Phylogenetic Framework, a New Species, and Two New Species Records for China in Rhizopogon
Abstract
1. Introduction
2. Materials and Methods
2.1. Morphological Characteristics Examination
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Phylogenetic Analyses
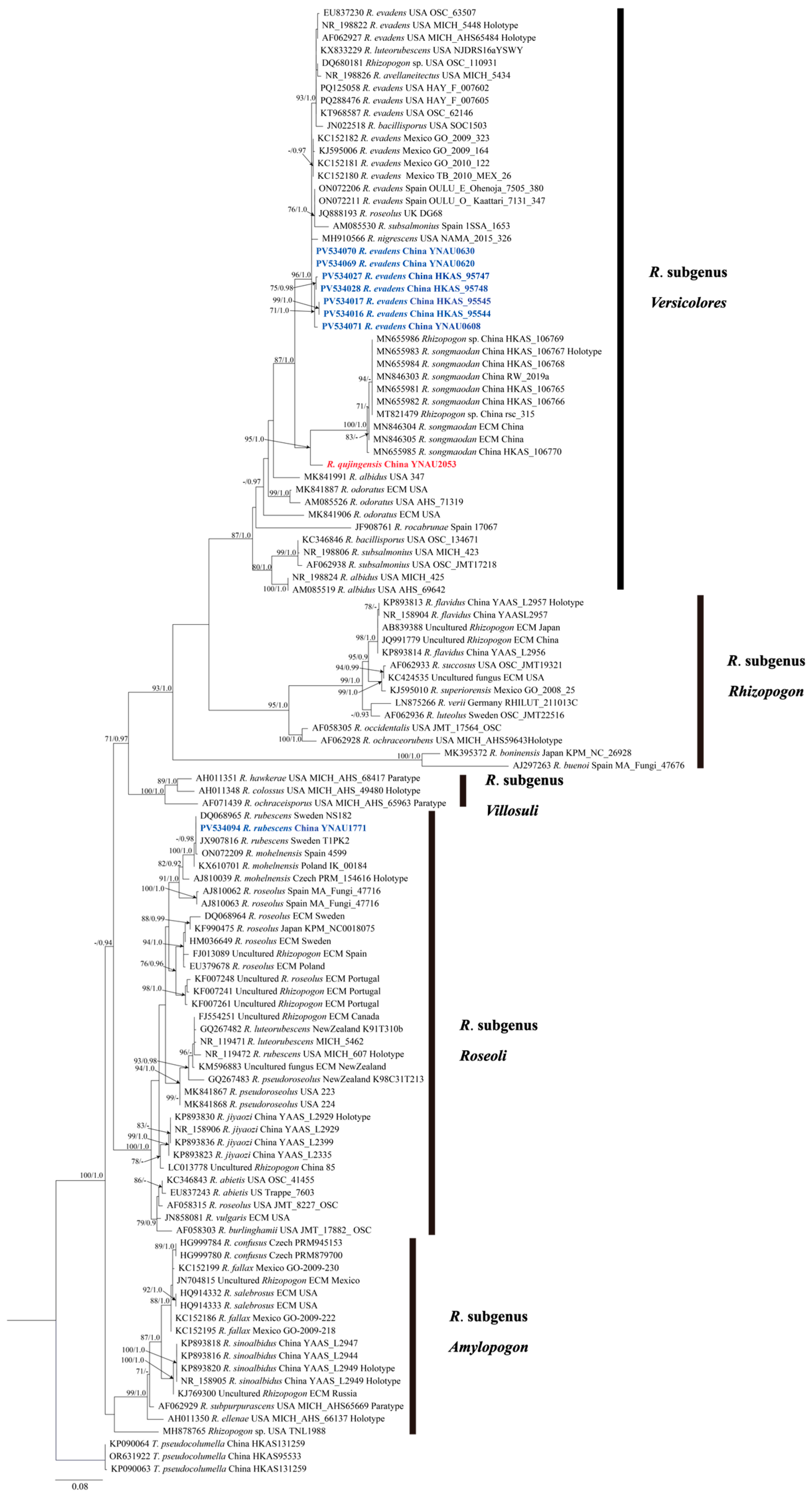
3. Results
3.1. Molecular Phylogenetics
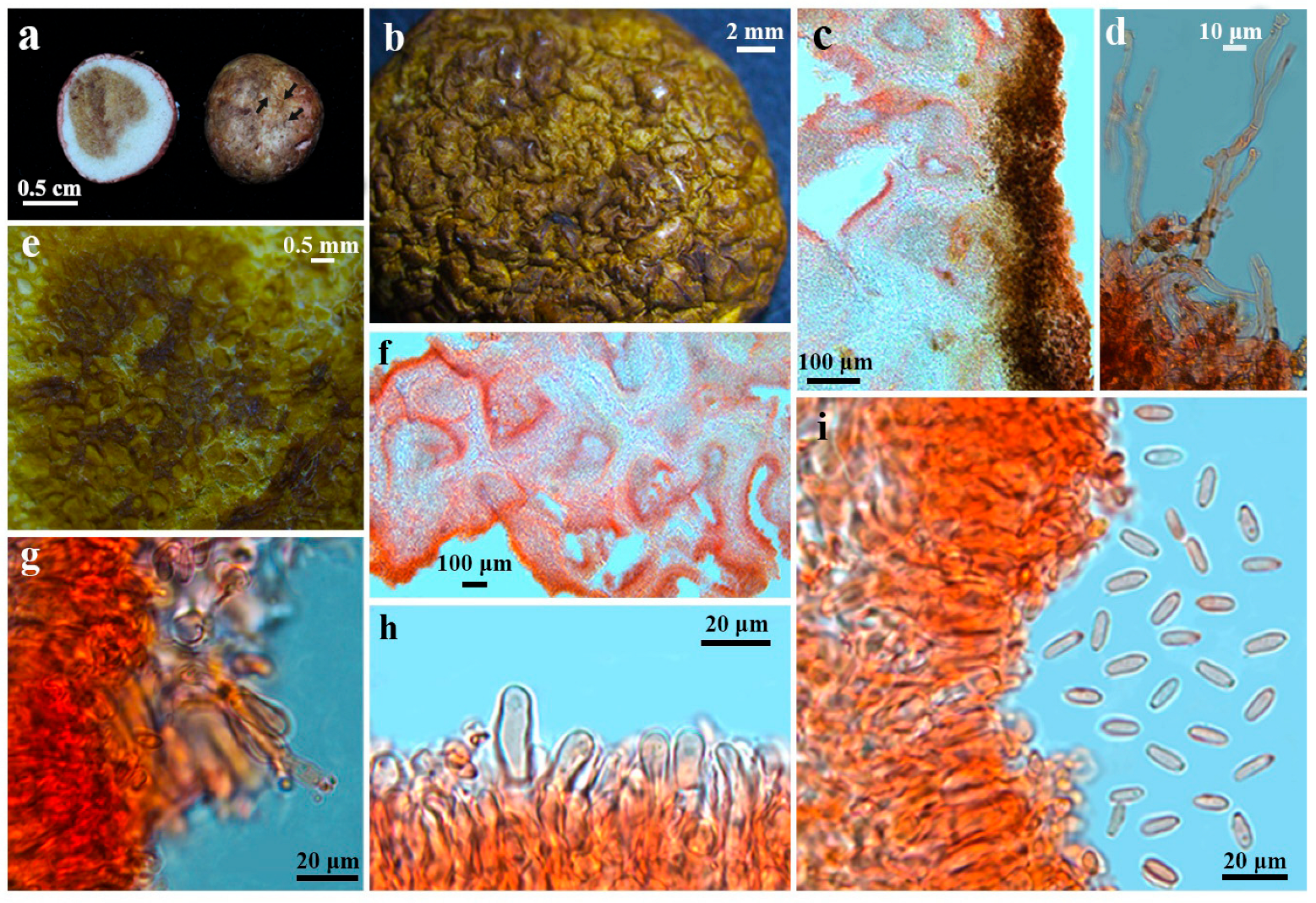
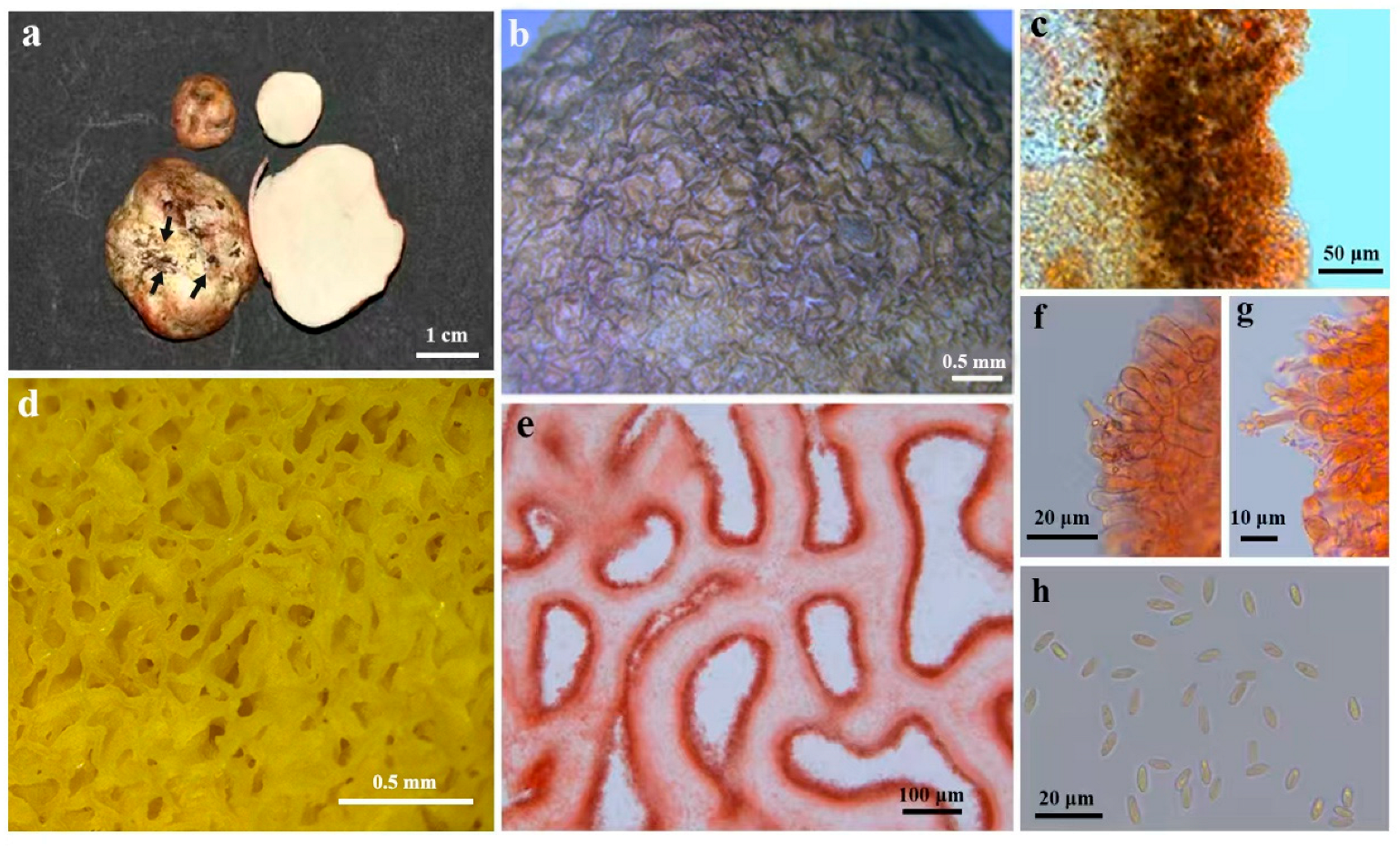
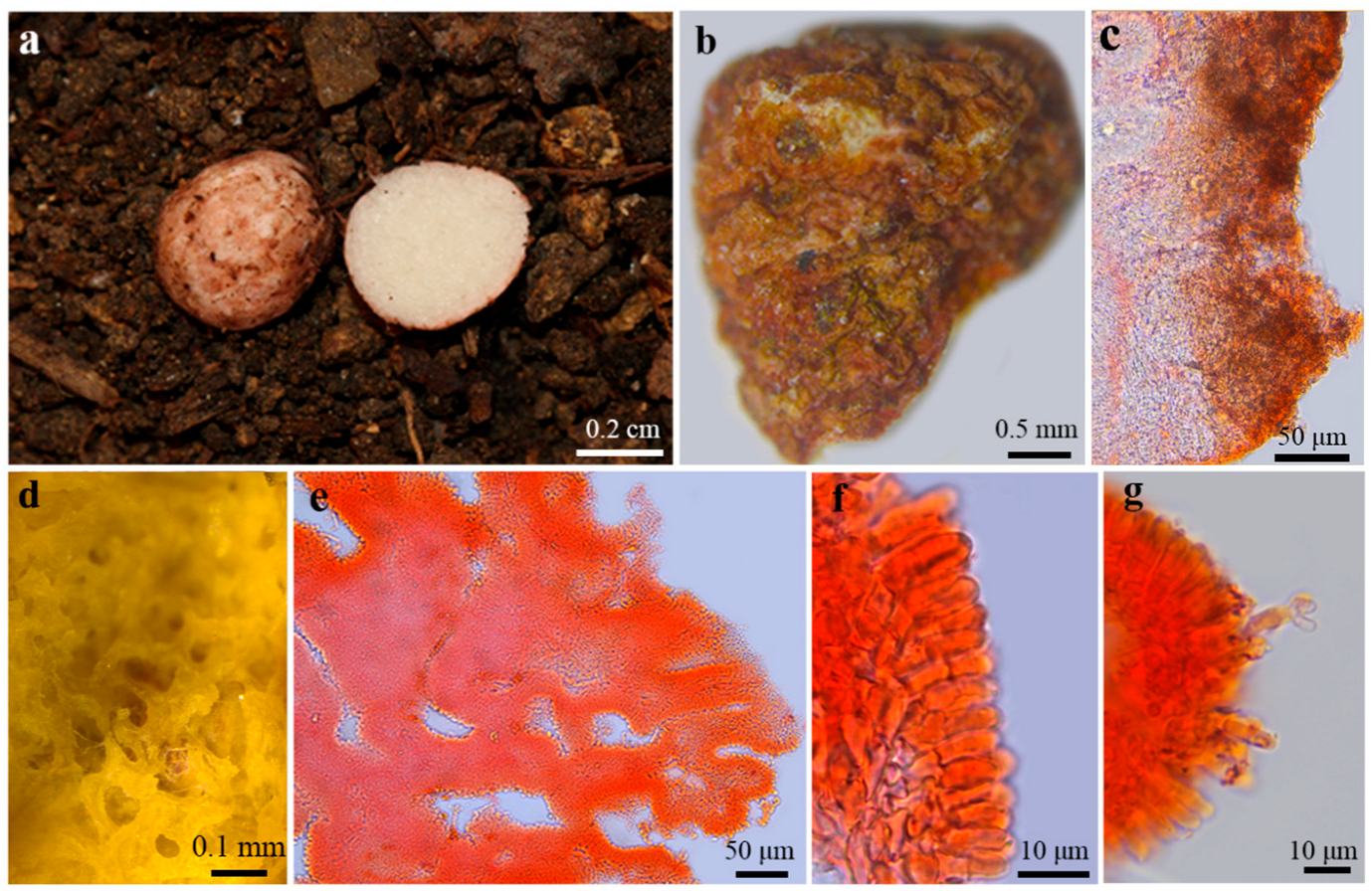
3.2. Taxonomy
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Species Name | Voucher/Isolation | Origin | GenBank No. of ITS | References |
|---|---|---|---|---|
| R. evadens | GO-2009-164 | Mexico | KJ595006 | Unpublished |
| Uncultured Rhizopogon | ECM | Russia | KJ769300 | [48] |
| R. superiorensis | GO-2008-25 | Mexico | KJ595010 | Unpublished |
| Uncultured R. roseolus | ECM | Portugal | KF007248 | Unpublished |
| Uncultured Rhizopogon | ECM | Portugal | KF007241 | Unpublished |
| R. roseolus | KPM-NC0018075 | Japan | KF990475 | [49] |
| Uncultured Rhizopogon | ECM | Portugal | KF007261 | Unpublished |
| R. roseolus | MA-Fungi 47716 | Spain | AJ810062 | [26] |
| R. roseolus | MA-Fungi 47716 | Spain | AJ810063 | [26] |
| Uncultured Rhizopogon | ECM | Spain | FJ013089 | Unpublished |
| Uncultured Rhizopogon | ECM | Canada | FJ554251 | [50] |
| R. roseolus | ECM | Poland | EU379678 | Unpublished |
| R. abietis | Trappe 7603 | US | EU837243 | Unpublished |
| R. evadens | OSC 63507 | USA | EU837230 | Unpublished |
| R. roseolus | ECM | Sweden | HM036649 | [51] |
| R. pseudoroseolus | K98C31T213 | New Zealand | GQ267483 | [52] |
| R. luteorubescens | K91T310b | New Zealand | GQ267482 | [52] |
| R. abietis | OSC: 41455 | USA | KC346843 | Unpublished |
| R. fallax | GO-2009-222 | Mexico | KC152186 | Unpublished |
| R. fallax | GO-2009-230 | Mexico | KC152199 | Unpublished |
| R. fallax | GO-2009-218 | Mexico | KC152195 | Unpublished |
| R. evadens | GO-2010-122 | Mexico | KC152181 | Unpublished |
| Uncultured fungus | ECM | USA | KC424535 | [53] |
| R. luteorubescens | NJDRS16aYSWY | USA | KX833229 | [54] |
| R. jiyaozi | YAAS-L2399 | China | KP893836 | [30] |
| R. sinoalbidus | YAAS-L2947 | China | KP893818 | [30] |
| Uncultured Rhizopogon | 85 | China | LC013778 | [55] |
| R. vulgaris | ECM | USA | JN858081 | [56] |
| R. bacillisporus | SOC1503 | USA | JN022518 | Unpublished |
| Uncultured Rhizopogon | ECM | Mexico | JN704815 | Unpublished |
| R. salebrosus | ECM | USA | HQ914333 | [57] |
| R. salebrosus | ECM | USA | HQ914332 | [57] |
| Uncultured Rhizopogon | ECM | Japan | AB839388 | [58] |
| Uncultured fungus | ECM | New Zealand | KM596883 | Unpublished |
| R. roseolus | DG68 | UK | JQ888193 | [59] |
| Uncultured Rhizopogon | ECM | China | JQ991779 | Unpublished |
| R. flavidus | YAAS: L2957 | China | NR_158904 | [30] |
| R. sinoalbidus | YAAS: L2949 (Holotype) | China | NR_158905 | [30] |
| R. jiyaozi | YAASL: 2929 | China | NR_158906 | [30] |
| R. succosus | OSC JMT19321 | USA | AF062933 | [22] |
| R. luteolus | OSC JMT22516 | Sweden | AF062936 | [22] |
| R. verii | RHILUT_211013C | Germany | LN875266 | [29] |
| R. songmaodan | HKAS-106768 | China | MN655984 | [10] |
| R. songmaodan | RW_2019a | China | MN846303 | [10] |
| R. nigrescens | NAMA 2015-326 | USA | MH910566 | Unpublished |
| R. pseudoroseolus | 223 | USA | MK841867 | Unpublished |
| R. albidus | 347 | USA | MK841991 | Unpublished |
| R. pseudoroseolus | 224 | USA | MK841868 | Unpublished |
| Rhizopogon sp. | Trappe 28417; SC 110931 | USA | DQ680181 | Unpublished |
| R. subsalmonius | 1SSA-1653 | Spain | AM085530 | Unpublished |
| R. subsalmonius | OSC JMT17218 | USA | AF062938 | [22] |
| R. albidus | AHS 69642 | USA | AM085519 | Unpublished |
| R. evadens | MICH AHS 65484 (Holotype) | USA | AF062927 | [22] |
| R. odoratus | AHS 71319 | USA | AM085526 | Unpublished |
| R. rocabrunae | 17067 | Spain | JF908761 | [60] |
| R. occidentalis | JMT 17564, OSC | USA | AF058305 | [22] |
| R. ochraceorubens | MICH AHS 59643 (Holotype) | USA | AF062928 | [22] |
| R. flavidus | YAAS-L2957 (Holotype) | China | KP893813 | [30] |
| R. flavidus | YAAS-L2956 | China | KP893814 | [30] |
| R. subpurpurascens | MICH AHS 65669 (Paratype) | USA | AF062929 | [22] |
| R. ellenae | MICH AHS 66137 (Holotype) | USA | AH011350 | [22] |
| R. sinoalbidus | YAAS-L2944 | China | KP893816 | [30] |
| R. sinoalbidus | YAAS-L2949 (Holotype) | China | KP893820 | [30] |
| R. colossus | MICH AHS 49480 (Holotype) | USA | AH011348 | [22] |
| R. hawkerae | MICH AHS 68417 (Paratype) | USA | AH011351 | [22] |
| R. ochraceisporus | MICH AHS 65963 (Paratype) | USA | AF071439 | [22] |
| R. jiyaozi | YAAS-L2335 | China | KP893823 | [30] |
| R. jiyaozi | YAASL-2929 (Holotype) | China | KP893830 | [30] |
| R. roseolus | JMT 8227, OSC | USA | AF058315 | [22] |
| R. burlinghamii | JMT 17882, OSC | USA | AF058303 | [22] |
| R. buenoi | MA-Fungi 47676 (Holotype) | Spain | AJ297263 | [61] |
| R. boninensis | KPM-NC 26928 | Japan | MK395372 | Unpublished |
| R. songmaodan | HKAS-106767 (Holotype) | China | MN655983 | [10] |
| R. songmaodan | HKAS-106766 | China | MN655982 | [10] |
| R. mohelnensis | IK-00184 | Poland | KX610701 | [62] |
| R. rubescens | NS182 | Sweden | DQ068965 | [63] |
| R. rubescens | T1PK2 | Sweden | JX907816 | [64] |
| R. mohelnensis | 4599 | Spain | ON072209 | [65] |
| R. albidus | AHS 69642 | USA | AM085519 | Unpublished |
| R. albidus | MICH: 425 | USA | NR_198824 | Unpublished |
| R. evadens | MICH: 5448 (Holotype) | USA | NR_198822 | Unpublished |
| R. avellaneitectus | MICH: 5434 | USA | NR_198826 | Unpublished |
| R. subsalmonius | MICH: 423 | USA | NR_198806 | Unpublished |
| R. odoratus | Mycorrhizal root | USA | MK841906 | Unpublished |
| R. odoratus | Mycorrhizal root | USA | MK841887 | Unpublished |
| R. bacillisporus | OSC: 134671 | USA | KC346846 | Unpublished |
| Rhizopogon sp. | TNL1988 | USA | MH878765 | Unpublished |
| R. confusus | PRM945153 | Czech | HG999784 | [2] |
| R. confusus | PRM879700 | Czech | HG999780 | [2] |
| R. songmaodan | rsc-315 | China | MT821479 | [10] |
| R. songmaodan | Synthesized ECM of Pinus armandii | China | MN846304 | [10] |
| R. songmaodan | Synthesized ECM of P. armandii | China | MN846305 | [10] |
| R. songmaodan | HKAS-106770 | China | MN655985 | [10] |
| R. songmaodan | HKAS-106769 | China | MN655986 | [10] |
| R. songmaodan | HKAS-106765 | China | MN655981 | [10] |
| R. evadens | HAY-F-007602 | USA | PQ125058 | Unpublished |
| R. evadens | OSC 62146 | USA | KT968587 | Unpublished |
| R. evadens | OULU: E.Ohenoja 7505-380 | Spain | ON072206 | [65] |
| R. evadens | OULU: O.Kaattari 7131-347 | Spain | ON072211 | [65] |
| R. evadens | TB-2010-MEX 26 | Mexico | KC152180 | Unpublished |
| R. evadens | GO-2009-323 | Mexico | KC152182 | Unpublished |
| R. evadens | HAY-F-007605 | USA | PQ288476 | Unpublished |
| R. rubescens | MICH: 607 (Holotype) | USA | NR_119472 | [26] |
| R. mohelnensis | PRM 154616 (Holotype) | Czech | AJ810039 | [26] |
| R. roseolus | ECM | Sweden | DQ068964 | [63] |
| R. luteorubescens | MICH: 5462 | USA | NR_119471 | [26] |
| T. pseudocolumella | HKAS131259 | China | KP090064 | Unpublished |
| T. pseudocolumella | HKAS131259 | China | KP090063 | Unpublished |
| T. pseudocolumella | HKAS95533 | China | OR631922 | Unpublished |
References
- Fries, E.M. Symbolae Gasteromycorum; Ex Officina Berlingiana: Lund, Sweden, 1817. [Google Scholar]
- Koukol, O.; Valda, S.; Gaisler, J.; Kunca, V.; Dowie, N.J. Rhizopogon confusus sp. nov., a correct name for a fungus previously recorded in Central Europe as the North American Rhizopogon salebrosus. Mycol. Prog. 2022, 21, 49. [Google Scholar] [CrossRef]
- Smith, A.H.; Zeller, S.M. A preliminary account of the North American species of Rhizopogon. Mem. N. Y. Bot. Gard. 1966, 14, 1–178. [Google Scholar]
- Massicotte, H.B.; Molina, R.; Luoma, D.L.; Smith, J.E. Biology of the ectomycorrhizal genus, Rhizopogon. 2. Patterns of host-fungus specificity following spore inoculation of diverse hosts grown in monoculture and dual culture. New Phytol. 1994, 126, 677–690. [Google Scholar]
- Molina, R.; Massicotte, H.B.; Trappe, J.M. Biology of the ectomycorrhizal genus, Rhizopogon.1. Host associations, host-specificity and pure culture syntheses. New Phytol. 1994, 126, 653–675. [Google Scholar]
- Trappe, J.M.; Molina, R.; Luoma, D.L.; Cázares, E.; Pilz, D.; Smith, J.E.; Castellano, M.A.; Miller, S.L.; Trappe, M.J. Diversity, Ecology, and Conservation of Truffle Fungi in Forests of the Pacific Northwest; US Department of Agriculture, Forest Service, Pacific Northwest Research Station: Corvallis, OR, USA, 2009; Volume 772, pp. 1–194. [Google Scholar]
- Bubriski, R.; Kennedy, P. A molecular and morphological analysis of the genus Rhizopogon subgenus Villosuli section Villosuli as a preface to ecological monitoring. Mycologia 2014, 106, 353–361. [Google Scholar] [CrossRef]
- Kawai, M.; Yamahara, M.; Ohta, A. Bipolar incompatibility system of an ectomycorrhizal basidiomycete, Rhizopogon rubescens. Mycorrhiza 2008, 18, 205–210. [Google Scholar] [CrossRef]
- Molina, R.; Trappe, J.M.; Grubisha, L.C.; Spatafora, J.W. Rhizopogon. In Ectomycorrhizal Fungi: Key Genera in Profile; Cairney, J.W.G., Chambers, S.M., Eds.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 129–161. [Google Scholar]
- Wang, R.; Yu, F.Q.; Jesús, P.M.; Carlos, C. A new edible Rhizopogon species from Southwest China, and its mycorrhizal synthesis with two native pines. Mycorrhiza 2020, 30, 85–92. [Google Scholar] [CrossRef]
- Shimomura, N.; Matsuda, M.; Ariyoshi, K.; Aimi, T. Development of mycelial slurries containing surfactant for cultivation of the edible ectomycorrhizal mushroom Rhizopogon roseolus (syn. Rhizopogon rubescens). Botany 2012, 90, 839–844. [Google Scholar] [CrossRef]
- Wang, Y.; Hall, I.R.; Dixon, C.; Hance-Halloy, M.; Strong, G.; Brass, P. The cultivation of Lactarius deliciosus (saffron milk cap) and Rhizopogon rubescens (shoro) in New Zealand. In Proceedings of the Second International Conference on Edible Mycorrhizal Mushrooms, Christchurch, New Zealand, 3–6 July 2001; Edible Mycorrhizal Mushrooms and Their Cultivation. Hall, I.R., Wang, Y., Danell, E., Zambonelli, A., Eds.; New Zealand Institute for Crop & Food Research: Christchurch, New Zealand, 2002. [Google Scholar]
- Pacioni, G. Italian hypogeous fungi. Mycologia 1984, 76, 988–997. [Google Scholar]
- Pietras, M.; Kolanowska, M. Predicted potential occurrence of the North American false truffle Rhizopogon salebrosus (AH Sm.) in Europe. Fungal Ecol. 2019, 39, 225–230. [Google Scholar] [CrossRef]
- Martín, M.P.; Calonge, F.D. Rhizopogon sect. Fulvigleba in Europe: A taxonomic revision. Mycotaxon 2006, 95, 229–240. [Google Scholar]
- Sugiyama, Y.; Murata, M.; Nara, K. A new Rhizopogon species associated with Pinus amamiana in Japan. Mycoscience 2018, 59, 176–180. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Maximov, T.C.; Sugimoto, A.; Nara, K. Discovery of Rhizopogon associated with Larix from northeastern Siberia: Insights into host shift of ectomycorrhizal fungi. Mycoscience 2019, 60, 274–280. [Google Scholar] [CrossRef]
- Kumar, A.; Tapwal, A.; Kumar, D.; Yadav, R. Ectomycorrhizas of Rhizopogon himalayensis on Cedrus deodara. J. Basic Microbiol. 2024, 64, 2300616. [Google Scholar] [CrossRef]
- Grubisha, L.C. Systematics of the Genus Rhizopogon Inferred from Nuclear Ribosomal DNA Large Subunit and Internal Transcribed Spacer Sequences. Master’s Thesis, Oregon State University, Corvallis, OR, USA, 1998. [Google Scholar]
- Martín, M.P.; Högberg, N.; Nylund, J.-E. Molecular analysis confirms morphological reclassification of Rhizopogon. Mycol. Res. 1998, 102, 855–858. [Google Scholar] [CrossRef]
- Grubisha, L.C.; Trappe, J.M.; Molina, R.; Spatafora, J.W. Biology of the ectomycorrhizal genus Rhizopogon. V. Phylogenetic relationships in the Boletales inferred from LSU rDNA sequences. Mycologia 2001, 93, 82–89. [Google Scholar] [CrossRef]
- Grubisha, L.C.; Trappe, J.M.; Molina, R.; Spatafora, J.W. Biology of the ectomycorrhizal genus Rhizopogon. VI. Re-examination of infrageneric relationships inferred from phylogenetic analyses of ITS sequences. Mycologia 2002, 94, 381–619. [Google Scholar] [CrossRef]
- Grubisha, L.C.; Trappe, J.M.; Beyerle, A.R.; Wheeler, D. NATS truffle and truffle-like fungi 12: Rhizopogon ater sp. nov. and R. brunsii sp. nov. (Rhizopogonaceae, Basidiomycota). Mycotaxon 2005, 93, 345–353. [Google Scholar]
- Kretzer, A.M.; Luoma, D.L.; Molina, R.; Spatafora, J.W. Taxonomy of the Rhizopogon vinicolor species complex based on analysis ofITS sequences and microsatellite loci. Mycologia 2003, 95, 480–487. [Google Scholar] [CrossRef]
- Binder, M.; Hibbett, D.S. Molecular systematics and biological diversification of Boletales. Mycologia 2006, 98, 971–981. [Google Scholar] [CrossRef]
- Martín, M.P.; García, M.A. How many species in the Rhizopogon roseolus group? Mycotaxon 2009, 109, 111–128. [Google Scholar] [CrossRef]
- Trappe, J.M. A revision of the genus Alpova with notes on Rhizopogon and the Melanogastraceae. Beih. Nova Hedwigia 1975, 51, 279–309. [Google Scholar]
- Martín, M.P. The Genus Rhizopogon in Europe; Edicions de la Universitat de Barcelona (BCG): Barcelona, Spain, 1996. [Google Scholar]
- Sulzbacher, M.A.; Grebenc, T.; Garcia, M.A.; Silva, B.D.B.; Silveira, A.; Antoniolli, Z.I.; Marinho, P.; Munzenberger, B.; Telleria, M.T.; Baseia, I.G.; et al. Molecular and morphological analyses confirm Rhizopogon verii as a widely distributed ectomycorrhizal false truffle in Europe, and its presence in South America. Mycorrhiza 2016, 26, 377–388. [Google Scholar] [CrossRef]
- Li, L.; Zhao, Y.C.; Zhou, D.Q.; Yu, F.Q.; Zheng, L.; Wang, Y.; Zhang, X.L.; Duan, Z.J.; Zhao, X.Y.; He, Z.H.; et al. Three new species of Rhizopogon from Southwest China. Phytotaxa 2016, 282, 151–163. [Google Scholar] [CrossRef]
- Liu, B. New species and new records of hypogous fungi from China I. Acta Mycol. Sin. 1985, 4, 84–89. [Google Scholar]
- Tao, K.; Chang, M.C. The hypogeous fungi on the Rhizopogon found in Shanxi (I). J. Shanxi Agric Univ. 1988, 8, 226–229. [Google Scholar]
- Yu, F.Q.; Liu, P.G. Species diversity of wild edible mushroom from Pinus yunnanensis forest and conservation strategies. Biodivers. Sci. 2005, 13, 58–69. [Google Scholar] [CrossRef]
- Dai, Y.C.; Yang, Z.L. A revised checklist of medicinal fungi in China. Mycosystema 2008, 27, 801–824. [Google Scholar]
- Dai, Y.C.; Zhou, L.W.; Yang, Z.L.; Wen, H.A.; Bau, T.; Li, T.H. A revised checklist of edible fungi in China. Mycosystema 2010, 29, 1–21. [Google Scholar]
- Shao, D.H.; Yang, X.P.; Zhang, X.D.; Bai, S.L.; Zheng, R.; Wang, J.G. Ectomycorrhiza formation on Pinus tabulaeformis through Rhizopogon luteolus infection. Chin. J. Ecol. 2013, 32, 78–81. [Google Scholar]
- Liu, P.G. Rhizopogon. In Flora Fungorum Sinicorum; Science Press: Beijing, China, 2005; Volume 17, pp. 1–183. [Google Scholar]
- Fan, L. Research Progress on Species Diversity and Phylogeny of Hypogeous Fungi in China. J. Fungal Res. 2023, 21, 65–81. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Nylander, J.A.A. MrModeltest, version 2.3; Program Distributed by the Author; Evolutionary Biology Center, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Stamatakis, A. RAxML-VI-HPC: Maximum-likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Zotti, M.; Di Piazza, S.; Vizzini, A. First records of Rhizopogon rocabrunae and R. pumilionum(Boletales) from Italy. Mycotaxon 2010, 113, 291–300. [Google Scholar]
- Martín, M.P.; Calonge, F.D. Rhizopogon aromaticus (Basidiomycotina) a new species found in Spain. Mycotaxon 2000, 76, 289–292. [Google Scholar]
- Trappe, M.; Evans, F.; Trappe, J. Field Guide to North American Truffles: Hunting, Identifying, and Enjoying the World’s Most Prized Fungi; Ten Speed Press: Berkeley, CA, USA, 2007. [Google Scholar]
- Malysheva, V.F.; Malysheva, E.F.; Kovalenko, A.E.; Pimenova, E.A.; Gromyko, M.N.; Bondarchuk, S.N. Ectomycorrhizal fungal diversity of Pinus koraiensis in the forests of the Central Sikhote-Alin based on rDNA sequence analysis of mycorrhizal tips. Mycol. Phytopathol. 2014, 48, 372–385. [Google Scholar]
- Orihara, T.; Okada, T.; Daiguji, T.; Takagi, N. Occurrence of a Truffle-like Fungus, Rhizopogon roseolus (Rhizopogonaceae, Boletales) in Kanagawa Prefecture, Japan. Kanagawa Kenritsu Hakubutsukan Kenkyu Hokoku Shizen Kagaku, 2014; in press. [Google Scholar]
- Hartmann, M.; Lee, S.; Hallam, S.J.; Mohn, W.W. Bacterial, archaeal and eukaryal community structures throughout soil horizons of harvested and naturally disturbed forest stands. Environ. Microbiol. 2009, 11, 3045–3062. [Google Scholar] [CrossRef]
- Menkis, A.; Vasaitis, R. Fungi in Roots of Nursery Grown Pinus sylvestris: Ectomycorrhizal Colonisation, Genetic Diversity and Spatial Distribution. Microb. Ecol. 2011, 61, 52–63. [Google Scholar] [CrossRef]
- Walbert, K.; Ramsfield, T.D.; Ridgway, H.J.; Jones, E.E. Ectomycorrhizal species associated with Pinus radiata in New Zealand including novel associations determined by molecular analysis. Mycorrhiza 2010, 20, 209–215. [Google Scholar]
- Cowden, C.C.; Peterson, C.J. Annual and Seasonal Dynamics of Ectomycorrhizal Fungi Colonizing White Pine (Pinus strobus) Seedlings Following Catastrophic Windthrow in Northern Georgia, USA. Can. J. For. Res. 2013, forthcoming. [Google Scholar] [CrossRef]
- Dowie, N.J.; Grubisha, L.C.; Burton, B.A.; Klooster, M.R.; Miller, S.L. Increased phylogenetic resolution within the ecologically important Rhizopogon subgenus Amylopogon using 10 anonymous nuclear loci. Mycologia 2017, 109, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Long, D.; Liu, J.; Han, Q.; Wang, X.; Huang, J. Ectomycorrhizal fungal communities associated with Populus simonii and Pinus tabuliformis in the hilly-gully region of the Loess Plateau, China. Sci. Rep. 2016, 6, 24336. [Google Scholar] [CrossRef] [PubMed]
- Peay, K.G.; Schubert, M.G.; Nguyen, N.H.; Bruns, T.D. Measuring Ectomycorrhizal Fungal Dispersal: Macroecological Patterns Driven by Microscopic Propagules. Mol. Ecol. 2012, 21, 4122–4136. [Google Scholar] [CrossRef]
- Dowie, N.J.; Hemenway, J.J.; Miller, S.L. Identity, genetic lineages and putative hybrids of an obligate mycobiont associated with the mycoheterotrophic plant Pterospora andromedea in the south-central Rocky Mountains. Fungal Ecol. 2012, 5, 137–146. [Google Scholar] [CrossRef]
- Huang, J.; Nara, K.; Zong, K.; Lian, C. Soil propagule banks of ectomycorrhizal fungi along forest development stages after mining. Microb. Ecol. 2015, 69, 768–777. [Google Scholar]
- Pickles, B.J.; Genney, D.R.; Anderson, I.C.; Alexander, I.J. Spatial analysis of ectomycorrhizal fungi reveals that root tip communities are structured by competitive interactions. Mol. Ecol. 2012, 21, 5110–5123. [Google Scholar] [CrossRef]
- Osmundson, T.W.; Robert, V.A.; Schoch, C.L.; Baker, L.J.; Smith, A.; Robich, G.; Mizzan, L.; Garbelotto, M.M. Filling Gaps in Biodiversity Knowledge for Macrofungi: Contributions and Assessment of an Herbarium Collection DNA Barcode Sequencing Project. PLoS ONE 2013, 8, e62419. [Google Scholar]
- Martín, M.P.; Calonge, F.D. Rhizopogon buenoi (Boletales, Basidiomycota) a new species from Spain. Mycotaxon 2001, 79, 101–105. [Google Scholar]
- Kalucka, I.L.; Jagodzinski, A.M.; Nowinski, M. Biodiversity of Ectomycorrhizal Fungi in Surface Mine Spoil Restoration Stands in Poland—First Time Recorded, Rare, and Red-Listed Species. Acta Mycol. 2016, 51, 1080. [Google Scholar] [CrossRef]
- Menkis, A.; Vasiliauskas, R.; Taylor, A.F.; Stenlid, J.; Finlay, R. Fungal communities in mycorrhizal roots of conifer seedlings in forest nurseries under different cultivation systems, assessed by morphotyping, direct sequencing and mycelial isolation. Mycorrhiza 2005, 16, 33–41. [Google Scholar] [CrossRef]
- Klavina, D.; Gaitnieks, T.; Menkis, A. Survival, Growth and Ectomycorrhizal Community Development of Container- and Bare-Root Grown Pinus sylvestris and Picea abies Seedlings Outplanted on a Forest Clear-Cut. Balt. For. 2013, 19, 39–49. [Google Scholar]
- Martín, M.P.; Huang, A.; Ortega, M.A. Additions to Rhizopogon (Boletales, Basidiomycota) in Finland and Europe based on molecular and morphological evidence. Ann. Bot. Fenn. 2023, 60, 67–70. [Google Scholar] [CrossRef]
| Species Name | Voucher/Isolation | Origin | GenBank No. of ITS |
|---|---|---|---|
| R. qujingensis | YNAU2053 | China | PV584203 |
| R. evadens | YNAU0620 | China | PV534069 |
| R. evadens | YNAU0630 | China | PV534070 |
| R. evadens | HKAS95544 | China | PV534016 |
| R. evadens | HKAS95545 | China | PV534017 |
| R. evadens | YNAU0608 | China | PV534071 |
| R. evadens | HKAS95747 | China | PV534027 |
| R. evadens | HKAS95746 | China | PV534028 |
| R. rubescens | YNAU1771 | China | PV534094 |
| R. sinoalbidus | Ectomycorrhiza3117-5 | China | PX368895 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Li, L.; Zheng, Y.; Pérez-Moreno, J.; Li, Y.; Yu, C.; Yu, F.; Wan, S. Phylogenetic Framework, a New Species, and Two New Species Records for China in Rhizopogon. Diversity 2025, 17, 683. https://doi.org/10.3390/d17100683
Zhang H, Li L, Zheng Y, Pérez-Moreno J, Li Y, Yu C, Yu F, Wan S. Phylogenetic Framework, a New Species, and Two New Species Records for China in Rhizopogon. Diversity. 2025; 17(10):683. https://doi.org/10.3390/d17100683
Chicago/Turabian StyleZhang, Huiwen, Lin Li, Yuhao Zheng, Jesús Pérez-Moreno, Yuenan Li, Chengjin Yu, Fuqiang Yu, and Shanping Wan. 2025. "Phylogenetic Framework, a New Species, and Two New Species Records for China in Rhizopogon" Diversity 17, no. 10: 683. https://doi.org/10.3390/d17100683
APA StyleZhang, H., Li, L., Zheng, Y., Pérez-Moreno, J., Li, Y., Yu, C., Yu, F., & Wan, S. (2025). Phylogenetic Framework, a New Species, and Two New Species Records for China in Rhizopogon. Diversity, 17(10), 683. https://doi.org/10.3390/d17100683






