Abstract
Grass shrimp of the genus Palaemon (Crustacea, Decapoda, Palaemonidae) occur worldwide in freshwater and saline wetlands. Palaemon species are frequently misidentified, and the genus itself has been reorganized several times. To clarify the intrageneric phylogenetic relationships and analyze the regional genetic diversity, we sequenced fragments of the mitochondrial cytochrome c oxidase subunit I (COI) and the nuclear Histone H3 (H3) genes from specimens collected along the northern Gulf of Mexico, where several morphologically similar Palaemon species reside. The generated sequences were combined with publicly available Palaemon sequences for phylogenetic and haplotype analyses. Our analyses indicate that the rostral formula is an unreliable character for species identification, that the Mississippi River does not act as a genetic barrier between the eastern and western populations, and that freshwater species are likely not derived from the saltwater species in the region.
1. Introduction
Grass shrimp in the genus Palaemon occur worldwide in wetlands ranging from freshwater to hypersaline. Several species are present on the east coast of North America, the northern Gulf of Mexico (nGoM), and within the nearby rivers [1]. Palaemon species play important roles in wetland ecology, contributing to the decomposition of organic material and passing energy to higher trophic levels [1]. Their annual productivity ranges from 9–16 g dry weight/m2/year, large enough for them to be ecologically significant [1]. The daggerblade shrimp, Palaemon pugio, can reduce the blockages of dead plant matter that form in wetland habitats, maintaining the ecosystem itself [2].
The family Palaemonidae was previously divided into several subfamilies, with Palaemon classified in the subfamily Palaemoninae until De Grave et al. [3] found evidence for paraphyly in the subfamilies and synonymized all of them with Palaemonidae. Similarly, Palaemon and Palaemononetes were long considered distinct genera, separated only by the lack of a mandibular palp in Palaemonetes [4], but, due to the inconsistency of this character, Palaemonetes is now considered a junior synonym of Palaemon [5]. On the other hand, more recent analysis of Palaemon detected polyphyly within the genus [6]. The taxonomy of Palaemon retains some ambiguities despite extensive research on the subject. Currently, Palaemon Weber, 1795 contains 95 recognized species [7].
One ambiguity within Palaemon exists among P. pugio (Holthuis, 1949), P. vulgaris Say, 1818, and P. mundusnovus De Grave and Ashelby, 2013 (the latter formerly known as Palaemonetes intermedius). These species have a sympatric distribution in the brackish marshes along the east and Gulf of Mexico coasts of the United States of America [8]. They are morphologically similar but also display some intraspecific variability [9]. This analysis includes a fourth species, Palaemon schmitti (Holthuis, 1950), which has not previously been reported from the nGoM. It is more commonly known from the Tropical Eastern Pacific [10,11,12], the Southern Caribbean, and South America [13,14,15].
Regarding the morphology, we focused on the rostral formula, which is often used for species identification and describes the arrangement of the teeth along the rostrum (Figure 1). The rostral formula is used here in accordance with [16] in the following format:
where a indicates the number of dorsal postorbital teeth on the rostrum, b the number of preorbital teeth, and c the number of ventral teeth.
a)b/c

Figure 1.
An example of shrimp rostral formula determination shown on Palaemon paludosus. For this specimen, the rostral formula would be written as 1)5/3. Images adapted from https://www.marylandbiodiversity.com/view/19699 (accessed on 1 August 2023).
Of the species mentioned above, P. pugio is the most widely studied. It is considered an r-selected species; i.e., it has fast growth, early sexual maturity, frequent reproduction, and high fecundity [8,17,18]. In the coastal wetlands of the southeast United States, it has been referred to as a “core species” [8]. P. vulgaris generally has smaller population sizes and is considered a “fringe species” in the region [8]. Further north on the US east coast where both species are common, P. pugio has a longer reproductive period with three or more spawning cycles per year compared to P. vulgaris with only two spawning cycles [19]. Additionally, P. pugio’s eggs are larger, potentially increasing the offspring’s fitness [19]. In both species, fecundity decreases with latitude, but the trend is more pronounced in P. vulgaris [19]. Comparatively less is known about the life cycle of P. mundusnovus, but the reproductive period is shorter than in both P. pugio and P. vulgaris, and the fecundity is lower than in P. pugio [20]. Although this study focuses on the relationships among saltwater Palaemon species, we also included representatives of several freshwater species (Table S1) to examine the relationships among the freshwater and saltwater species on the coast of the nGoM.
To provide additional clarity to the relationships within the Palaemon genus, specifically P. pugio, P. vulgaris, and P. mundusnovus, we conducted an analysis of fragments of the mitochondrial cytochrome c oxidase subunit I (COI) and nuclear Histone H3 (H3) genes on specimens from across the northern Gulf of Mexico (GoM) and the US east coast. As the most common “DNA barcoding” sequence for animals, COI is commonly used as a tool for species identification, and a large number of reference sequences from multiple Palaemon species were already publicly available. However, the marker is too variable to provide a solid resolution at deeper taxonomic levels. We therefore supplemented the COI dataset with a more conserved nuclear marker (H3), for which a limited number of reference sequences were also publicly available.
In the GoM, the Mississippi River discharges a massive amount of freshwater and sediment [21], which may act as a dispersal barrier for coastal species, including grass shrimp. At least 15 marine taxa within the GoM are separated into pairs of genetically distinct species, subspecies, or populations that converge in a vicariance zone near the Mississippi River [22]. Thus, we hypothesize that the Palaemon populations on the west and east sides of the Mississippi River would be genetically distinct from each other.
2. Materials and Methods
Live shrimp were collected with dip nets from tidal streams and marshes along the Texas, Florida, and Louisiana coasts. Most of the specimens were found near hard substrates or shaded areas. These collected samples were combined with preserved samples from the Texas A&M Biodiversity Research and Teaching Collections. Shrimp species were identified using dichotomous keys in [4]. Sampling locations are shown in Figure 2 (see also Table S1).
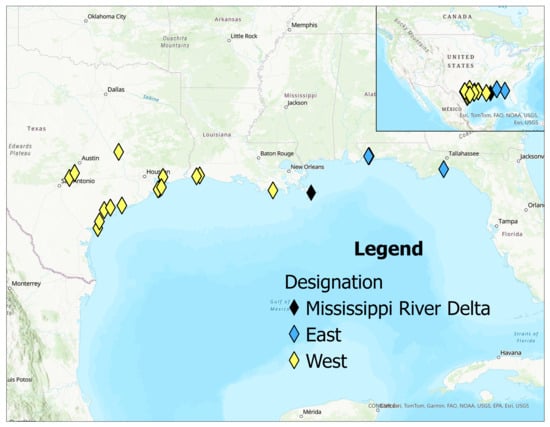
Figure 2.
Map of collection sites of Palaemon spp. used in this study. The Mississippi River Delta is indicated to show the dividing point set for this study, and other collection sites are labeled as coming from east or west of this point.
Shrimp were preserved in 95% ethanol, then had their rostral formulas recorded. Abdominal tissue was extracted from each selected shrimp sample for DNA extraction, using the Qiagen Dneasy Blood & Tissue kit, following the manufacturer’s instructions. PCR was then performed to amplify the target gene using the OneTaq kit (New England Biolabs, Ipswich, MA, USA) in 25 microliter (µL) reactions. Each reaction contained 12.5 µL of the OneTaq Master Mix, 1 µL each of the forward and reverse primers at 10 μM, 1 µL of template DNA, and 9.5 µL of water. Multiple COI sequences were collected from each sampling location to define major clades within Palaemon. Singular Histone H3 sequences were sequenced from each of the sampling locations COI sequences were collected from to resolve deeper branches and increase branch support. COI sequences used the primers LCO1490 and HCO2198 [23], while Histone H3 sequences used the primers H3aF and H3aR [24]. The PCR cycle used for COI consisted of (i) 94 °C for 30 s, (ii) denaturation at 94 °C for 30 s, (iii) annealing at 48 °C for 30 s, (iv) extension at 68 °C for 1 min, (v) a total of 30 cycles of steps ii through iv, and (vi) a final extension at 68 °C for 5 min ending with a hold at 4 °C. The PCR cycle for H3 consisted of (i) 94 °C for 7 min, (ii) denaturation at 94 °C for 40 s, (iii) annealing at 57 °C for 40 s, (iv) extension at 72 °C for 1 min, (v) a total of 45 cycles of steps ii through iv, and (vi) a final extension at 72 °C for 7 min ending with a hold at 4 °C.
Successful amplification was confirmed via gel electrophoresis in a 1% agarose gel with ethidium bromide. PCR products were enzymatically cleaned with EXOSAP-IT (ThermoFisher Scientific, Waltham, MA, USA) according to the product’s listed protocol of 15 min at 37 °C followed by 15 min at 80 °C. Product concentration was checked using the Qubit DNA HS protocol, with either 1 or 2 µL of PCR product. Sanger sequencing was performed in both directions at Genewiz/Azenta using the same primers as for the PCR reactions.
Forward and reverse sequences were assembled, followed by trimming of primer sequences in CodonCode Aligner (CodonCode Corporation, Centerville, MA, USA). Resulting consensus sequences were aligned with publicly available Palaemon sequences in MEGA11 [25] using the MUSCLE Multiple Sequences Alignment (MSA) program. Four different sequence sets were analyzed: (1) COI sequences from North and Central America only (COI-NCA tree: 124 sequences, length: 585 bp); (2) H3 sequences from North and Central America (H3-NCA tree: 38 sequences, length 286 bp; (3) concatenated COI and H3 sequences from North America (COI + H3 NA tree: 47 sequences, length: 286 bp H3 + 591 bp COI = 877 bp). Only a single H3 sequence was generated for each location in which Palaemon specimens were collected; all the COI sequences from that location were then combined with the same H3 sequence; and (4) all available COI sequences from global locations (global tree: 231 sequences, length: 539 bp) (Table S2). Average genetic distances were calculated within and among the North American freshwater and saltwater clades, from in MEGA 11 [25], using the Kimura-2-Parameter (K2P) model. We chose the K2P model because it is the most commonly used to calculate genetic distances for marine invertebrates, making our values comparable to other studies.
After running Modeltest on the alignments to determine the most appropriate model of evolution, Maximum Likelihood (ML) trees were formed for the multiple sequence alignment (MSA) using 500 bootstrap replicates for the NCA tree and 1000 bootstraps for the global tree. Any duplicate sequences were automatically removed during the tree formation. If the optimal evolution model suggested by Modeltest was available in RaxMLGUI, it was used; if not, the GTR substitution model with Gamma substitution rates with an ML estimate of invariant sites was employed instead. Macrobrachium nipponense was used as an outgroup for rooting purposes for both COI trees. For the COI + H3 tree, the outgroup was Macrobrachium nipponense.
A haplotype network was generated with only the sequences from NCA saltwater species using the minimum spanning network method in PopArt [26] with an epsilon value of zero. More than 5% of the sites in this analysis contained undefined states and were automatically masked by the PopArt v1.7 for Windows software.
3. Results
3.1. Rostral Formula
The rostral formulae were variable (Table S1) and only corresponded to a limited degree to the identified morphospecies or to the clades identified in the phylogenetic analyses (Figure 3). One consistent feature is that all the individuals in Clade 4 of the COI-NCA tree (Figure 3) have two postrostral teeth as opposed to a single postrostral tooth in most of the other studied samples. The only other two individuals with two postrostral teeth are the two samples identified as P. schmitti belonging to Clade 3.
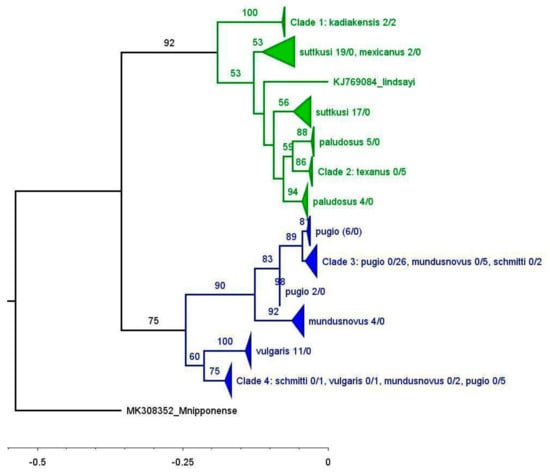
Figure 3.
Maximum Likelihood tree of publicly available Palaemon COI sequences and COI sequences generated for this project (COI−NCA tree) from North/Central America. Triangles and flat lines indicate compressed subtrees; values within parentheses indicate the number of collected and public sequences in each subtree in the particular format (publicly available/generated in this study). Saltwater Palaemon species are colored blue; freshwater species are shown in green. The scale indicates relative node ages. Values at the nodes indicate bootstrap support (500 replicates).
3.2. COI-NCA Tree and Haploptype Network
The phylogenetic analysis of all the available Palaemon sequences from North and Central America resulted in a tree with thirteen primary lineages (Figure 3), separated into freshwater and saltwater clades, with the freshwater clade having higher bootstrap support (92%) than the saltwater clade (75%). The average K2P distance for COI within the freshwater clade is 0.08, and 0.09 within the saltwater clade. The average K2P distance between the two clades is 0.228. Of the thirteen primary lineages, ten correspond to a single identified morphospecies, while the remaining three include more than one species. One of the latter comprised the freshwater species P. suttkusi and P. mexicanus. These were publicly available sequences for which no vouchers were available for morphological characterization. Further analysis will focus on the clades labeled Clade 3 and Clade 4, which each include multiple saltwater species.
The haplotype analysis was based on 67 individual sequences and resulted in a total of 26 haplotypes, of which 11 were shared among several individuals and 15 were unique to single individuals (Figure 4; Table S1). Clade 3 (Figure 3) includes three morphospecies (P. pugio, P. mundusnovus, and P. schmitti), which group into four shared haplotypes (H1, H2, H3, and H4, Table S1) and four singleton haplotypes, without any clear correlation between haplotype assignment and species identification (Figure 4A). H1 is by far the most common and widespread haplotype, including three morphospecies (P. pugio, P. mundusnovus, and P. schmitti) and spanning the entire northern Gulf of Mexico from Florida to Texas. Clade 4 (Figure 3) includes the shared haplotypes H7 and H8, as well as two individual sequences for P. schmitti and P. pugio, respectively. H7 comprises two specimens identified as P. mundusnovus from Texas. H8 includes four sequences of P. pugio from Florida and one individual of P. vulgaris from Louisiana. Despite the overlap in morphospecies, Clades 3 and 4 are separated by 62 mutations (Figure 4B). When colored by location (Figure 4C), it also becomes obvious that no phylogeographic break exists between the eastern and western nGoM. The east coast samples fall into two groups: one comprising P. vulgaris and the second one P. mundusnovus and P. pugio.

Figure 4.
Haplotype networks formed from the North American sequences of Palaemon species pugio, vulgaris, mundusnovus, and schmitti. (A) marks each sequence by species, (B) marks them by Clade position in Figure 3, and (C) marks them by collection region. Shared haplotypes are marked with the formula HX (see Table S1 for haplotype assignments). The numbers in parentheses indicate the number of mutations separating each node. Collected sequences are annotated in the format ID_species_State. Publicly available sequences are annotated with their GenBank number, followed by species.
3.3. H3 Analysis of North and Central American Palaemon spp. (H3-NCA)
The analysis of the H3 also supports a separation into freshwater and saltwater species (Figure 5), although the freshwater clade does not appear as monophyletic. Similar to the COI-NCA tree (Figure 3), one of the freshwater clades includes the two morphospecies P. suttkusi and P. mexicanus. Likewise, there is no one-to-one correspondence between the morphospecies and clade for the saltwater species. The average K2P distance within the freshwater group is 0.03, and 0.01 within the saltwater clade. The average K2P distance between the two species groups is 0.0742.
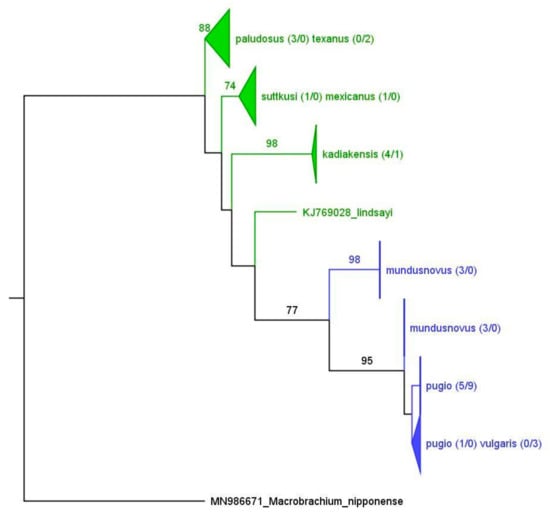
Figure 5.
Maximum likelihood tree containing publicly available Palaemon H3 sequences and H3 sequences generated for this project from Central and North America (H3-NCA). Triangles and flat lines indicate compressed subtrees, and the values within parentheses indicate the number of collected and public sequences in each subtree in the particular format (publicly available/generated in this study). Saltwater Palaemon species are colored blue; freshwater species are shown in green. Values at the nodes indicate bootstrap support (500 replicates).
3.4. Analysis of Concatenated COI and H3 Sequences from North America (COI-H3-NA)
The combination of the limited H3 sequences with COI (Figure 6) revealed the freshwater species as monophyletic, increased the bootstrap support for the saltwater clade, and maintained the lack of correspondence between the saltwater morphospecies and the clades defined by molecular analysis.
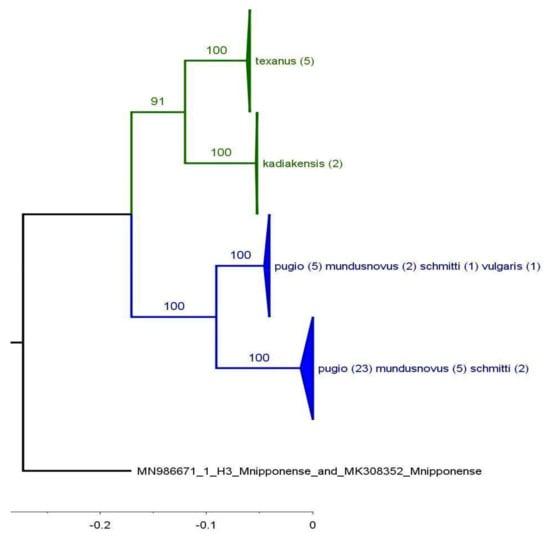
Figure 6.
Maximum likelihood tree using concatenated COI and H3 sequences. Triangles indicate compressed subtrees, and the annotations on triangles indicate the species, locales, and number of collected sequences within. Each individual sequence was formed by pasting a COI sequence to the end of an H3 sequence. Saltwater Palaemon species are colored blue; freshwater species are shown in green. The scale indicates relative node ages. Values at the nodes indicate bootstrap support (500 replicates).
3.5. Analysis of Global Palaemon Species (Global Tree)
The analysis of the global COI Palaemon species resulted in 21 primary lineages (Figure 7), each assigned to one of six broad regions: (1) Caribbean/Gulf of Mexico/northwest Atlantic; (2) southwest Atlantic; (3) Mediterranean/northeast Atlantic; (4) Indo-Pacific; (5) northeast Pacific; and (6) South Africa. As in the other analyses, the American freshwater and saltwater lineages assigned to the Caribbean/GoM/NW Atlantic region are clearly separated from each other. The freshwater species form a well-supported monophyletic clade, whereas the monophyly among the tree saltwater lineages has poor bootstrap support; additionally, the clade includes one Indo-Pacific lineage (P. debilis).
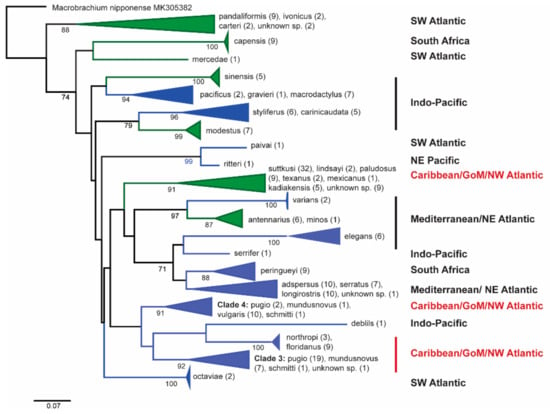
Figure 7.
Global ML tree generated from publicly available Palaemon sequences from global locations and sequences generated for this project. Subtree colors indicate the environments containing Palaemon species: green for freshwater and blue for saltwater only. Only bootstrap values above 70 are marked.
4. Discussion
Our data reveal that the taxonomy of the Palaemon species from the northern Gulf of Mexico remains poorly resolved, at least when using the taxonomic key in [4]. As shown in the NCA phylogenetic tree (Figure 3) and the haplotype network (Figure 4A), none of the identified saltwater morphospecies from the nGoM form cohesive units. P. vulgaris from the US east coast forms a well-supported clade, but the single P. vulgaris from the nGoM shares its haplotype with several P. pugio individuals (Figure 3 and Figure 4A; Table S1). Similarly, all the P. mundusnovus samples from the east coast form a monophyletic group (Figure 3, Figure 4C and Figure 5), whereas the nGoM samples of P. mundusnovus group more closely with P. vulgaris or P. pugio.
According to [4], P. vulgaris and P. schmitti have two postorbital teeth, whereas only one postorbital tooth is present in P. pugio and P. mundusnovus. However, the rostral formula can be highly variable [9]. Given that Clade 4 appears as the sister group to the east coast P. vulgaris (Figure 3) and all its members have two postorbital teeth, it is possible that all the specimens in Clade 4 should actually be classified as P. vulgaris. Likewise, as Clade 3 is nested within a group of P. pugio from the east coast, all its members might be correctly classified as P. pugio. Clade 3 includes two specimens originally identified as P. schmitti, both of them with two postorbital teeth, but this is possibly a misidentification. The two postorbital teeth in the specimens identified as P. schmitti may be a further indication for the intraspecific variability of the rostral formula. The incorrect identification is further supported by the fact that the species has not previously been reported from the nGoM, and its type locality is on the Pacific side of the Panama Canal, whereas the other three species (P. pugio, P. vulgaris, and P. mundusnovus) have type localities on the US east coast [4]. One possible reason why the species identification is challenging is hybridization among the species, a phenomenon that has been documented in other Palaemon lineages based on mitochondrial/nuclear genomic and karyotype analyses [27,28].
The nGoM saltwater samples show no discernible differentiation between the coasts east and west of the Mississippi River Delta (Figure 4C), indicating high population connectivity along the entire nGoM. Previous studies of P. pugio found a lack of genetic breaks along the coasts of Georgia [25] and Texas [17], but ours is the first to examine the genetic differentiation across the Mississippi River Delta. This finding is remarkable considering the preference of P. pugio for estuarine habitats, which are more likely to be fragmented than open water, and the fact that P. pugio reproduces by direct sperm transfer on a seasonal basis rather than broadcast spawning [26]. It is likely that the population connectivity of P. pugio is maintained by their larvae, which can tolerate slightly higher salinity levels and have the potential for greater distribution [18]. Another possible explanation for the high population connectivity is anthropogenic transport through shipping traffic, for example in fouling communities of ships and barges, via the Intracoastal Waterway, which extends along the entire nGoM coast. Assessing the impact of shipping on the distribution of Palaemon species would require the sequencing of Palaemon specimens from a vessel’s fouling communities and their putative source and sink populations, as well as knowledge of the vessel’s travel and cleaning schedule. Palaemon shrimp might be one of many taxa being transported across presumed dispersal barriers in this way. If anthropogenic transport is determined to be a significant factor for the transport of Palaemon species, assessments of their ecological roles (e.g., interspecific competition and predator-prey interactions) in multiple locations along their range would be warranted.
The separation of the saltwater and freshwater Palaemon species of the nGoM is another interesting finding (Figure 3, Figure 5, Figure 6 and Figure 7). In fact, the North/Central American freshwater species do not appear to be derived from the saltwater species but rather have colonized the freshwater habitats independently. Our COI datasets revealed average genetic distances of 22.8% between the North/Central American freshwater and saltwater species, supporting their divergence at a deep level within the genus. Based on a wide range of DNA barcoding data for decapods, this value falls well within the range of intrageneric divergence in Palaemonidae [29]. The relationships among the freshwater species are currently under investigation, with an emphasis on P. texanus and P. paludosus.
5. Conclusions
The results of this study demonstrated that a commonly used morphological feature, the rostral formula, is of limited value for the identification of Palaemon species in the nGoM. Given the taxonomic uncertainties within Palaemon, and the history of frequent rearrangements of the genus and the Palaemonidae as a whole, we suggest a shift in focus from the taxonomy to the broader patterns of evolution, population connectivity, morphological, and perhaps physiological and developmental plasticity. Even if careful morphological work may reveal unambiguous species-specific characters, it is unlikely that these will be practical for use without significant investments of time and effort in advanced microscopic methods. Taxonomic names are still the basic units for biodiversity assessments but should be supplemented with additional information about the population characteristics whenever possible.
Our analyses further revealed high population connectivity among the Palaemon populations along the entire nGoM coast, even across the Mississippi River Delta. Whether this high connectivity results from the life history characteristics of grass shrimps or from anthropogenic transport remains to be determined.
Lastly, our data indicate that the saltwater and freshwater species have radiated separately in the region, suggesting that the habitat preferences of Palaemon species are more strongly determined by physiological limits rather than geographic vicinity, even though transitions between saltwater and freshwater have occurred repeatedly within the evolution of the genus. Further research into the environmental tolerances of the different life history stages would lend additional support to this hypothesis.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/d16090543/s1, Table S1: Palaemon spp. specimens examined for this study with their GenBank accession numbers for the sequenced gene regions of cytochrome c oxidase subunit I (COI) and Histone H3 (H3), clade and shared haplotype assignments, species ID, rostral formula, and collection information. Table S2: Cytochrome c subunit I (COI) sequences used to generate the global tree (Figure 7), their species identification, assigned geographic region, habitat (SW: saltwater; FW: freshwater), clade, and haplotype assignment.
Author Contributions
Conceptualization, A.S. and M.K.W.; methodology D.D.F., A.S. and M.K.W.; analysis, D.D.F. and A.S.; data curation, D.D.F. and A.S.; writing—original draft preparation, D.D.F.; writing—review and editing, D.D.F., A.S. and M.K.W.; visualization, D.D.F. and A.S.; supervision, A.S. and M.K.W.; project administration, A.S.; funding acquisition, A.S. and M.K.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by a Panther Research & Innovation for Scholarly Excellence (PRISE) Grant Program to Texas A&M University and Prairie View A&M University: Diversity of grass shrimp (Palaemon spp.) and their parasites (Microphallus spp.) in Gulf of Mexico wetlands (2021).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The DNA sequence data generated for this project are publicly available in GenBank (National Library of Medicine, Bethesda, MD, USA; https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 26 June 2024)) under accession numbers PP989494–PP989543 (COI) and PP990521–PP990533 (H3). Alignments used to generate phylogenetic trees are available on Figshare (London, UK): COI-NCA: https://doi.org/10.6084/m9.figshare.26264672.v1; H3-NCA: https://doi.org/10.6084/m9.figshare.26264702.v1; COI-H3-NA: https://doi.org/10.6084/m9.figshare.26264399.v4; global COI: https://doi.org/10.6084/m9.figshare.26264789.v1.
Acknowledgments
We would like to thank Stephanie Archer at the Louisiana Universities Marine Consortium (LUMCON) for assistance with specimen collections and identifications.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Key, P.B.; Wirth, E.F.; Fulton, M.H. A review of grass shrimp, Palaemonetes spp., as a bioindicator of anthropogenic impacts. Environ. Bioindic. 2006, 1, 115–128. [Google Scholar] [CrossRef]
- Welsh, B.L. The role of grass shrimp, Palaemonetes pugio, in a tidal marsh wcosystem. Ecology 1975, 56, 513–530. [Google Scholar] [CrossRef]
- De Grave, S.; Fransen, C.H.; Page, T.J. Let’s be pals again: Major systematic changes in Palaemonidae (Crustacea: Decapoda). PeerJ 2015, 3, e1167. [Google Scholar] [CrossRef] [PubMed]
- Holthuis, L.B. A General Revision of the Palaemonidae (Crustacea Decapoda Natantia) of the Americas; University of Southern California Press: Los Angeles, CA, USA, 1951; Volume 11. [Google Scholar]
- De Grave, S.; Ashelby, C.W. A re-appraisal of the systematic status of selected genera in Palaemoninae (Crustacea: Decapoda: Palaemonidae). Zootaxa 2013, 3734, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, F.L.; De Grave, S.; Mantelatto, F.L. An integrative approach to the evolution of shrimps of the genus. Palaemon (Decapoda, Palaemonidae). Zool. Scr. 2017, 46, 473–485. [Google Scholar] [CrossRef]
- DecaNet. Palaemon Weber. 1795. Available online: https://decanet.info/aphia.php?p=taxdetails&id=107032 (accessed on 26 June 2024).
- Pennington, P.L.; Daugomah, J.W.; Key, P.B.; DeLorenzo, M.E. Grass Shrimp Biomonitoring at Two Sites in Coastal South Carolina 2001–2016: Long-Term Trends and Associations with Water Quality Variables; NOAA National Ocean Service, National Centers for Coastal Ocean Science: Silver Spring, MD, USA, 2024. [CrossRef]
- Jordán-Hernández, M.; Rodríguez-Almaráz, G.; Favela-Lara, S. Delimitation of sympatric Palaemon (Decapoda, Palaemonidae) species of the Laguna Madre, Mexico. Zool. Scr. 2019, 48, 667–678. [Google Scholar] [CrossRef]
- Rodríguez-Almaraz, G.A.; Leija-Tristán, A.; Mendoza, R. Records of caridean shrimps (Crustacea: Decapoda) from the coasts of the Mexican Pacific Ocean, Gulf of Mexico and Mexican Caribbean. Bull. Mar. Sci. 2000, 67, 857–868. [Google Scholar]
- Wicksten, M.K.; Hendrickx, M.E. An updated checklist of benthic marine and brackish water shrimps (Decapoda: Penaeoidea, Stenopodidea, Caridea) from the Eastern Tropical Pacific. Contrib. Study East. Pac. Crustac. 2003, 2, 49–76. [Google Scholar]
- Ashelby, C.W.; Page, T.J.; De Grave, S.; Hughes, J.M.; Johnson, M.L. Regional scale speciation reveals multiple invasions of freshwater in Palaemoninae (Decapoda). Zool. Scr. 2012, 41, 293–306. [Google Scholar] [CrossRef]
- Almeida, A.; Fransozo, A.; Teixeira, G.; Hiroki, K.A.N.; Furlan, M.; Bertini, G. Ecological distribution of the shrimp Nematopalaemon schmitti (Crustacea: Decapoda: Caridea) in three bays on the south-eastern coast of Brazil. Afr. J. Mar. Sci. 2012, 34, 93–102. [Google Scholar] [CrossRef]
- Fransozo, V.; Castilho, A.L.; Freire, F.A.M.; Furlan, M.; De Almeida, A.C.; Teixeira, G.M.; Baeza, J.A. Spatial and temporal distribution of the shrimp Nematopalaemon schmitti (Decapoda: Caridea: Palaemonidae) at a subtropical enclosed bay in South America. J. Mar. Biol. Assoc. United Kingd. 2009, 89, 1581–1587. [Google Scholar] [CrossRef]
- Ramiro Herrera, D.; Maia Davanso, T.; Caetano da Costa, R. Relative growth and morphological sexual maturity of the caridean shrimp Nematopalaemon schmitti (Decapoda: Caridea: Palaemonidae) in an upwelling region in the Western Atlantic. Invertebr. Reprod. Dev. 2018, 62, 56–62. [Google Scholar] [CrossRef]
- Chace, F.A., Jr.; Bruce, A. The Caridean Shrimps (Crustacea: Decapoda) of the Albatross Philippine Expedition 1907–1910, Part 6: Superfamily Palaemonoidea; Smithsonian Institution Press: Washington, DC, USA, 1993. [Google Scholar]
- Cházaro-Olvera, S. Growth, mortality, and fecundity of Palaemonetes pugio from a lagoon system inlet in the southwestern Gulf of Mexico. J. Crustac. Biol. 2009, 29, 201–207. [Google Scholar] [CrossRef]
- Espinoza, G.J.; Alvarado Bremer, J.R. Comparative phylogeography, historical demography, and population genetics of three common coastal fauna in Spartina marshes of the northwestern Gulf of Mexico. Diversity 2023, 15, 792. [Google Scholar] [CrossRef]
- Ganz, H.H.; Knowlton, R.E. Reproductive differences among Delmarva grass shrimp (Palaemonetes pugio and P. vulgaris) populations. Va. J. Sci. 2002, 53, 3. [Google Scholar]
- Anderson, G. Species Profiles: Life Histories and Environmental Requirements (Gulf of Mexico)—Grass Shrimp; US Fish and Wildlife Service Biological Report 82 (11:35), U.S. Army Corps of Engineers, TR EL 82-4; U.S. Army Corps of Engineers: Washington, DC, USA, 1985; 19p.
- Dagg, M.J.; Breed, G.A. Biological effects of Mississippi River nitrogen on the northern Gulf of Mexico—A review and synthesis. J. Mar. Syst. 2003, 43, 133–152. [Google Scholar] [CrossRef]
- Portnoy, D.S.; Gold, J.R. Evidence of multiple vicariance in a marine suture-zone in the Gulf of Mexico. J. Biogeogr. 2012, 39, 1499–1507. [Google Scholar] [CrossRef]
- Folmer, O.; Hoeh, W.; Black, M.; Vrijenhoek, R. Conserved primers for PCR amplification of mitochondrial DNA from different invertebrate phyla. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Colgan, D.J.; McLauchlan, A.; Wilson, G.D.; Livingston, S.; Edgecombe, G.; Macaranas, J.; Cassis, G.; Gray, M. Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution. Aust. J. Zool. 1998, 46, 419–437. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D.; Nakagawa, S. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Garcia, D.K.; Davis, S.K. Evidence for a mosaic hybrid zone in the grass shrimp Palaemonetes kadiakensis (Palaemonidae) as revealed by multiple genetic markers. Evolution 1994, 48, 376–391. [Google Scholar] [CrossRef] [PubMed]
- Torrecilla, Z.; Martínez-Lage, A.; Perina, A.; González-Ortegón, E.; González-Tizón, A.M. Comparative cytogenetic analysis of marine Palaemon species reveals a X1X1X2X2/X1X2Y sex chromosome system in Palaemon elegans. Front. Zool. 2017, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Matzen da Silva, J.; Creer, S.; Dos Santos, A.; Costa, A.C.; Cunha, M.R.; Costa, F.O.; Carvalho, G.R. Systematic and evolutionary insights derived from mtDNA COI barcode diversity in the Decapoda (Crustacea: Malacostraca). PLoS ONE 2011, 6, e19449. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).