Comparative Genomics of Limosilactobacillus pontis Strains: Niche-Specific Variations and Adaptations
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains
2.2. Main Instruments and Reagents
2.3. Strain Culture and Genomic DNA Extraction
2.4. Genome Sequencing and Assembly
2.5. Comparative Genomics Analysis
2.5.1. Average Nucleotide Identity (ANI) Value Calculation
2.5.2. Construction of Core Gene Set and Accessory Gene Set Analysis
2.5.3. Phylogenetic Tree Construction
2.5.4. Functional Gene Annotation
2.5.5. CAZy Annotation
2.5.6. Bacteriocin
2.6. Uploading Strain Genome
3. Results
3.1. General Genomic Characteristics
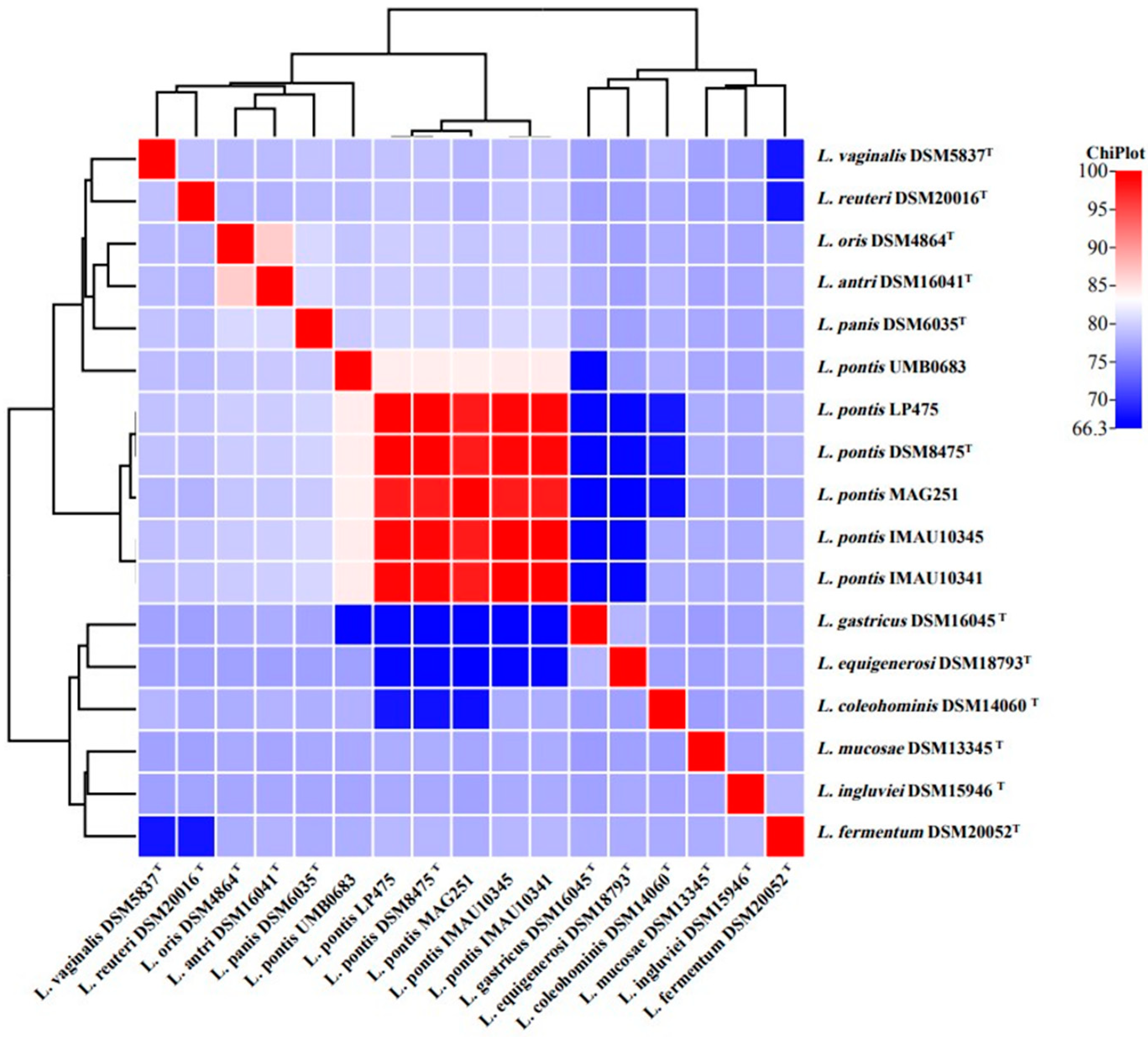
3.2. ANI Analysis
3.3. Core Genome and Pangenome Analysis
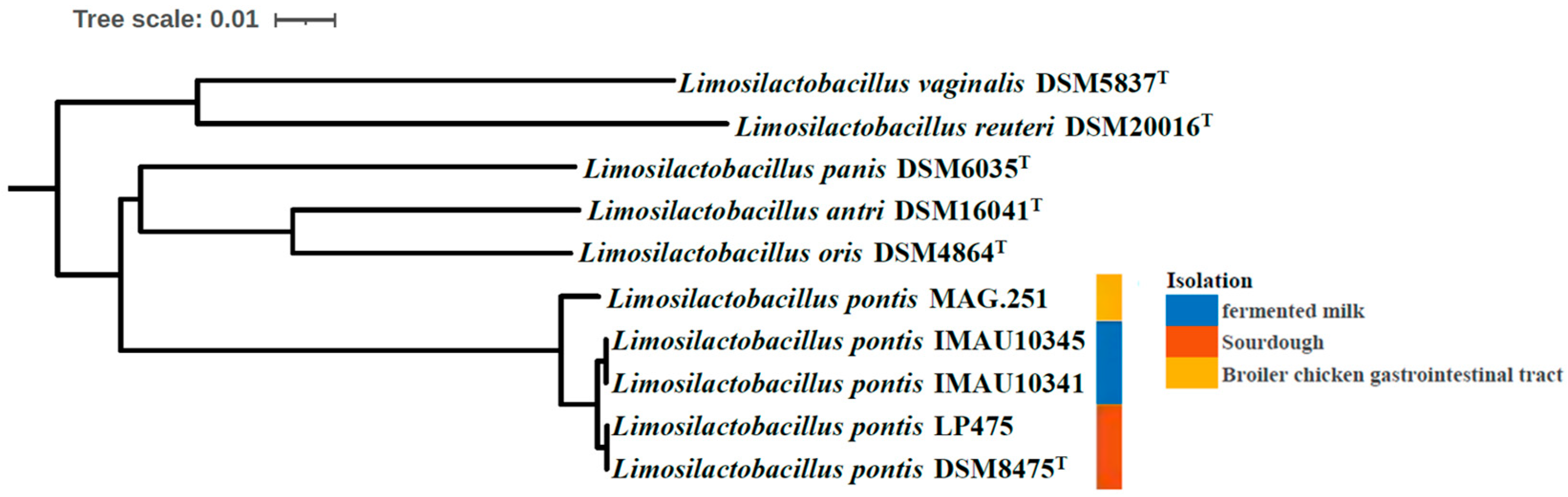
3.4. Phylogenetic Tree
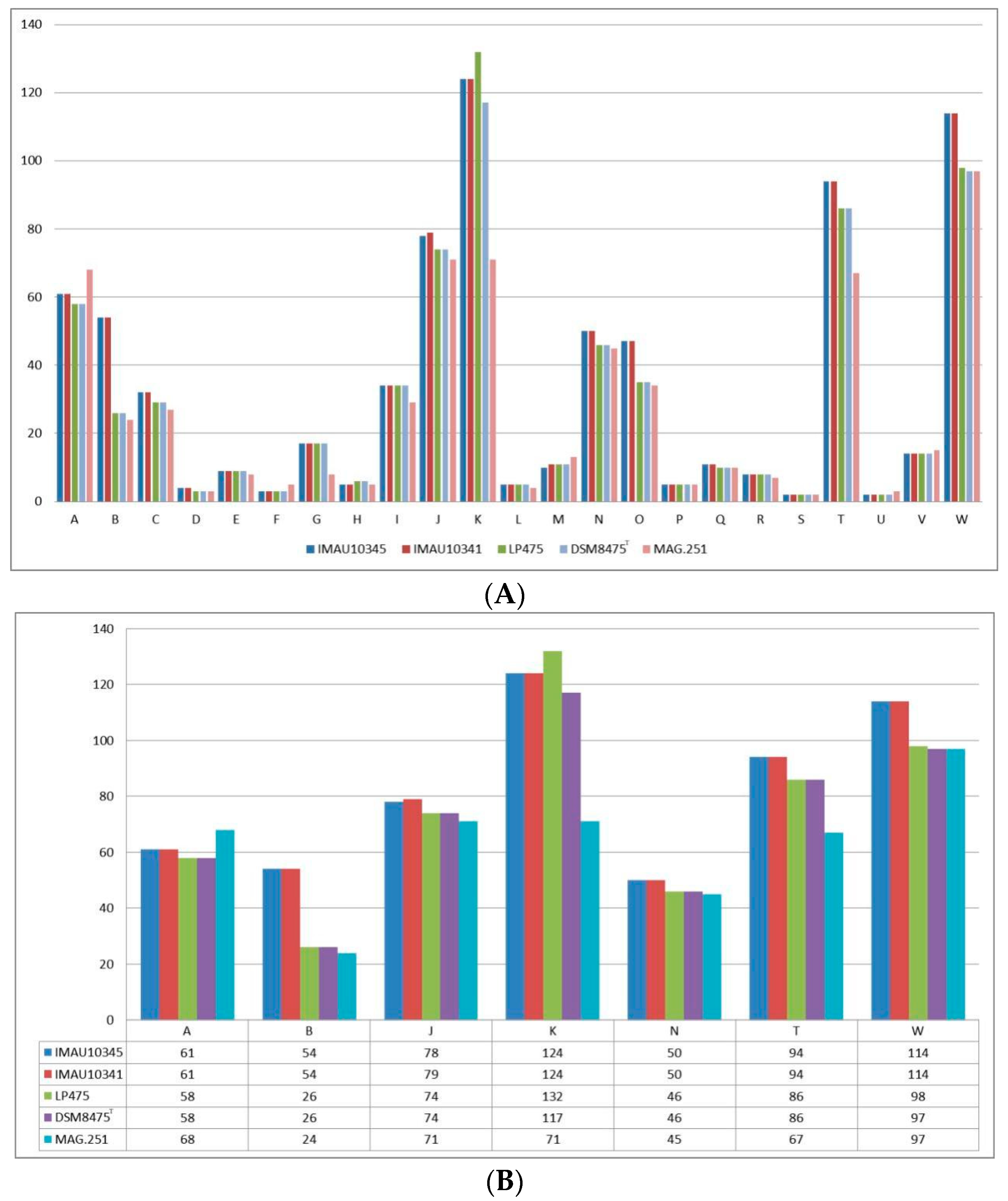
3.5. Functional Gene Analysis
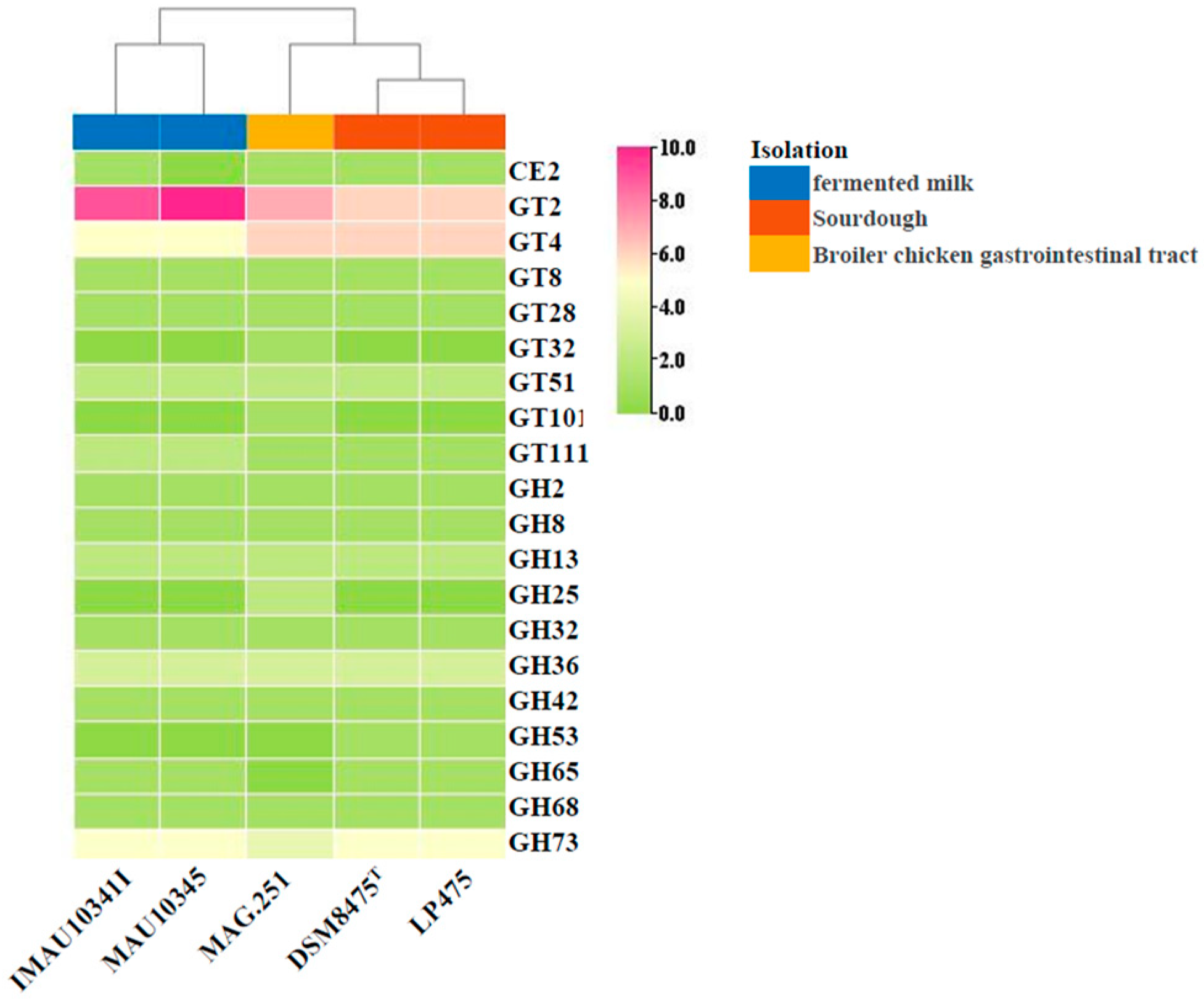
3.6. Carbohydrate-Active Enzyme Analysis
3.7. Bacteriocin
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Vogel, R.F.; BöCKER, G.; Stolz, P.; Ehrmann, M.; Fanta, D.; Ludwig, W.; Pot, B.; Kersters, K.; Schleifer, K.H.; Hammes, W.P. Identification of Lactobacilli from sourdough and description of Lactobacillus pontis sp. nov. Int. J. Syst. Evol. Microbiol. 1994, 44, 223–229. [Google Scholar] [CrossRef][Green Version]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.; Harris, H.M.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Maya-Barrios, A.; Lira-Hernandez, K.; Jiménez-Escobar, I.; Hernández, L.; Ortiz-Hernandez, A.; Jiménez-Gutiérrez, C.; López-Velázquez, G.; Gutiérrez-Castrellón, P. Limosilactobacillus reuteri ATCC PTA 5289 and DSM 17938 as adjuvants to improve evolution of pharyngitis/tonsillitis in children: Randomised controlled trial. Benef. Microbes 2021, 12, 137–145. [Google Scholar] [CrossRef]
- Bao, Y.; Zhang, Y.; Zhang, Y.; Liu, Y.; Wang, S.; Dong, X.; Wang, Y.; Zhang, H. Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products. Food Control 2010, 21, 695–701. [Google Scholar] [CrossRef]
- Shazadi, K.; Arshad, N. Evaluation of inhibitory and probiotic properties of lactic acid bacteria isolated from vaginal microflora. Folia Microbiol. 2022, 67, 427–445. [Google Scholar] [CrossRef]
- Saviano, A.; Brigida, M.; Migneco, A.; Gunawardena, G.; Zanza, C.; Candelli, M.; Franceschi, F.; Ojetti, V. Lactobacillus Reuteri DSM 17938 (Limosilactobacillus reuteri) in diarrhea and constipation: Two sides of the same coin? Medicina 2021, 57, 643. [Google Scholar] [CrossRef] [PubMed]
- Talarico, T.; Casas, I.; Chung, T.C.; Dobrogosz, W. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 1988, 32, 1854–1858. [Google Scholar] [CrossRef]
- Arqués, J.L.; Fernández, J.; Gaya, P.; Nuñez, M.; Rodríguez, E.; Medina, M. Antimicrobial activity of reuterin in combination with nisin against food-borne pathogens. Int. J. Food Microbiol. 2004, 95, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Dobrogosz, W.J.S.E. 5413960 Antibiotic reuterin: Dobrogosz Walter J.; Lindgren Sven E Raleigh, NC, United States Assigned to Biogaia AB. Biotechnol. Adv. 1996, 14, 288. [Google Scholar] [CrossRef]
- Feyereisen, M.; Mahony, J.; Kelleher, P.; Roberts, R.J.; O’Sullivan, T.; Geertman, J.-M.A.; van Sinderen, D. Comparative genome analysis of the Lactobacillus brevis species. BMC Genom. 2019, 20, 416. [Google Scholar] [CrossRef]
- Francis, F.; Kim, J.; Ramaraj, T.; Farmer, A.; Rush, M.C.; Ham, J.H. Comparative genomic analysis of two Burkholderia glumae strains from different geographic origins reveals a high degree of plasticity in genome structure associated with genomic islands. Mol. Genet. Genom. 2013, 288, 195–203. [Google Scholar] [CrossRef]
- Oh, P.L.; Benson, A.K.; Peterson, D.A.; Patil, P.B.; Moriyama, E.N.; Roos, S.; Walter, J. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 2010, 4, 377–387. [Google Scholar] [CrossRef]
- Jiang, J.Y.; Li, K.; Xiao, Y.; Zhong, A.; Tang, J.J.; Duan, Y.F.; Li, Z.J. Limosilactobacillus reuteri Regulating Intestinal Function: A Review. Fermentation 2022, 9, 19. [Google Scholar] [CrossRef]
- Tyagi, A.; Yeon, S.J.; Chelliah, R.; Oh, D.H. Draft genome sequence of Limosilactobacillus reuteri, isolated from human breast milk. Microbiol. Resour. Announc. 2023, 12, 112822. [Google Scholar] [CrossRef]
- Liu, W.; Bao, Q.; Jirimutu; Qing, M.; Siriguleng; Chen, X.; Sun, T.; Li, M.; Zhang, J.; Yu, J. Isolation and identification of lactic acid bacteria from Tarag in Eastern Inner Mongolia of China by 16S rRNA sequences and DGGE analysis. Microbiol. Res. 2012, 167, 110–115. [Google Scholar] [CrossRef]
- Yu, J.; Zhao, J.; Song, Y.; Zhang, J.; Yu, Z.; Zhang, H.; Sun, Z. Comparative genomics of the herbivore gut symbiont Lactobacillus reuteri reveals genetic diversity and lifestyle adaptation. Front. Microbiol. 2018, 9, 1151. [Google Scholar] [CrossRef]
- Tanaka, T.; Sakai, R.; Kobayashi, R.; Hatakeyama, K.; Matsunaga, T. Contributions of phosphate to DNA adsorption/desorption behaviors on aminosilane-modified magnetic nanoparticles. Langmuir 2009, 25, 2956–2961. [Google Scholar] [CrossRef]
- Modi, A.; Vai, S.; Caramelli, D.; Lari, M. The Illumina Sequencing Protocol and the NovaSeq 6000 System. Methods Mol. Biol. 2021, 2242, 15–42. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 2012, 1, 2047-217X-1-18. [Google Scholar] [CrossRef]
- Xu, M.; Guo, L.; Gu, S.; Wang, O.; Zhang, R.; Peters, B.A.; Fan, G.; Liu, X.; Xu, X.; Deng, L. TGS-GapCloser: A fast and accurate gap closer for large genomes with low coverage of error-prone long reads. GigaScience 2020, 9, 94. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Chen, J.; Yang, X.; Chen, J.; Cen, Z.; Guo, C.; Jin, T.; Cui, Y. SISP: A Fast Species Identification System for Prokaryotes Based on Total Nucleotide Identity of Whole Genome Sequences. Infect. Dis. Transl. Med. 2015, 1, 30–55. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Duranti, S.; Milani, C.; Lugli, G.A.; Mancabelli, L.; Turroni, F.; Ferrario, C.; Mangifesta, M.; Viappiani, A.; Sánchez, B.; Margolles, A. Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis. Sci. Rep. 2016, 6, 23971. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, 233–238. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, 278–281. [Google Scholar] [CrossRef]
- Johnson, B.; Shneiderman, B. Tree-Maps: A Space-Filling Approach to the Visualization of Hierarchical Information Structures; IEEE: San Diego, CA, USA, 1991; pp. 284–291. [Google Scholar] [CrossRef]
- Wang, S.; Yang, B.; Ross, R.P.; Stanton, C.; Zhao, J.; Zhang, H.; Chen, W. Comparative genomics analysis of Lactobacillus ruminis from different niches. Genes 2020, 11, 70. [Google Scholar] [CrossRef]
- Balakirev, E.S.; Krupnova, T.N.; Ayala, F.J. DNA variation in the phenotypically-diverse brown alga Saccharina japonica. BMC Plant Biol. 2012, 108, 1471–2229. [Google Scholar] [CrossRef]
- Accetto, T.; Avguštin, G. Polysaccharide utilization locus and CAZYme genome repertoires reveal diverse ecological adaptation of Prevotella species. Syst. Appl. Microbiol. 2015, 38, 453–461. [Google Scholar] [CrossRef]
- Cao, H.; Ekstrom, A.; Yin, Y. Plant Carbohydrate Active Enzyme (CAZyme) Repertoires: A Comparative Study; Springer International Publishing: New York, NY, USA, 2015; pp. 115–134. [Google Scholar] [CrossRef]
- Vilma, K.; Mantas, S.; Kristina, B.; Naujokaitytė, G.; Mulkytė, K.; Malakauskas, M.; Maruška, A. Current state of purification, isolation and analysis of bacteriocins produced by lactic acid bacteria. Appl. Microbiol. Biotechnol. 2017, 101, 1323–1335. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Montalban-Lopez, M.; Mu, D.; Kuipers, O. Bacteriocins of lactic acid bacteria: Extending the family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef]
- Sinéad, C.; Li, Y.; Riedel, C.U.; O’Toole, P.W.; Hill, C.; Gahan, C.G.M. From the Cover: Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. USA 2007, 104, 761. [Google Scholar] [CrossRef]
- Bobay, L.-M.; Ochman, H. Impact of recombination on the base composition of bacteria and archaea. Mol. Biol. Evol. 2017, 34, 2627–2636. [Google Scholar] [CrossRef]
- Musto, H.; Naya, H.; Zavala, A.; Romero, H.; Alvarez-Valín, F.; Bernardi, G. Correlations between genomic GC levels and optimal growth temperatures in prokaryotes. FEBS Lett. 2004, 573, 73–77. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Mining genome traits that determine the different gut colonization potential of Lactobacillus and Bifidobacterium species. Microb. Genom. 2021, 7, 000581. [Google Scholar] [CrossRef]
- Maldonado-Gómez, M.X.; Martínez, I.; Bottacini, F.; O’Callaghan, A.; Ventura, M.; van Sinderen, D.; Hillmann, B.; Vangay, P.; Knights, D.; Hutkins, R.W. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe 2016, 20, 515–526. [Google Scholar] [CrossRef]
- Lerat, E.; Ochman, H. Recognizing the pseudogenes in bacterial genomes. Nucleic Acids Res. 2005, 33, 3125–3132. [Google Scholar] [CrossRef]
- Jung, J.; Kim, K.; Yoo, D.; Lee, C.; Kang, J.; Cho, K.; Kang, D.-K.; Kwak, W.; Yoon, S.H.; Sohn, H. Comparative genomic analysis of Lactobacillus plantarum GB-LP4 and identification of evolutionarily divergent genes in high-osmolarity environment. Genes Genom. 2018, 40, 217–223. [Google Scholar] [CrossRef]
- Walter, J.; Britton, R.A.; Roos, S. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc. Natl. Acad. Sci. USA 2011, 108, 4645–4652. [Google Scholar] [CrossRef]
- El Kafsi, H.; Binesse, J.; Loux, V.; Buratti, J.; Boudebbouze, S.; Dervyn, R.; Kennedy, S.; Galleron, N.; Quinquis, B.; Batto, J.M. Lactobacillus delbrueckii ssp. lactis and ssp. bulgaricus: A chronicle of evolution in action. BMC Genom. 2014, 15, 407. [Google Scholar] [CrossRef]


| Strain | Isolation Source | CDS | Size (Mb) | G + C Content (%) | rRNA | tRNA | Accession Number |
|---|---|---|---|---|---|---|---|
| L. pontis IMAU10341 | fermented milk | 1787 | 1.77 | 52.49 | 6 | 64 | GCF_026236575.1 |
| L. pontis IMAU10345 | fermented milk | 1786 | 1.76 | 52.49 | 6 | 64 | GCF_026236595.1 |
| L. pontis DSM8475T | rye sourdough | 1539 | 1.67 | 53.40 | 3 | 29 | GCF_001435345.1 |
| L. pontis LP475 | rye sourdough | 1561 | 1.71 | 53.30 | 18 | 67 | GCF_009428965.1 |
| L. pontis UMB0683 | catheter | 1698 | 1.87 | 48.70 | 7 | 59 | GCF_002940945.1 |
| L. pontis MAG.251 | broiler chicken gastrointestinal tract | 1551 | 1.60 | 53.60 | 1 | 32 | GCF_017888165.1 |
| L. antri DSM16041T | stomach mucosa | 2038 | 2.30 | 51.00 | 2 | 45 | GCF_000160835.1 |
| L. oris DSM4864T | saliva | 1861 | 2.03 | 50.00 | 2 | 51 | GCF_001434465.1 |
| L. panis DSM6035T | sourdough | 1797 | 2.01 | 48.10 | 2 | 33 | GCF_001435935.1 |
| L. reuteri DSM20016T | intestine of adult | 1889 | 2.00 | 38.90 | 18 | 68 | GCF_000016825.1 |
| L. vaginalis DSM5837T | vaginal swab | 1681 | 1.79 | 40.50 | 3 | 24 | GCF_001435915.1 |
| L. equigenerosi DSM18793T | thoroughbred horses | 1528 | 1.60 | 42.70 | 1 | 41 | GCF_001435245.1 |
| L. gastricus DSM16045T | gastric biopsies, stomach mucosa | 1761 | 1.85 | 41.60 | 3 | 37 | GCF_001434365.1 |
| L. fermentum DSM20052T | fermented beets | 1719 | 1.89 | 52.50 | 15 | 59 | GCF_013394085.1 |
| L. coleohominis DSM14060T | vagina | 1715 | 1.72 | 40.80 | 3 | 29 | GCF_001435055.1 |
| L. ingluviei DSM15946T | crop | 2055 | 2.15 | 49.90 | 2 | 69 | GCF_001435775.1 |
| L. mucosae DSM13345T | small intestine | 1990 | 2.25 | 46.40 | 1 | 42 | GCF_001436025.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, X.; Liu, Q.; Li, W.; Li, Y.; Zhao, L.; Liu, W. Comparative Genomics of Limosilactobacillus pontis Strains: Niche-Specific Variations and Adaptations. Diversity 2024, 16, 380. https://doi.org/10.3390/d16070380
Lei X, Liu Q, Li W, Li Y, Zhao L, Liu W. Comparative Genomics of Limosilactobacillus pontis Strains: Niche-Specific Variations and Adaptations. Diversity. 2024; 16(7):380. https://doi.org/10.3390/d16070380
Chicago/Turabian StyleLei, Xueyan, Qing Liu, Weicheng Li, Yu Li, Lixia Zhao, and Wenjun Liu. 2024. "Comparative Genomics of Limosilactobacillus pontis Strains: Niche-Specific Variations and Adaptations" Diversity 16, no. 7: 380. https://doi.org/10.3390/d16070380
APA StyleLei, X., Liu, Q., Li, W., Li, Y., Zhao, L., & Liu, W. (2024). Comparative Genomics of Limosilactobacillus pontis Strains: Niche-Specific Variations and Adaptations. Diversity, 16(7), 380. https://doi.org/10.3390/d16070380







