C.M.; Duncan, R.P. Does Size Matter? An Experimental Evaluation of the Relative Abundance and Decay Rates of Aquatic Environmental DNA. Environ. Sci. Technol. 2018, 52, 6408–6416. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Buhay, J.E.; Whiting, M.F.; Crandall, K.A. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Natl. Acad. Sci. USA 2008, 105, 13486–13491. [Google Scholar] [CrossRef] [PubMed]
- Magnacca, K.N.; Brown, M.J. Mitochondrial heteroplasmy and DNA barcoding in Hawaiian Hylaeus (Nesoprosopis) bees (Hymenoptera: Colletidae). BMC Evol. Biol. 2010, 10, 174. [Google Scholar] [CrossRef]
- Sultana, S.; Ali, M.E.; Hossain, M.A.M.; Asing Naquiah, N.; Zaidul, I.S.M. Universal mini COI barcode for the identification of fish species in processed products. Food Res. Int. 2018, 105, 19–28. [Google Scholar] [CrossRef]
- Palumbo, F.; Scariolo, F.; Vannozzi, A.; Barcaccia, G. NGS-based barcoding with mini-COI gene target is useful for pet food market surveys aimed at mislabelling detection. Sci. Rep. 2020, 10, 17767. [Google Scholar] [CrossRef]
- Hajibabaei, M.; McKenna, C. DNA mini-barcodes. Methods Mol. Biol. 2012, 858, 339–353. [Google Scholar]
- Yuan, H.; Han, J.; Yang, M.; Chen, S.; Pang, X. DNA Metabarcoding: Current Applications and Challenges. J. Agric. Food Chem. 2025, 73, 20616–20632. [Google Scholar] [CrossRef]
- Creedy, T.J.; Andujar, C.; Meramveliotakis, E.; Noguerales, V.; Overcast, I.; Papadopoulou, A.; Morlon, H.; Vogler, A.P.; Emerson, B.C.; Arribas, P. Coming of age for COI metabarcoding of whole organism community DNA: Towards bioinformatic harmonisation. Mol. Ecol. Resour. 2022, 22, 847–861. [Google Scholar] [CrossRef] [PubMed]
- Burki, F.; Sandin, M.M.; Jamy, M. Diversity and ecology of protists revealed by metabarcoding. Curr. Biol. 2021, 31, R1267–R1280. [Google Scholar] [CrossRef] [PubMed]
- Miya, M. Environmental DNA Metabarcoding: A Novel Method for Biodiversity Monitoring of Marine Fish Communities. Ann. Rev. Mar. Sci. 2022, 14, 161–185. [Google Scholar] [CrossRef] [PubMed]
- Francioli, D.; Lentendu, G.; Lewin, S.; Kolb, S. DNA Metabarcoding for the Characterization of Terrestrial Microbiota-Pitfalls and Solutions. Microorganisms 2021, 9, 361. [Google Scholar] [CrossRef]
- Harr, B.; Zangerl, B.; Schlotterer, C. Removal of microsatellite interruptions by DNA replication slippage: Phylogenetic evidence from Drosophila. Mol. Biol. Evol. 2000, 17, 1001–1009. [Google Scholar] [CrossRef]
- Shinde, D.; Lai, Y.; Sun, F.; Arnheim, N. Taq DNA polymerase slippage mutation rates measured by PCR and quasi-likelihood analysis: (CA/GT)n and (A/T)n microsatellites. Nucleic Acids Res. 2003, 31, 974–980. [Google Scholar] [CrossRef]
- Hosseinzadeh-Colagar, A.; Haghighatnia, M.J.; Amiri, Z.; Mohadjerani, M.; Tafrihi, M. Microsatellite (SSR) amplification by PCR usually led to polymorphic bands: Evidence which shows replication slippage occurs in extend or nascent DNA strands. Mol. Biol. Res. Commun. 2016, 5, 167–174. [Google Scholar]
- Morin, P.A.; Luikart, G.; Wayne, R.K.; the SNP workshop group. SNPs in ecology, evolution and conservation. Trends Ecol. Evol. 2004, 19, 208–216. [Google Scholar] [CrossRef]
- Fang, Z.W.; Li, L.; Zhou, J.F.; You, A.Q.; Gao, L.F.; Li, T.T.; Chen, H.; Han, R.X.; Cui, Y.H.; Chen, L.H.; et al. Multiple nucleotide polymorphism DNA markers for the accurate evaluation of genetic variations. bioRxiv 2021. 03:09.434561. [Google Scholar] [CrossRef]
- Liu, J.; Wang, H.; Fan, X.; Zhang, Y.; Sun, L.; Liu, C.; Fang, Z.; Zhou, J.; Peng, H.; Jiang, J. Establishment and application of a Multiple nucleotide polymorphism molecular identification system for grape cultivars. Sci. Hortic. 2024, 325, 112642. [Google Scholar] [CrossRef]
- Ling, Y.Y.; Zhang, M.Z.; Ling, Z.L.; Cao, B.; Wu, X.P.; Peng, H.; Wang, Z.R.; Zhao, R.L. Evolutionary relationship and a novel method of efficient identification of Lentinula edodes cultivars in China. Mycosphere 2023, 13, 56–85. [Google Scholar] [CrossRef]
- Liu, F.; Wang, S.H.; Jia, D.H.; Tan, H.; Wang, B.; Zhao, R.L. Development of Multiple Nucleotide Polymorphism Molecular Markers for Enoki Mushroom (Flammulina filiformis) Cultivars Identification. J. Fungi 2023, 9, 330. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.F.; Zhao, Q.; Li, T.T.; Teng, C.L.; Peng, H.; Yao, Z.G.; Fang, Z.W.; Zhou, J.F.; Yang, X.H.; Qiao, J.X.; et al. Availability Evaluation and Application of MNP (Multiple Nucleotide Polymorphism) Markers in Variety Identification of Chrysanthemum. Horticulturae 2024, 10, 845. [Google Scholar] [CrossRef]
- Yi, Y.; Jiang, Z.; Ma, L.; Hou, X.; Li, L.; Ye, D.; Du, J.; Peng, H.; Han, G.; Li, H.; et al. Simultaneous identification of multiple animal-derived components in meat and meat products by using MNP marker based on high-throughput sequencing. Food Sci. Hum. Wellness 2025, 14, 9250178. [Google Scholar] [CrossRef]
- Ye, H.M.; Li, P.C.; Wu, J.M.; Shen, L.; Li, j.Y.; Du, H. Habitat distribution characteristics and key influencing factors analysis of artificially stocked juvenile Yangtze sturgeon (Acipenser dabryanus) in the Luzhou section of the Yangtze River. Acta Hydrobiol. Sin. 2025, 2025, 1–10. (In Chinese) [Google Scholar]
- Liu, W.C.; Li, J.Y.; Wu, J.M.; Ye, H.M.; Li, P.C.; Shen, L.; Du, H. Effectiveness assessment of Yangtze sturgeon (Acipenser dabryanus) stock enhancement in the upper Yangtze River during the initial fishing ban period. Acta Hydrobiol. Sin. 2025, 49, 26–33. (In Chinese) [Google Scholar]
- Wang, Y.; Hu, Y.; Zhang, T. Current status and perspective of RAD-seq in genomic research. Yi Chuan 2014, 36, 41–49. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Gao, L.; Li, L.; Fang, B.; Fang, Z.; Xiang, Y.; Zhang, M.; Zhou, J.; Song, H.; Chen, L.; Li, T.; et al. Carryover Contamination-Controlled Amplicon Sequencing Workflow for Accurate Qualitative and Quantitative Detection of Pathogens: A Case Study on SARS-CoV-2. Microbiol. Spectr. 2023, 11, e0020623. [Google Scholar] [CrossRef]
- Chen, K.; Hu, Z.; Xia, Z.; Zhao, D.; Li, W.; Tyler, J.K. The Overlooked Fact: Fundamental Need for Spike-In Control for Virtually All Genome-Wide Analyses. Mol. Cell Biol. 2015, 36, 662–667. [Google Scholar] [CrossRef]
- Wilhelm, J.; Pingoud, A.; Hahn, M. Real-time PCR-based method for the estimation of genome sizes. Nucleic Acids Res. 2003, 31, e56. [Google Scholar] [CrossRef]
- Blackburn, J.; Wong, T.; Madala, B.S.; Barker, C.; Hardwick, S.A.; Reis, A.L.M.; Deveson, I.W.; Mercer, T.R. Use of synthetic DNA spike-in controls (sequins) for human genome sequencing. Nat. Protoc. 2019, 14, 2119–2151. [Google Scholar] [CrossRef] [PubMed]
- Du, C.Z.; Jin, G.F.; Wu, L.; Que, P.Z.; Lu, X.Y.; Lu, H.H.; Sun, C.; Gu, Z.X. Evaluation of the proliferation and release effect of Schizothorax in high-altitude areas based on labeling technology. Environ. Ecol. 2025, 7, 46–52. (In Chinese) [Google Scholar]
- Stewart, D.R.; Hafen, T.; Hendrickson, D.A.; Taylor, A.T.; Varela-Romero, A.; Mason, D.H.; Dysthe, J.C.; Franklin, T.W.; Young, M.K.; McKelvey, K.S.; et al. Environmental DNA mitochondrial markers to assess potential occupancy of Endangered Yaqui catfish in the Yaqui River basin, Mexico. Endanger. Species Res. 2024, 53, 569–586. [Google Scholar] [CrossRef]
- Crossman James, A.; Flores, A.M.; Messmer, A.; Nelson, R.J.; McAdam Steve, O.; Johnson, P.; Reece, P.; Koop Ben, F. Development of eDNA Protocols for Detection of Endangered White Sturgeon (Acipenser transmontanus) in the Wild. Environ. DNA 2024, 6, e70006. [Google Scholar] [CrossRef]
- Singh, N.; Choudhury, D.R.; Singh, A.K.; Kumar, S.; Srinivasan, K.; Tyagi, R.K.; Singh, N.K.; Singh, R. Comparison of SSR and SNP markers in estimation of genetic diversity and population structure of Indian rice varieties. PLoS ONE 2013, 8, e84136. [Google Scholar] [CrossRef]
- Kalita, M.C.; Mohapatra, T.; Dhandapani, A.; Yadavad, D.K.; Srinivasanb, K.; Mukherjeee, A.K.; Sharmab, R.P. Comparative Evaluation of RAPD, ISSR and Anchored-SSR Markers in the Assessment of Genetic Diversity and Fingerprinting of Oilseed Brassica Genotypes. J. Plant Biochem. Biotechnol. 2007, 16, 41–48. [Google Scholar] [CrossRef]
- Gettings, K.B.; Kiesler, K.M.; Vallone, P.M. Performance of a next generation sequencing SNP assay on degraded DNA. Forensic Sci. Int. Genet. 2015, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.M.; Coble, M.D.; Vallone, P.M. STRs vs. SNPs: Thoughts on the future of forensic DNA testing. Forensic Sci. Med. Pathol. 2007, 3, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Jerde, C.L.; Olds, B.P.; Shogren, A.J.; Andruszkiewicz, E.A.; Mahon, A.R.; Bolster, D.; Tank, J.L. Influence of Stream Bottom Substrate on Retention and Transport of Vertebrate Environmental DNA. Environ. Sci. Technol. 2016, 50, 8770–8779. [Google Scholar] [CrossRef]
- Andruszkiewicz Allan, E.; Zhang, W.G.; CLavery, A.; FGovindarajan, A. Environmental DNA shedding and decay rates from diverse animal forms and thermal regimes. Environ. DNA 2020, 3, 492–514. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Y.; Gong, Q.; Lai, J.; Du, J.; Deng, X. Paternity assignment in the polyploid Acipenser dabryanus based on a novel microsatellite marker system. PLoS ONE 2017, 12, e0185280. [Google Scholar] [CrossRef]
- Zhang, S.M.; Zhang, Y.P.; Zheng, X.Z.; Chen, Y.J.; Deng, H.; Wang, D.Q.; Wei, Q.W.; Zhang, Q.W.; Nie, L.; Wu, Q.J. Sequence variation of the mitochondrial DNA ND4L-ND4 gene and molecular systematics of 12 species of Acipenseriformes. Sci. China (Ser. C) 1999, 29, 608–614. (In Chinese) [Google Scholar]
- Zhang, S.M.; Wu, Q.J.; Zhang, Y.P. Tandem repeat sequences in the control region of mtDNA and their evolutionary significance in Chinese sturgeon (Acipenser sinensis) and related species. Acta Biochim. Biophys. Sin. 2000, 32, 458–461. (In Chinese) [Google Scholar]
- Zhang, S.M.; Wu, Q.J.; Zhang, Y.P. On the taxonomic status of Yangtze sturgeon, Asian and American green sturgeon based on mitochondrial control region sequences. Acta Zool. Sin. 2001, 47, 632–639. (In Chinese) [Google Scholar]
- Guillera-Arroita, G.; Lahoz-Monfort, J.J.; van Rooyen, A.R.; Weeks, A.R.; Tingley, R.; McCrea, R. Dealing with false-positive and false-negative errors about species occurrence at multiple levels. Methods Ecol. Evol. 2017, 8, 1081–1091. [Google Scholar] [CrossRef]
- Lahoz-Monfort, J.J.; Guillera-Arroita, G.; Tingley, R. Statistical approaches to account for false-positive errors in environmental DNA samples. Mol. Ecol. Resour. 2016, 16, 673–685. [Google Scholar] [CrossRef]
- Hansen, B.K.; Bekkevold, D.; Clausen, L.W.; Nielsen, E.E. The sceptical optimist: Challenges and perspectives for the application of environmental DNA in marine fisheries. Fish Fish. 2018, 19, 751–768. [Google Scholar] [CrossRef]
- Shu, L.; Ludwig, A.; Peng, Z. Standards for Methods Utilizing Environmental DNA for Detection of Fish Species. Genes 2020, 11, 296. [Google Scholar] [CrossRef]
- Ai, Q.; Yuan, H.; Wang, Y.; Li, C. Estimation of Species Abundance Based on the Number of Segregating Sites Using Environmental DNA (eDNA). Mol. Ecol. Resour. 2025, 25, e14076. [Google Scholar] [CrossRef]
- Liu, W.E.; Tan, D.; Zhang, Z. Serum HBV DNA detected by polymerase chain reaction with dUTP/uracil-DNA glycosylase. Hunan Yi Ke Da Xue Xue Bao 1998, 23, 278–280. [Google Scholar]
- Song, D.; Xu, C.; Sang, P.; Liu, Y.; Huang, X. Rapid and contamination-free detection of cucumber green mottle mosaic virus as a viral indicator in wastewater via UDG-RT-LAMP combined with CRISPR/Cas12a. J. Hazard. Mater. 2025, 497, 139571. [Google Scholar] [CrossRef]
- Xu, L.X.; Zhou, L.; Wei, Q.W. Stock status and conservation dilemma of species of Acipenseriformes in the Yangtze River and relevant suggestions. J. Fish. China 2023, 47, 029304. (In Chinese) [Google Scholar]
- Ding, Y.; Ren, Z.J.; Xu, X.; Lu, Y.J.; Lou, F.R. Evaluation of stock enhancement effect of Oplegnathus fasciatus in Yinshan Bay based on microsatellite markers. J. Yantai Univ. (Nat. Sci. Eng. Ed.) 2025. (In Chinese) [Google Scholar] [CrossRef]
- Qiu, J.Y.; Lu, D.; Hu, Y.L.; Wang, W.J.; Lu, G.Z.; Shan, X.J. Evaluation of the proliferation and release effect of Fenneropenaeus chinensis in high-altitude areas based on molecular marker technology. J. Fish. Sci. China 2025, 32, 362–371. (In Chinese) [Google Scholar]
- Sy, Z. Evaluation of Callista chinensis stock enhancement effect based on microsatellite markers. J. Fish. Sci. China 2025, 52–55. (In Chinese) [Google Scholar]
- Fazekas, G.; Kovács, G.; Sándor, Z.J.; Bogár, K.; Géczi, A.; Kovács, B. Genetic Characterization of Endangered Sterlet (Acipenser ruthenus, Linnaeus 1758) Gene Bank Broodstock, Natural and Cultured Populations in Hungary. Fishes 2024, 9, 201. [Google Scholar] [CrossRef]
- Yang, S.; Zhao, Z.; Xu, Z.; Liu, Y.; Jiang, M.; Fu, L.; Zhang, J.; Jing, Z.; Pang, X.; Shao, W.; et al. Identification of Hybrid Sturgeon (Acipenser baerii × Acipenser schrenckii) from Their Parents Using Germplasm. Animals 2025, 15, 907. [Google Scholar] [CrossRef] [PubMed]
- Chevrinais, M.; Bourret, A.; Cote, G.; Faille, G.; Gagne, N.; Parent, G.J. Improving an endangered marine species distribution using reliable and localized environmental DNA detections combined with trawl captures. Sci. Rep. 2025, 15, 11926. [Google Scholar] [CrossRef] [PubMed]

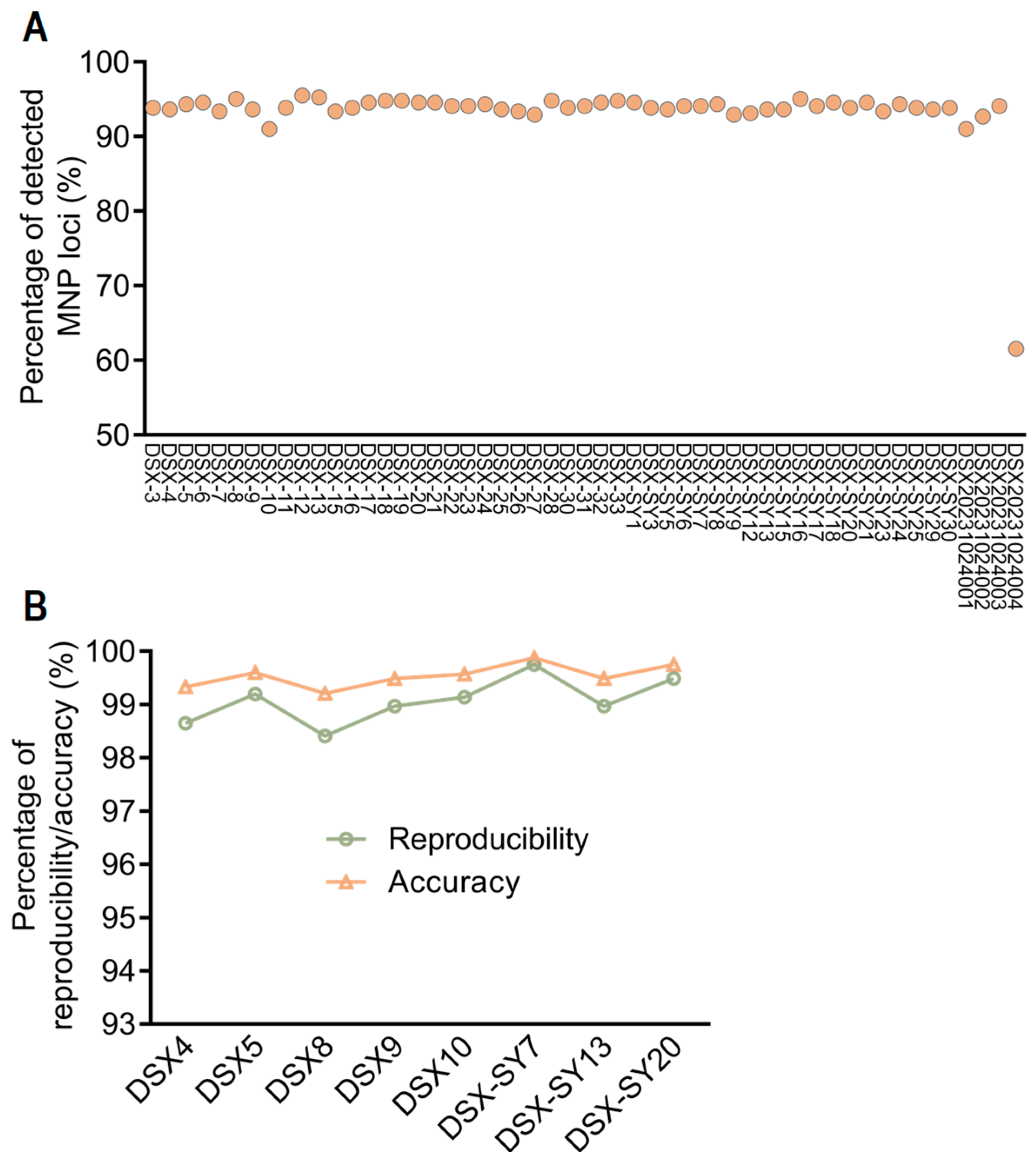
| Sample Name | Library 1 | Library 2 | Number of Common MNP Markers | Number of Different MNP Markers | Reproducibility | Accuracy |
|---|---|---|---|---|---|---|
| DSX4 | DSX4-1 | DSX4-2 | 370 | 5 | 98.65% | 99.33% |
| DSX5 | DSX5-1 | DSX5-2 | 374 | 3 | 99.20% | 99.60% |
| DSX8 | DSX8-1 | DSX8-2 | 377 | 6 | 98.41% | 99.21% |
| DSX9 | DSX9-1 | DSX9-2 | 389 | 4 | 98.97% | 99.49% |
| DSX10 | DSX10-1 | DSX10-2 | 350 | 3 | 99.14% | 99.57% |
| DSX-SY7 | DSX-SY7-1 | DSX-SY7-2 | 394 | 1 | 99.75% | 99.88% |
| DSX-SY13 | DSX-SY13-1 | DSX-SY13-2 | 388 | 4 | 98.97% | 99.49% |
| DSX-SY20 | DSX-SY20-1 | DSX-SY20-2 | 391 | 2 | 99.49% | 99.75% |
| Water Sample | DNA Spike-In-Based Copy Number | Number of MNP Loci | Number of MNP Loci |
|---|---|---|---|
| NCK-1 | 4 | 3 | 0 |
| NCK-2 | 0 | 0 | 0 |
| NCK-3 | 3 | 1 | 0 |
| W-L3 | 2658 | 9 | 30 |
| W-M2 | 8 | 4 | 33 |
| BX-L2 | 63 | 47 | 3 |
| BX-M2 | 35 | 26 | 2 |
| BX-R1 | 8125 | 8 | 35 |
| T2-M1 | 1092 | 16 | 18 |
| T1-L2 | 216 | 3 | 5 |
| HJ2-M3 | 1250 | 19 | 0 |
| HJ2-R3 | 1394 | 7 | 0 |
| HJ1-M1 | 650 | 10 | 1 |
| HJ1-R3 | 15 | 4 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, L.; Jiang, W.; Fang, Z.; Peng, H.; Chen, H.; Wan, R.; Gao, L.; Zhang, B.; Xiao, Z.; Li, S.; et al. Genetic Monitoring of the Endangered Acipenser dabryanus Using a High-Resolution MNP System. Diversity 2025, 17, 704. https://doi.org/10.3390/d17100704
Cai L, Jiang W, Fang Z, Peng H, Chen H, Wan R, Gao L, Zhang B, Xiao Z, Li S, et al. Genetic Monitoring of the Endangered Acipenser dabryanus Using a High-Resolution MNP System. Diversity. 2025; 17(10):704. https://doi.org/10.3390/d17100704
Chicago/Turabian StyleCai, Lu, Wei Jiang, Zhiwei Fang, Hai Peng, Hao Chen, Renjing Wan, Lifen Gao, Baolong Zhang, Zilan Xiao, Sha Li, and et al. 2025. "Genetic Monitoring of the Endangered Acipenser dabryanus Using a High-Resolution MNP System" Diversity 17, no. 10: 704. https://doi.org/10.3390/d17100704
APA StyleCai, L., Jiang, W., Fang, Z., Peng, H., Chen, H., Wan, R., Gao, L., Zhang, B., Xiao, Z., Li, S., Li, L., Chen, L., Song, H., Li, T., & Zhou, J. (2025). Genetic Monitoring of the Endangered Acipenser dabryanus Using a High-Resolution MNP System. Diversity, 17(10), 704. https://doi.org/10.3390/d17100704







