Whole-Genome Resequencing Reveals Population Genetic Structure and Selection Signatures in the Golden Wild Yak
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and Genome Resequencing
2.3. Sequence Quality Control and SNP Calling
2.4. Genetic Parameters Estimation
2.5. Population Structure Analysis
2.6. Selective Sweep Analysis
3. Results
3.1. Resequencing Statistics and SNP Annotation
3.2. Population Genetic Diversity
3.3. Population Structure Stratification
3.4. Selective Signals and Gene Function Annotation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FHom | Inbreeding coefficient |
| Fst | Fixation index |
| He | Expected heterozygosity |
| Ho | Observed heterozygosity |
| LD | Linkage disequilibrium |
| PCA | Principal component analysis |
| PSG | Positively selected gene |
| QTP | Qinghai–Tibet Plateau |
| ROH | Runs of homozygosity |
| SNP | Single-nucleotide polymorphism |
| WGR | Whole-genome resequencing |
| XP-CLR | Cross-population composite likelihood ratio |
| π | Nucleotide diversity |
References
- Chen, M.; Sun, Y.; Yang, C.; Zeng, G.; Li, Z.; Zhang, J. The road to wild yak protection in China. Science 2018, 360, 866. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.; Schaller, G.B.; Cheng, E.; Kang, A.; Krebs, M.; Li, L.; Hebblewhite, M. Legacies of past exploitation and climate affect mammalian sexes differently on the roof of the world—The case of wild yaks. Sci. Rep. 2015, 5, 8676. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Cheng, H.; Wang, N.; Bai, L.; Chen, X.; Liu, X.; Qiao, B. Identifying climate refugia for wild yaks (Bos mutus) on the Tibetan Plateau. J. Environ. Manag. 2024, 366, 121655. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, N.; Cheng, H.; Wang, Y.; Liu, X.; Qiao, B.; Zhao, L. Mapping conservation priorities for wild yak (Bos mutus) habitats on the Tibetan Plateau, China. Sci. Total Environ. 2024, 914, 169803. [Google Scholar] [CrossRef]
- Kusi, N.; Manandhar, P.; Senn, H.; Joshi, J.; Ghazali, M.; Hengaju, K.D.; Suwal, S.P.; Lama, T.L.; Poudyal, L.P.; Thapa, M.; et al. Phylogeographical analysis shows the need to protect the wild yaks’ last refuge in Nepal. Ecol. Evol. 2021, 11, 8310–8318. [Google Scholar] [CrossRef]
- Liu, X.; Liu, W.; Lenstra, J.A.; Zheng, Z.; Wu, X.; Yang, J.; Li, B.; Yang, Y.; Qiu, Q.; Liu, H.; et al. Evolutionary origin of genomic structural variations in domestic yaks. Nat. Commun. 2023, 14, 5617. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Gangwar, M.; Kumar, A.; Kumar, A.; Dige, M.S.; Jha, G.K.; Gaur, G.K.; Dutt, T. Dissecting genomes of multiple yak populations: Unveiling ancestry and high-altitude adaptation through whole-genome resequencing analysis. BMC Genom. 2025, 26, 214. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.; Lu, H.; Liu, F.; Li, D.; Feng, J. Preliminary analysis on taxonomic status of golden wild yak in Tibet. Acta Theriol. Sin. 2015, 35, 48–54. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Q.; Yang, L.; Chen, X.; Wang, C.; Zhang, Y.; Zeng, Y.; Xu, L.; Lu, C.; Zeng, C.; et al. Characterization of the complete mitochondrial genome sequence of golden wild yak and revealed its phylogenetic relationship with 9 yak subspecies. Mitochondrial DNA Part B 2019, 4, 660–661. [Google Scholar] [CrossRef]
- Lee, J.; Jung, J.H.; Oh, S.-H. Enhancing animal breeding through quality control in genomic data—A review. J. Anim. Sci. Technol. 2024, 66, 1099–1108. [Google Scholar] [CrossRef]
- Li, G.; Luo, J.; Wang, F.; Xu, D.; Ahmed, Z.; Chen, S.; Li, R.; Ma, Z. Whole-genome resequencing reveals genetic diversity, differentiation, and selection signatures of yak breeds/populations in Qinghai, China. Front. Genet. 2022, 13, 1034094. [Google Scholar] [CrossRef]
- Li, B.; Yang, J.; Liu, Y.; Jiang, M. Genome Variation Map of Domestic Qinghai-Tibet Plateau Yaks by SLAF-Seq Reveals Genetic Footprint during Artificial Selection. Animals 2023, 13, 2963. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Jin, H.; Yang, Q.; Poyarkov, A.; Korablev, M.; Rozhnov, V.; Shao, J.; Fu, Q.; Hernandez-Blanco, J.A.; Zhan, X.; et al. Genomic evidence for low genetic diversity but purging of strong deleterious variants in snow leopards. Genome Biol. 2025, 26, 94. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wei, F.; Zhan, X.; Fan, H.; Zhao, P.; Huang, G.; Chang, J.; Lei, Y.; Hu, Y.; Takahashi, A. Evolutionary Conservation Genomics Reveals Recent Speciation and Local Adaptation in Threatened Takins. Mol. Biol. Evol. 2022, 39, msac111. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, S.; Wang, Y.F.; Li, S.; Wu, S.X.; Yan, R.G.; Zhang, Y.W.; Wan, R.D.; He, Z.; Song, R.D.; et al. Long read genome assemblies complemented by single cell RNA-sequencing reveal genetic and cellular mechanisms underlying the adaptive evolution of yak. Nat. Commun. 2022, 13, 4887. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, W.; Liu, X.; Du, X.; Zhang, K.; Zhang, Y.; Song, Y.; Zi, Y.; Qiu, Q.; Lenstra, J.A.; et al. Structural Variants Selected during Yak Domestication Inferred from Long-Read Whole-Genome Sequencing. Mol. Biol. Evol. 2021, 38, 3676–3680. [Google Scholar] [CrossRef]
- Leslie, D.M.; Schaller, G.B. Bos grunniens and Bos mutus (Artiodactyla: Bovidae). Mamm. Species 2009, 836, 1–17. [Google Scholar] [CrossRef]
- Peng, W.; Fu, C.; Shu, S.; Wang, G.; Wang, H.; Yue, B.; Zhang, M.; Liu, X.; Liu, Y.; Zhang, J.; et al. Whole-genome resequencing of major populations revealed domestication-related genes in yaks. BMC Genom. 2024, 25, 69. [Google Scholar] [CrossRef]
- Wang, T.; Ma, X.; Ma, C.; Wu, X.; ZhaXi, T.; Yin, L.; Li, W.; Li, Y.; Liang, C.; Yan, P. Whole genome resequencing-based analysis of plateau adaptation in Meiren yak (Bos grunniens). Anim. Biotechnol. 2024, 35, 2298406. [Google Scholar] [CrossRef]
- Cai, B.; Wu, X.; Shi, Y.; Kang, Y.; Ding, Z.; Guo, S.; Cao, M.; Hu, L.; Zhang, B.; Wang, X.; et al. Whole-Genome Sequencing Unveils the Uniqueness of Yushu Yaks (Bos grunniens). Int. J. Mol. Sci. 2025, 26, 3879. [Google Scholar] [CrossRef]
- Guo, S.; Yu, T.; Wang, X.; Zhao, S.; Zhao, E.; Ainierlitu; Ba, T.; Gan, M.; Dong, C.; Naerlima; et al. Whole-genome resequencing reveals the uniqueness of Subei yak. J. Anim. Sci. 2024, 102, skae152. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, gix120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Behr, A.A.; Liu, K.Z.; Liu-Fang, G.; Nakka, P.; Ramachandran, S. pong: Fast analysis and visualization of latent clusters in population genetic data. Bioinformatics 2016, 32, 2817–2823. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Chen, H.; Patterson, N.; Reich, D. Population differentiation as a test for selective sweeps. Genome Res. 2010, 20, 393–402. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Wang, T.R.; Meng, H.H.; Wang, N.; Zheng, S.S.; Jiang, Y.; Lin, D.Q.; Song, Y.G.; Kozlowski, G. Adaptive divergence and genetic vulnerability of relict species under climate change: A case study of Pterocarya macroptera. Ann. Bot. 2023, 132, 241–254. [Google Scholar] [CrossRef]
- Angst, P.; Ameline, C.; Haag, C.R.; Ben-Ami, F.; Ebert, D.; Fields, P.D. Genetic Drift Shapes the Evolution of a Highly Dynamic Metapopulation. Mol. Biol. Evol. 2022, 39, msac264. [Google Scholar] [CrossRef]
- Woolfit, M. Effective population size and the rate and pattern of nucleotide substitutions. Biol. Lett. 2009, 5, 417–420. [Google Scholar] [CrossRef]
- Xue, Y.; Prado-Martinez, J.; Sudmant, P.H.; Narasimhan, V.; Ayub, Q.; Szpak, M.; Frandsen, P.; Chen, Y.; Yngvadottir, B.; Cooper, D.N.; et al. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science 2015, 348, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Nan, Z.; Prete, D. Protecting wild yak (Bos mutus) species and preventing its hybrid in China. J. Arid Land 2016, 8, 811–814. [Google Scholar] [CrossRef][Green Version]
- Casacci, L.P.; Barbero, F.; Balletto, E. The “Evolutionarily Significant Unit” concept and its applicability in biological conservation. Ital. J. Zool. 2013, 81, 182–193. [Google Scholar] [CrossRef]
- Howard-McCombe, J.; Jamieson, A.; Carmagnini, A.; Russo, I.M.; Ghazali, M.; Campbell, R.; Driscoll, C.; Murphy, W.J.; Nowak, C.; O’Connor, T.; et al. Genetic swamping of the critically endangered Scottish wildcat was recent and accelerated by disease. Curr. Biol. 2023, 33, 4761–4769.e4765. [Google Scholar] [CrossRef]
- Nei, M.; Maruyama, T.; Chakraborty, R. The Bottleneck Effect and Genetic Variability in Populations. Evolution 1975, 29, 1–10. [Google Scholar] [CrossRef]
- Frankham, R.; Bradshaw, C.J.A.; Brook, B.W. Genetics in conservation management: Revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol. Conserv. 2014, 170, 56–63. [Google Scholar] [CrossRef]
- Kobayashi, T.; Imokawa, G.; Bennett, D.C.; Hearing, V.J. Tyrosinase Stabilization by Tyrp1 (the brown Locus Protein). J. Biol. Chem. 1998, 273, 31801–31805. [Google Scholar] [CrossRef]
- Nakamura, H.; Fukuda, M. Establishment of a synchronized tyrosinase transport system revealed a role of Tyrp1 in efficient melanogenesis by promoting tyrosinase targeting to melanosomes. Sci. Rep. 2024, 14, 2529. [Google Scholar] [CrossRef]
- Lyons, L.A.; Foe, I.T.; Rah, H.C.; Grahn, R.A. Chocolate coated cats: TYRP1 mutations for brown color in domestic cats. Mamm. Genome 2005, 16, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Mao, H.; Zhang, Z.; Xiao, S.; Ding, N.; Huang, L. A 6-bp deletion in the TYRP1 gene causes the brown colouration phenotype in Chinese indigenous pigs. Heredity 2010, 106, 862–868. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y.; Shen, L.; Du, J.; Luo, J.; Liu, C.; Pu, Q.; Yang, R.; Li, X.; Bai, L.; et al. A 6-bp deletion in exon 8 and two mutations in introns of TYRP1 are associated with blond coat color in Liangshan pigs. Gene 2016, 578, 132–136. [Google Scholar] [CrossRef]
- Peterson, S.M.; Watowich, M.M.; Renner, L.M.; Martin, S.; Offenberg, E.; Lea, A.; Montague, M.J.; Higham, J.P.; Snyder-Mackler, N.; Neuringer, M.; et al. Genetic variants in melanogenesis proteins TYRP1 and TYR are associated with the golden rhesus macaque phenotype. G3 Genes Genomes Genet. 2023, 13, jkad168. [Google Scholar] [CrossRef]
- Jung, H.; Lee, K.S.; Choi, J.K. Comprehensive characterisation of intronic mis-splicing mutations in human cancers. Oncogene 2021, 40, 1347–1361. [Google Scholar] [CrossRef]
- Claringbould, A.; Zaugg, J.B. Enhancers in disease: Molecular basis and emerging treatment strategies. Trends Mol. Med. 2021, 27, 1060–1073. [Google Scholar] [CrossRef]
- Dong, K.; Bai, Z.; He, X.; Zhang, L.; Hu, G.; Yao, Y.; Cai, C.L.; Zhou, J. Generation of a novel constitutive smooth muscle cell-specific Myh11-driven Cre mouse model. J. Mol. Cell. Cardiol. 2025, 202, 144–152. [Google Scholar] [CrossRef]
- Liddiard, K.; Aston-Evans, A.N.; Cleal, K.; Hendrickson, E.A.; Baird, D.M. POLQ suppresses genome instability and alterations in DNA repeat tract lengths. NAR Cancer 2022, 4, zcac020. [Google Scholar] [CrossRef] [PubMed]
- Gardner, C.C.; Abele, J.A.; Winkler, T.J.; Reckers, C.N.; Anas, S.A.; James, P.F. Common as well as unique methylation-sensitive DNA regulatory elements in three mammalian SLC9C1 genes. Gene 2024, 893, 147897. [Google Scholar] [CrossRef] [PubMed]
- Nagarkatti-Gude, D.R.; Jaimez, R.; Henderson, S.C.; Teves, M.E.; Zhang, Z.; Strauss, J.F., 3rd. Spag16, an axonemal central apparatus gene, encodes a male germ cell nuclear speckle protein that regulates SPAG16 mRNA expression. PLoS ONE 2011, 6, e20625. [Google Scholar] [CrossRef]
- Oura, S.; Kazi, S.; Savolainen, A.; Nozawa, K.; Castaneda, J.; Yu, Z.; Miyata, H.; Matzuk, R.M.; Hansen, J.N.; Wachten, D.; et al. Cfap97d1 is important for flagellar axoneme maintenance and male mouse fertility. PLoS Genet. 2020, 16, e1008954. [Google Scholar] [CrossRef]
- Cacina, C.; Turan Sürmen, S.; Arıkan, S.; Pençe, S.; Yaylım, İ. The influence of CASP8 D302H gene variant in colorectal cancer risk and prognosis. Turk. J. Biochem. 2023, 48, 234–238. [Google Scholar] [CrossRef]
- Yu, S.; Long, F.; Wei, X.; Gu, H.; Hao, Z. Myeloid PGGT1B Deficiency Promotes Psoriasiform Dermatitis by Promoting the Secretion of Inflammatory Factors. Int. J. Mol. Sci. 2025, 26, 4901. [Google Scholar] [CrossRef]
- Alloza, I.; Salegi, A.; Mena, J.; Navarro, R.T.; Martin, C.; Aspichueta, P.; Salazar, L.M.; Carpio, J.U.; Cagigal, P.D.; Vega, R.; et al. BIRC6 Is Associated with Vulnerability of Carotid Atherosclerotic Plaque. Int. J. Mol. Sci. 2020, 21, 9387. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Qiao, Z.; Duan, Q.; Nevo, E. Adaptation of mammals to hypoxia. Anim. Model. Exp. Med. 2021, 4, 311–318. [Google Scholar] [CrossRef] [PubMed]
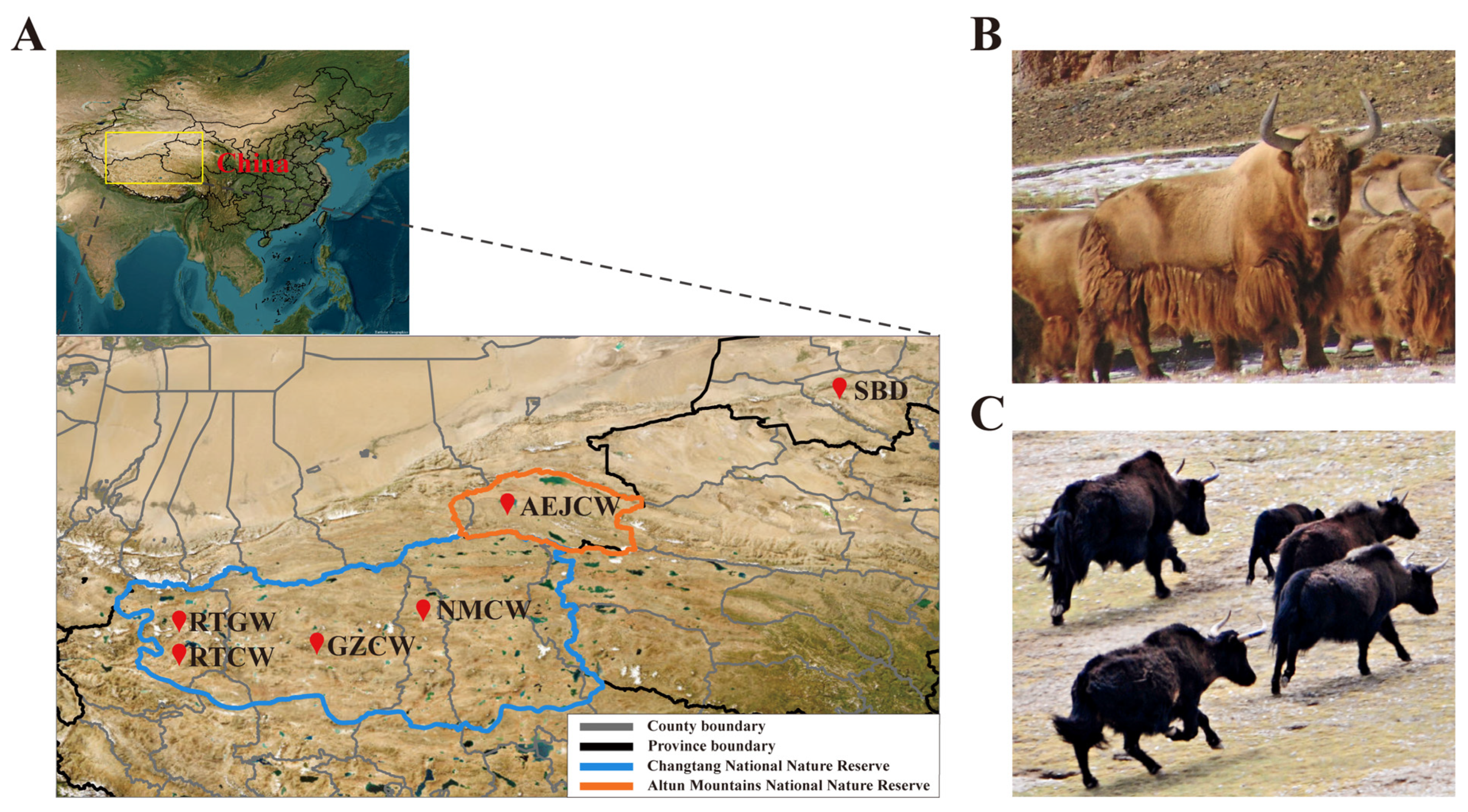
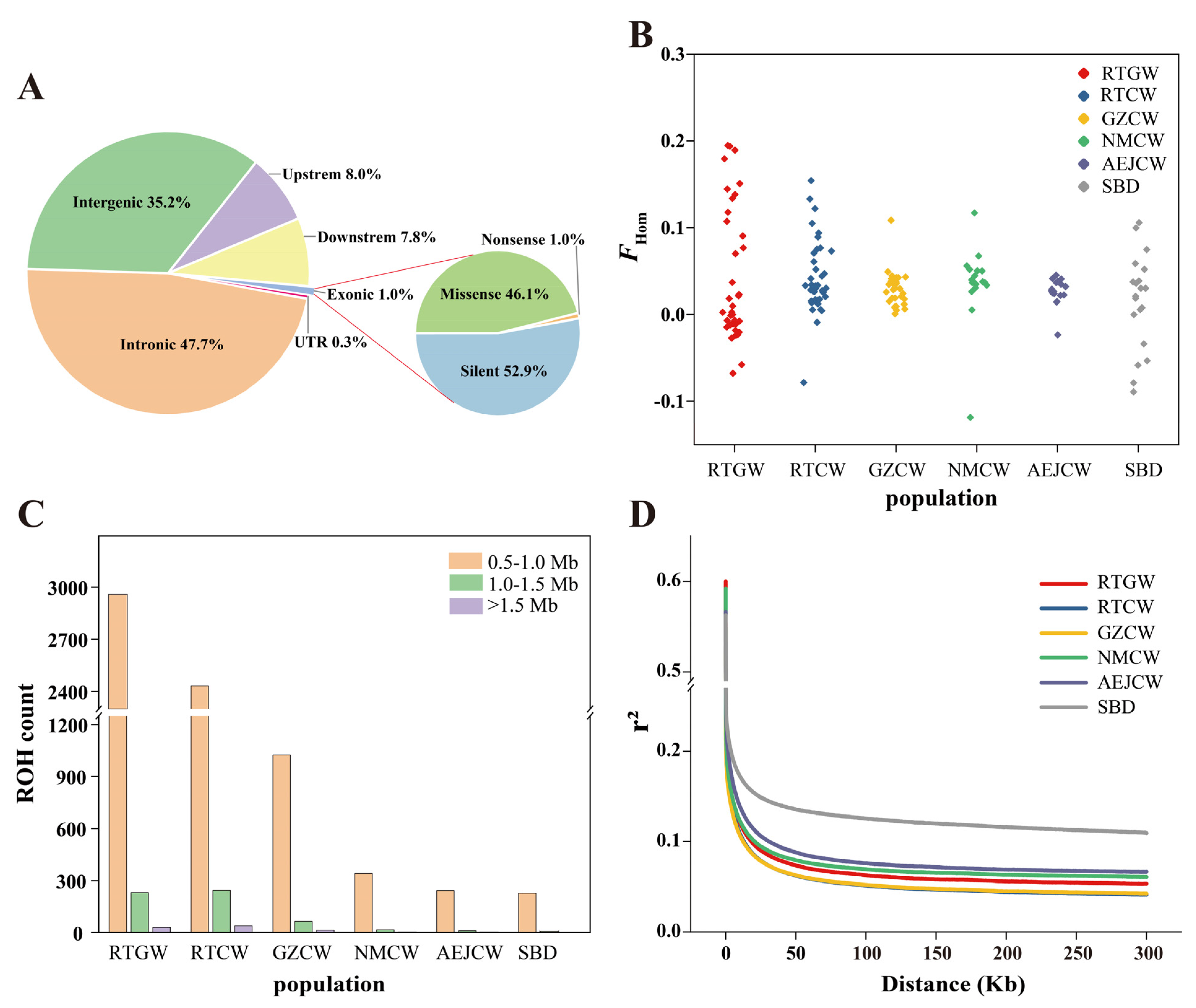
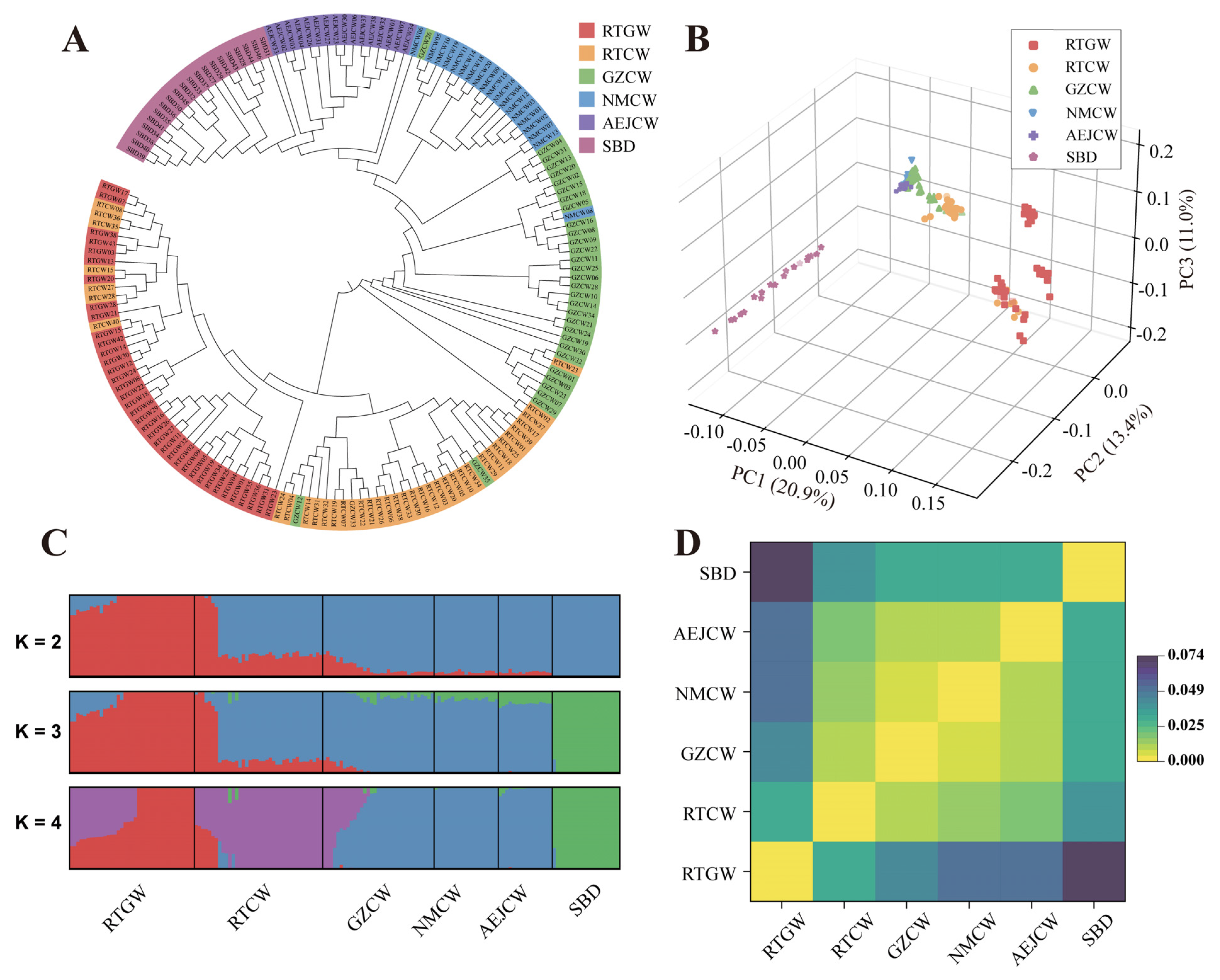
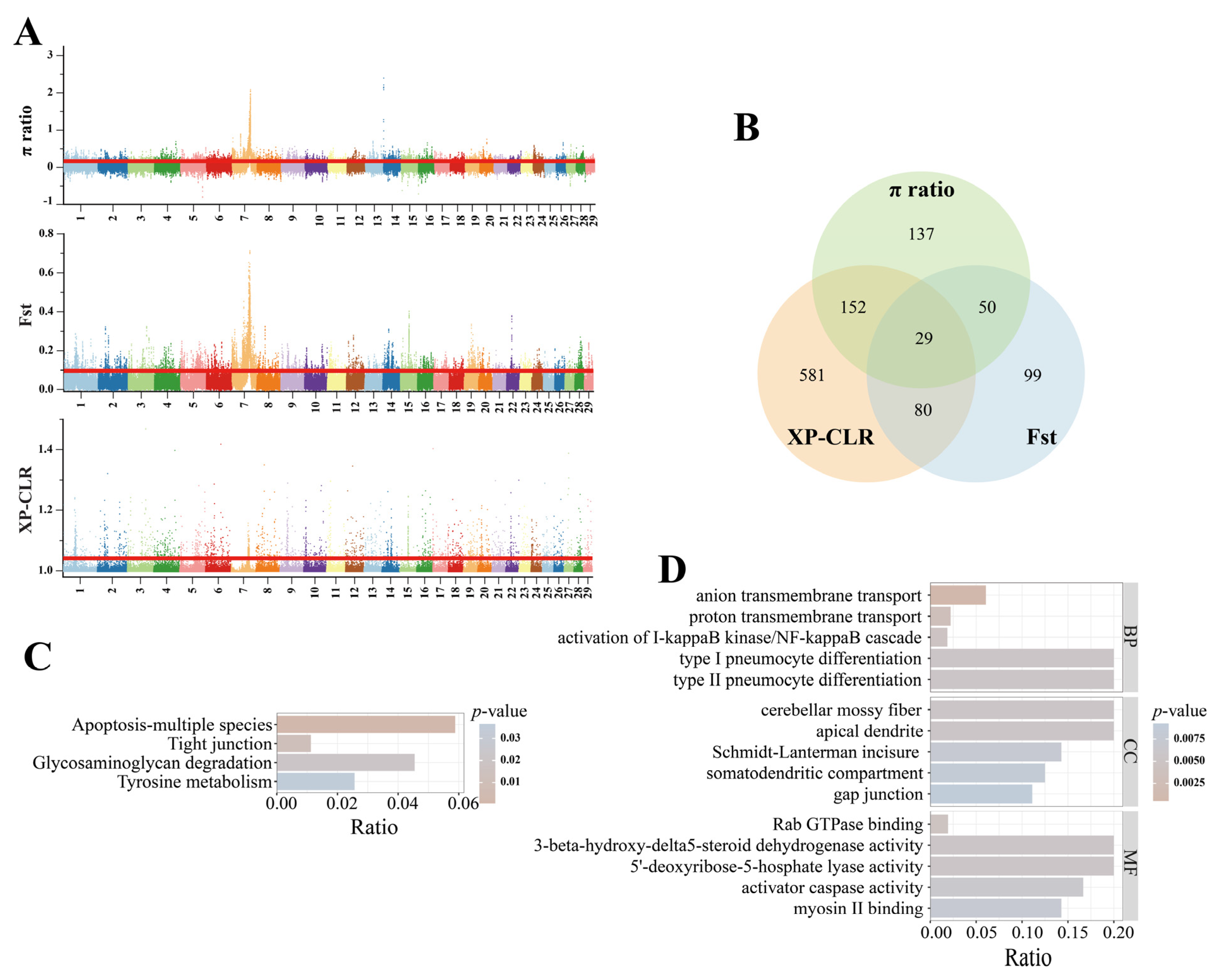
| Population | Sample Size | π | Ho | He | FHom |
|---|---|---|---|---|---|
| RTGW | 37 | 0.00148 | 0.213 | 0.222 | 0.043 |
| RTCW | 38 | 0.00156 | 0.205 | 0.214 | 0.042 |
| GZCW | 33 | 0.00160 | 0.201 | 0.207 | 0.029 |
| NMCW | 19 | 0.00162 | 0.215 | 0.223 | 0.035 |
| AEJCW | 16 | 0.00162 | 0.238 | 0.244 | 0.027 |
| SBD | 20 | 0.00157 | 0.225 | 0.229 | 0.015 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Cong, W.; Li, X.; Wang, L.; Jin, K.; Zhang, Y. Whole-Genome Resequencing Reveals Population Genetic Structure and Selection Signatures in the Golden Wild Yak. Diversity 2025, 17, 687. https://doi.org/10.3390/d17100687
Yu J, Cong W, Li X, Wang L, Jin K, Zhang Y. Whole-Genome Resequencing Reveals Population Genetic Structure and Selection Signatures in the Golden Wild Yak. Diversity. 2025; 17(10):687. https://doi.org/10.3390/d17100687
Chicago/Turabian StyleYu, Jianhua, Wei Cong, Xiuming Li, Lu Wang, Kun Jin, and Yuguang Zhang. 2025. "Whole-Genome Resequencing Reveals Population Genetic Structure and Selection Signatures in the Golden Wild Yak" Diversity 17, no. 10: 687. https://doi.org/10.3390/d17100687
APA StyleYu, J., Cong, W., Li, X., Wang, L., Jin, K., & Zhang, Y. (2025). Whole-Genome Resequencing Reveals Population Genetic Structure and Selection Signatures in the Golden Wild Yak. Diversity, 17(10), 687. https://doi.org/10.3390/d17100687







