Phylogeography of Dolichophis Populations in the Aegean Region (Squamata: Colubridae) with Taxonomic Remarks
Abstract
1. Introduction
2. Material and Methods
2.1. Sampling
2.2. DNA Extraction, Molecular Markers, PCR Amplification and Sequencing
| Species | Code/Voucher Number (Museum) | Locality | N | E | Country | GenBank Number Cyt b/C-mos/BDNF/Rag1 | Source |
|---|---|---|---|---|---|---|---|
| D. jugularis | 492 (CUHC) | Palio Pyli, Kos Island | 36.8457 | 27.1885 | Greece | PP378899/-/-/- | This study |
| D. jugularis | 493 (CUHC) | Tigaki, Kos Island | 36.8888 | 27.2697 | Greece | PP378900/PP378937/PP378942/PP378947 | This study |
| D. jugularis | 12199 (I. Strachinis) | Alyki, Kos Island | 36.8779 | 27.1757 | Greece | PP378926/-/-/- | This study |
| D. jugularis | 12200 (I. Strachinis) | Zia, Kos Island | 36.8522 | 27.2316 | Greece | PP378927/-/-/- | This study |
| D. jugularis | 12204 (I. Strachinis) | Gennadi, Rhodes Island | 36.0298 | 27.9236 | Greece | PP378928/PP378940/PP378941/PP378948 | This study |
| D. jugularis | 10568 (ADU) | Kızılot, Antalya Province | 36.6864 | 31.6048 | Türkiye | PP378913/PP378938/PP378944/PP378949 | This study |
| D. jugularis | 10608 (ADU) | Bafa Lake, Muğla Province | 37.4859 | 27.5435 | Türkiye | PP378918/-/-/- | This study |
| D. jugularis | 10621 (ADU) | Sabuca Village-Koçarlı, Aydın Province | 37.7623 | 27.6765 | Türkiye | PP378920/-/-/- | This study |
| D. jugularis | 10627 (ADU) | Madran Village-Bozdoğan, Aydın Province | 37.6744 | 28.2866 | Türkiye | PP378921/-/-/- | This study |
| D. jugularis | 10629 (ADU) | Topçam-Çine, Aydın Province | 37.6891 | 28.0148 | Türkiye | PP378922/-/-/- | This study |
| D. jugularis | 10630 (ADU) | Güneyköy-Bozdoğan, Aydın Province | 37.7136 | 28.2114 | Türkiye | PP378923/-/-/- | This study |
| D. jugularis | MVZ 230242 | 2 km NE Finike, Antalya Province | 36.3238 | 30.1672 | Türkiye | AY486917/AY486941/- | [13] |
| D. jugularis | ZMHRU 2012/50 | Kocaaliler, Burdur Province | 37.3166 | 30.6833 | Türkiye | AY486917/-/- | [22] |
| D. jugularis | ZMHRU 2012/51 | Kızılseki, Burdur Province | 37.2666 | 30.7500 | Türkiye | AY486917/-/- | [22] |
| D. jugularis | - | - | - | - | Jordan | -/AY376798/- | [12] |
| D. caspius | 216 (ZMUP) | Apollonia | 40.6441 | 23.4561 | Greece | PP378890/-/-/- | This study |
| D. caspius | 231 (ZMUP) | Farsala | 39.2564 | 22.3159 | Greece | PP378891/-/-/- | This study |
| D. caspius | 418 (ZMUP) | Tinos Island | 37.5741 | 25.1501 | Greece | PP378892/-/-/- | This study |
| D. caspius | 427 (ZMUP) | Tinos Island | 37.5957 | 25.1535 | Greece | PP378893/-/-/- | This study |
| D. caspius | 430 (ZMUP) | Karpathos Island | 35.5235 | 27.1103 | Greece | PP378894/-/-/- | This study |
| D. caspius | 431 (ZMUP) | Karpathos Island | 35.5641 | 27.0993 | Greece | PP378895/-/-/- | This study |
| D. caspius | 458 (ZMUP) | Serifos Island | 37.1303 | 24.4876 | Greece | PP378896/-/-/- | This study |
| D. caspius | 459 (ZMUP) | Serifos Island | 37.1440 | 24.4485 | Greece | PP378897/-/-/- | This study |
| D. caspius | 489 (ZMUP) | Tigaki, Kos Island | 36.8756 | 27.1853 | Greece | PP378898/PP378936/PP378943/PP378946 | This study |
| D. caspius | 763 (ZMUP) | Loutros | 40.8770 | 26.0468 | Greece | PP378901/-/-/- | This study |
| D. caspius | 6146 (CUHC) | Geoponiki | 40.3080 | 23.0940 | Greece | PP378902/-/-/- | This study |
| D. caspius | 6618 (CUHC) | Iztok | 41.9908 | 27.5436 | Bulgaria | PP378903/-/-/- | This study |
| D. caspius | 7761/80.3.35.51 (NHMC) | Stratoni | 40.5141 | 23.8238 | Greece | PP378904/-/-/- | This study |
| D. caspius | 9895 (CUHC) | Metamorfosi | 40.5615 | 21.3069 | Greece | PP378905/-/-/- | This study |
| D. caspius | 9908 (CUHC) | Prinos | 39.5900 | 21.6188 | Greece | PP378906/-/-/- | This study |
| D. caspius | 9921 (CUHC) | Deskati | 39.9230 | 21.8209 | Greece | PP378907/-/-/- | This study |
| D. caspius | 9928 (CUHC) | Svoronos | 40.2645 | 22.4487 | Greece | PP378908/-/-/- | This study |
| D. caspius | 9929 (CUHC) | Lofos | 40.2526 | 22.4046 | Greece | PP378909/-/-/- | This study |
| D. caspius | 10551/231/2013 (ZDEU) | Çukurca, Domaniç, Kütahya Province | 39.7908 | 29.6895 | Türkiye | PP378910/-/-/- | This study |
| D. caspius | 10552/179/2014 (ZDEU) | İhsaniye, Eyüb, İstanbul Province | 41.2411 | 28.8048 | Türkiye | PP378911/-/-/- | This study |
| D. caspius | 10557/232/2013 (ZDEU) | Yörükakçayır, Eskişehir Province | 39.7438 | 30.3389 | Türkiye | PP378912/-/-/- | This study |
| D. caspius | 10573 (ADU) | Between Çalı and Atlas, Bursa Province | 40.1506 | 28.9042 | Türkiye | PP378914/-/-/- | This study |
| D. caspius | 10584 (ADU) | Kepez, Aydın Province | 37.8611 | 27.8540 | Türkiye | PP378915/PP378939/PP378945/- | This study |
| D. caspius | 10595 (ADU) | Pamukkale, Denizli Province | 37.9296 | 29.1332 | Türkiye | PP378916/-/-/- | This study |
| D. caspius | 10601 (ADU) | Between Simav and Demirci, Kütahya Province | 39.1249 | 28.8580 | Türkiye | PP378917/-/-/- | This study |
| D. caspius | 10614 (ADU) | Kuşadası, Aydın Province | 37.6866 | 27.1604 | Türkiye | PP378919/-/-/- | This study |
| D. caspius | 12195 (I. Strachinis) | Alykes, Lesbos Island | 39.2260 | 26.2429 | Greece | PP378924/-/-/- | This study |
| D. caspius | 12197 (I. Strachinis) | Antimachia, Kos Island | 36.8144 | 27.1086 | Greece | PP378925/-/-/- | This study |
| D. caspius | 12205 (I. Strachinis) | Agios Efstratios Island | 39.5348 | 24.9940 | Greece | PP378929/-/-/- | This study |
| D. caspius | 12206 (I. Strachinis) | Agios Efstratios Island | 39.5396 | 24.9962 | Greece | PP378930/-/-/- | This study |
| D. caspius | 12207 (I. Strachinis) | Kos Island | 36.7669 | 27.0901 | Greece | PP378931/-/-/- | This study |
| D. caspius | 12208 (I. Strachinis) | Samothraki Island | 40.5087 | 25.5579 | Greece | PP378932/-/-/- | This study |
| D. caspius | 12209 (I. Strachinis) | Samothraki Island | 40.4265 | 25.5311 | Greece | PP378933/-/-/- | This study |
| D. caspius | 12211 (I. Strachinis) | Samothraki Island | 40.4433 | 25.4977 | Greece | PP378934/-/-/- | This study |
| D. caspius | 12214 (I. Strachinis) | Repanidi, Limnos Island | 39.9242 | 25.3147 | Greece | PP378935/-/-/- | This study |
| D. caspius | NHMW KCC1 | Serifos Island | 37.1608 | 24.4810 | Greece | AY376739/AY376797/-/- | [12] |
| D. caspius | - | Andros Island | 37.8467 | 24.8961 | Greece | AY376739/-/-/- | [12] |
| D. caspius | - | Şile, Istanbul Province | 41.1670 | 29.5923 | Türkiye | HM210777/-/-/- | [10] |
| D. caspius | - | Samos Island | 37.7135 | 26.8166 | Greece | HM210778/-/-/- | [10] |
| D. caspius | - | Samos Island | 37.7135 | 26.8166 | Greece | HM210779/-/-/- | [10] |
| D. caspius | - | Euboea (Evvoia) Island | 38.5346 | 23.8197 | Greece | HM210780/-/-/- | [10] |
| D. caspius | - | Thasos Island | 40.6768 | 24.6439 | Greece | HM210782/-/-/- | [10] |
| D. caspius | - | Prespes | 40.7428 | 21.1597 | Greece | HM210782/-/-/- | [10] |
| D. caspius | - | Veles | 41.7081 | 21.7759 | North Macedonia | HM210782/-/-/- | [10] |
| D. caspius | - | Brest | 43.2759 | 21.7287 | Serbia | HM210782/-/-/- | [10] |
| D. caspius | - | Zlot | 44.0119 | 21.9880 | Serbia | HM210782/-/-/- | [10] |
| D. caspius | Kolets, Haskovo | 41.9207 | 25.5469 | Bulgaria | HM210783/-/-/- | [10] | |
| D. caspius | - | Shumen | 43.2491 | 26.9427 | Bulgaria | HM210783/-/-/- | [10] |
| D. caspius | - | Hagieni, Constanta | 43.7829 | 28.4842 | Romania | HM210783/-/-/- | [10] |
| D. caspius | - | Tekirdağ, Tekirdağ Province | 40.9845 | 27.5020 | Türkiye | HM210785/-/-/- | [10] |
| D. caspius | - | Samothraki Island | 40.4482 | 25.5848 | Greece | HM210787/-/-/- | [10] |
| D. schmidti | ZISP 27777 | Khumlakh | 39.2347 | 47.0514 | Armenia | AY376743/AY376801/-/- | [12] |
| D. schmidti | CAS 182953 | Mt. Dushak, Mary Province | 37.1300 | 60.0200 | Turkmenistan | -/AY486947/-/- | [13] |
| D. andreanus | ICSTZM.7H.1154 | Darreh Shahr, Ilam Province | 33.1444 | 47.3828 | Iran | MN531565/-/-/- | [23] |
| E. collaris | ZISP 27859 | Arax Fluss, n. Megri | 38.8852 | 46.2589 | Armenia | AY376766/-/-/- | [12] |
2.3. Sequence Alignment and Data Analysis
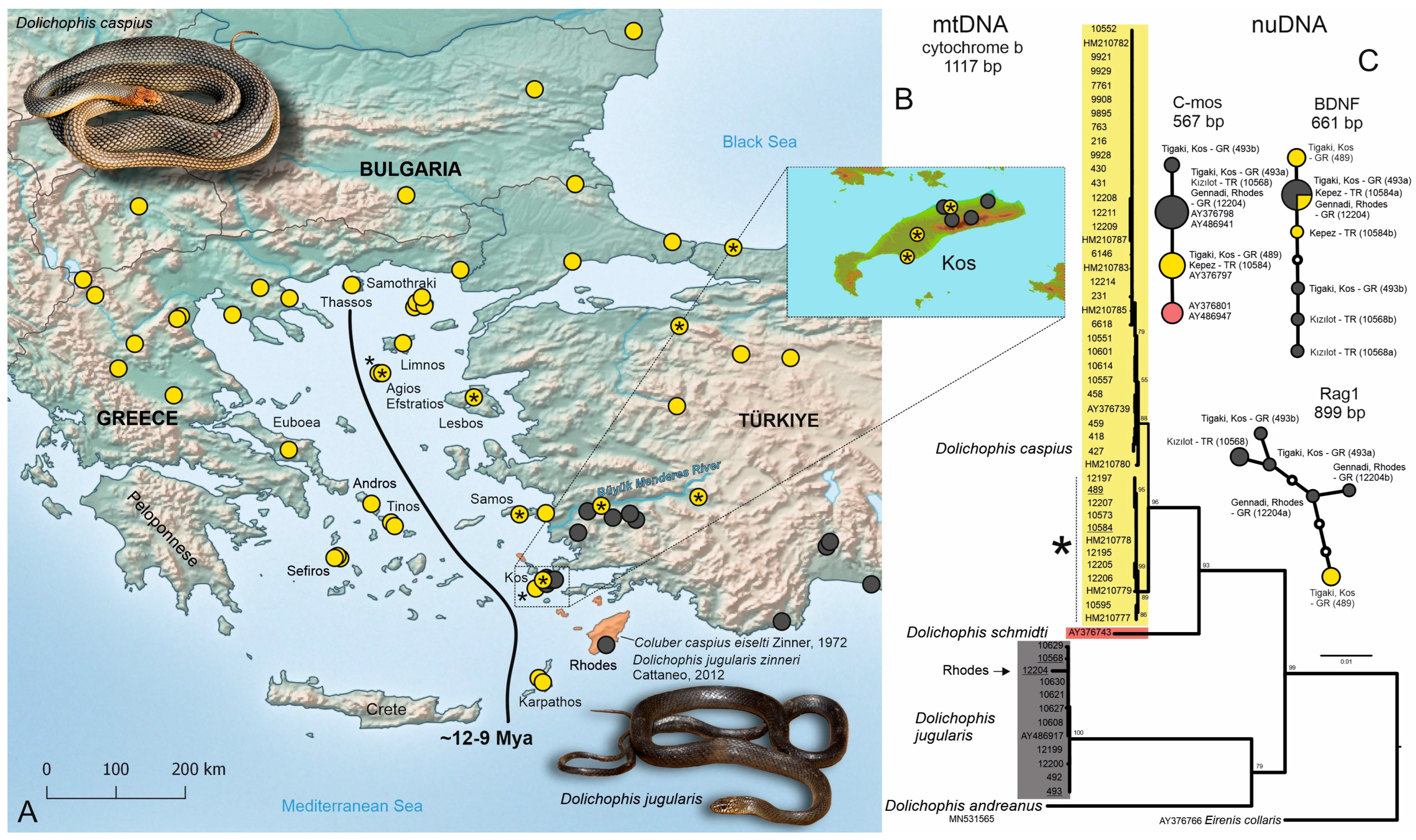
3. Results
3.1. Mitochondrial DNA Data
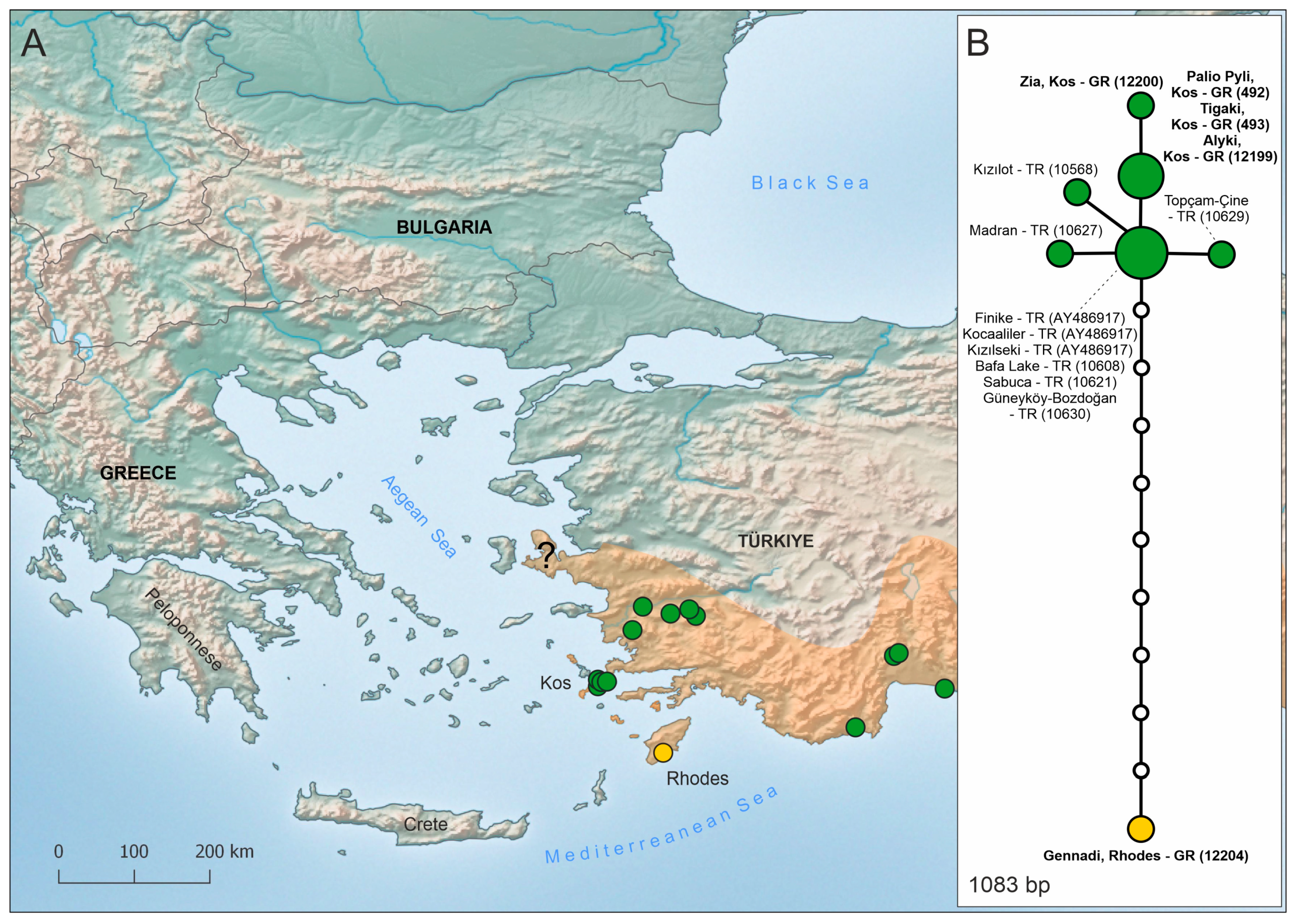
3.2. Nuclear DNA Data
4. Discussion
4.1. In Situ Evolution and Natural Dispersal
4.2. Human Mediated Introductions
4.3. Dolichophis jugularis and Island Endemism
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poulakakis, N.; Kapli, P.; Lymberakis, P.; Trichas, A.; Vardinoyiannis, K.; Sfenthourakis, S.; Mylonas, M. A review of phylogeographic analyses of animal taxa from the Aegean and surrounding regions. J. Zool. Syst. Evol. Res. 2015, 53, 18–32. [Google Scholar] [CrossRef]
- Dufresnes, C.; Probonas, N.M.; Strachinis, I. A reassessment of the diversity of green toads (Bufotes) in the circum-Aegean region. Integr. Zool 2021, 16, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Feiner, N.; Pinho, C.; While, G.M.; Kaliontzopoulou, A.; Harris, D.J.; Salvi, D.; Uller, T. Extensive introgression and mosaic genomes of Mediterranean endemic lizards. Nat. Commun. 2021, 12, 2762. [Google Scholar] [CrossRef] [PubMed]
- Jablonski, D.; Tzoras, E.; Panagiotopoulos, A.; Asztalos, M.; Fritz, U. Genotyping the phenotypic diversity in Aegean Natrix natrix moreotica (Bedriaga, 1882) (Reptilia, Serpentes, Natricidae). ZooKeys 2023, 1169, 87–94. [Google Scholar] [CrossRef]
- Kyriazi, P.; Kornilios, P.; Nagy, Z.T.; Poulakakis, N.; Kumlutaş, Y.; Ilgaz, Ҫ.; Avcı, A.; Göçmen, B.; Lymberakis, P. Comparative phylogeography reveals distinct colonization patterns of Cretan snakes. J. Biogeogr. 2013, 40, 1143–1155. [Google Scholar] [CrossRef]
- Thanou, E.; Kornilios, P.; Lymberakis, P.; Leaché, A.D. Genomic and mitochondrial evidence of ancient isolations and extreme introgression in the four-lined snake. Curr. Zool. 2020, 66, 99–111. [Google Scholar] [CrossRef]
- Asztalos, M.; Ayaz, D.; Bayrakci, Y.; Afsar, M.; Tok, C.V.; Kindler, C.; Jablonski, D.; Fritz, U. It takes two to tango—Phylogeography, taxonomy and hybridization in grass snakes and dice snakes (Serpentes: Natricidae: Natrix natrix, N. tessellata). Vertebr. Zool. 2021, 71, 813–834. [Google Scholar] [CrossRef]
- Sindaco, R.; Venchi, A.; Grieco, C. The Reptiles of the Western Palearctic 2. Annotated checklist and Distributional Atlas of the Snakes of Europe, North Africa, the Middle East and Central Asia, with an Update to the Volume 1; Societas Herpetologica Italica: Latina, Italy, 2013; p. 543. [Google Scholar]
- Baran, İ.; Avcı, A.; Kumlutaş, Y.; Olgun, K.; Ilgaz, Ç. Türkiye Amfibi ve Sürüngenleri; Palme Yayınevi: Ankara, Turkey, 2021; p. 230. ISBN 978-605-282-611-619. [Google Scholar]
- Nagy, Z.T.; Bellaagh, M.; Wink, M.; Paunović, A.; Korsós, Z. Phylogeography of the Caspian whipsnake in Europe with emphasis on the westernmost populations. Amphibia-Reptilia 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Mahtani-Williams, S.; Fulton, W.; Desvars-Larrive, A.; Lado, S.; Elbers, J.P.; Halpern, B.; Herczeg, D.; Babocsay, G.; Lauš, B.; Nagy, Z.T.; et al. Landscape Genomics of a Widely Distributed Snake, Dolichophis caspius (Gmelin, 1789) across Eastern Europe and Western Asia. Genes 2020, 11, 1218. [Google Scholar] [CrossRef]
- Nagy, Z.T.; Schmidtler, J.F.; Joger, U.; Wink, M. Systematik der Zwergnattern (Reptilia: Colubridae: Eirenis) und verwandter Gruppen anhand von DNA-Sequenzen und morphologischen Daten. Salamandra 2003, 39, 149–168. [Google Scholar]
- Nagy, Z.T.; Lawson, R.; Joger, U.; Wink, M. Molecular systematics of racers, whipsnakes and relatives (Reptilia: Colubridae) using mitochondrial and nuclear markers. J. Zool. Syst. Evol. Res. 2004, 42, 223–233. [Google Scholar] [CrossRef]
- Bader, T.; Riegler, C.; Grillitsch, H. The herpetofauna of the Island of Rhodes (Dodecanese, Greece). Herpetozoa 2009, 21, 147–169. [Google Scholar]
- Cattaneo, A. Il Colubro gola rossa dell’arcipelago di rodi: Dolichophis jugularis zinneri subsp. nova (Reptilia Serpentes). Nat. Sicil. 2012, 36, 77–103. [Google Scholar]
- Cattaneo, A. Morpho-ecology of the black whip snake Dolichophis jugularis (Linnaeus, 1758) of the southeast Aegean area (Dodecanese, SW Turkey) including previously unpublished growth rate data (Reptilia Serpentes). Nat. Sicil. 2018, 42, 3–24. [Google Scholar]
- Cattaneo, A.; Cattaneo, C.; Grano, M. Update on the herpetofauna of the Dodecanese Archipelago (Greece). Biodivers. J. 2020, 11, 69–84. [Google Scholar] [CrossRef]
- Cattaneo, A.; Cattaneo, C.; Grano, M. The herpetofauna of the Dodecanese Islands. In Life on Islands. 2. Zoological Diversity of the Aegean Archipelago. Studies dedicated to Norma Chapmann; Masseti, M., Ed.; Edizioni Danaus: Palermo, Italy, 2023; pp. 37–58. [Google Scholar]
- Szczerbak, N.N.; Böhme, W. Coluber caspius—Kaspische Pfeilnatter oder Springnatter. In Handbuch der Reptilien und Amphibien Europas, Band 3/I., Schlangen (Serpentes) I; Böhme, W., Ed.; Aula-Verlag: Wiesbaden, Germany, 1993; pp. 83–96. [Google Scholar]
- Zinner, H. Systematics and Evolution of the Species Group Coluber jugularis Linnaeus, 1758—Coluber caspius Gmelin, 1789 (Reptilia, Serpentes). PhD. Thesis, The Hebrew University of Jerusalem, Jerusalem, Israel, 1972; p. 62. [Google Scholar]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Göçmen, B.; Nagy, Z.T.; Ҫiçek, K.; Akman, B. An unusual juvenile coloration of the whip snake (Linnaeus, 1758) observed in Southwestern Anatolia, Turkey. Herpetol. Notes 2015, 8, 531–533. [Google Scholar]
- Rajabizadeh, M.; Pyron, R.A.; Nazarov, R.; Poyarkov, N.A.; Adriaens, D.; Herrel, A. Additions to the phylogeny of colubrine snakes in southwestern Asia, with description of a new genus and species (Serpentes: Colubridae: Colubrinae). PeerJ 2020, 8, C9016. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post analysis of large phylogenies. Bioinformatics 2019, 30, 1312–1313. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. Partitionfinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. Popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [PubMed]
- Fraser, D.J.; Bernatchez, L. Adaptive evolutionary conservation: Towards a unified concept for defining conservation units. Mol. Ecol. 2001, 10, 2741–2752. [Google Scholar] [CrossRef] [PubMed]
- Stephens, M.; Smith, N.J.; Donnelly, P.A. New statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef]
- Flot, J.F. Seqphase: A web tool for interconverting phase input/output fles and fasta sequence alignments. Mol. Ecol. Resour. 2010, 10, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Kaşot, N. Field notes on trophic spectrum of Dolichophis jugularis from Northern Cyprus. Biharean Biol. 2016, 10, 153–155. [Google Scholar]
- Schätti, B.; Utiger, U. Hemerophis, a new genus for Zamenis socotrae Günther, and a contribution to the phylogeny of Old World racers, whip snakes, and related genera (Reptilia: Squamata: Colubrinae). Rev. Suisse Zool. 2001, 108, 919–948. [Google Scholar] [CrossRef]
- Mertens, R.; Wermuth, H. Die Amphibien und Reptilien Europas (Dritte Liste, nach dem Stand vom 1. Januar 1960); Verlag Waldemar Kramer: Frankfurt am Main, France, 1960; p. XI + 264. [Google Scholar]
- Afrasiab, S.R.; Mohammad, M.K.; Hussein, A.M. Description of a new species of the genus Dolichophis Gitstel from the upper Mesopotamian Plain-Iraq (Reptilia-Serpentes-Colubridae). J. Biodivers. Environ. Sci. 2016, 8, 15–19. [Google Scholar]
- Sindaco, R.; Kornilios, P.; Sacchi, R.; Lymberakis, P. Taxonomic reassessment of Blanus strauchi (Bedriaga, 1884) (Squamata: Amphisbaenia: Blanidae), with the description of a new species from south-east Anatolia (Turkey). Zootaxa 2014, 3795, 311–326. [Google Scholar] [CrossRef]
- Karakasi, D.; Ilgaz, Ҫ.; Kumlutaş, K.; Güçlü, Ö.; Kankılıç, T.; Beşer, N.; Sindaco, R.; Lymberakis, P.; Poulakakis, N. More evidence of cryptic diversity in Anatololacerta species complex Arnold, Arribas and Carranza, 2007 (Squamata: Lacertidae) and re-evaluation of its current taxonomy. Amphibia-Reptilia 2021, 42, 201–216. [Google Scholar] [CrossRef]
- Hofmann, S.; Mebert, K.; Schulz, K.D.; Helfenberger, N.; Göçmen, B.; Böhme, W. A new subspecies of Zamenis hohenackeri (Strauch, 1873) (Serpentes: Colubridae) based on morphological and molecular data. Zootaxa 2018, 4471, 137–153. [Google Scholar] [CrossRef] [PubMed]
- Warnecke, H. Telescopus fallax (Fleischmann, 1831) auf den ozeanischen Strophaden-Inseln? Salamandra 1988, 24, 16–19. [Google Scholar]
- Insacco, G.; Spadola, F.; Russotto, S.; Scaravelli, D. Eryx jaculus (Linnaeus, 1758): A new species for the Italian herpetofauna (Squamata: Erycidae). Acta Herpetol. 2015, 10, 149–153. [Google Scholar] [CrossRef]
- Chondropoulos, B.P. A checklist of Greek reptiles. II. The snakes. Herpetozoa 1989, 2, 3–36. [Google Scholar]
- Tzoras, E.; Foufopoulos, J.; Volz, M.; Troidl, S.; Troidl, A.; Jablonski, D. Lost in the Cyclades: Genetic affiliation of the Yellow-bellied Toad, Bombina variegata (Anura: Bombinatoridae), from Paros Island, Greece. Salamandra 2023, 59, 92–95. [Google Scholar]
- Utiger, U.; Schätti, B. Morphology and phylogenetic relationships of the Cyprus racer, Hierophis cypriensis, and the systematic status of Coluber gemonensis gyarosensis Mertens (Reptilia: Squamata: Colubrinae). Rev. Suisse Zool. 2004, 111, 225–238. [Google Scholar] [CrossRef]
- Wettstein, O. Herpetologia aegaea. Sitzungsberichte der Akademie der Wissenschaften mathematisch-naturwissenschaftliche Klasse; Springer: Berlin/Heidelberg, Germany, 1953; Volume 162, pp. 651–833. [Google Scholar]
- Schätti, B.; Sigg, H. Die Herpetogauna der Insel Zypern. Teil 1: Die herpetologische Erforschung/Amphibien. Herpetofauna 1989, 11, 9–18. [Google Scholar]
- Göçmen, B.; Yildiz, M.Z. Snakes and their relations with humans. Kibr. Bilim (Cyprus Sci.) 2006, 2, 28–31. [Google Scholar]
- Kletečki, E.; Lanszki, J.; Trócsányi, B.; Mužinić, J.; Purger, J.J. First record of Dolichophis caspius (Gmelin, 1789), (Reptilia: Colubridae) on the island of Olib, Croatia. Nat. Croat. 2009, 18, 437–442. [Google Scholar]
- Beerli, P.; Hotz, H.; Uzzell, T. Geologically dated sea barriers calibrate a protein clock for aegean water frogs. Evolution 1996, 50, 1676–1687. [Google Scholar] [CrossRef]
- Dufresnes, C.; Poyarkov, N.; Jablonski, D. Acknowledging more biodiversity without more species. Proc. Natl. Acad. Sci. USA 2023, 120, e2302424120. [Google Scholar] [CrossRef] [PubMed]
- Kindler, C.; Böhme, W.; Corti, C.; Gvoždík, V.; Jablonski, D.; Jandzik, D.; Metallinou, M.; Široký, R.; Fritz, U. Mitochondrial phylogeography, contact zones and taxonomy of grass snakes (Natrix natrix, N. megalocephala). Zool. Scr. 2013, 42, 458–472. [Google Scholar] [CrossRef]
- Freitas, I.; Ursenbacher, S.; Mebert, K.; Zinenko, O.; Schweiger, S.; Wüster, W.; Brito, J.C.; Crnobrnja-Isailović, J.; Halpern, B.; Fahd, S.; et al. Evaluating taxonomic inflation: Towards evidence-based species delimitation in Eurasian vipers (Serpentes: Viperidae). Amphib. -Reptil. 2020, 41, 285–311. [Google Scholar] [CrossRef]
- Kalaentzis, K.; Kazilas, C.; Strachinis, I. Two cases of melanism in the Ring-headed Dwarf Snake Eirenis modestus (Martin, 1838) from Kastellorizo, Greece (Serpentes: Colubridae). Herpetol. Notes 2018, 11, 175–178. [Google Scholar]
- Schneider, B. Eine melanistische Schlanknatter, Coluber najadum kalymnensis n. subsp. (Colubridae, Serpentes), von der insel Kalymnos (Dodekanes, Ägäis). Bonn. Zool. Beitr. 1979, 30, 380–384. [Google Scholar]
- Kazilas, C.; Kalaentzis, K.; Strachinis, I. A case of piebaldism in Anatolian Worm Lizard Blanus strauchi (Bedriaga, 1884) (Squamata: Blanidae) from Kastellorizo, Greece. Herpetol. Notes 2018, 11, 527–529. [Google Scholar] [CrossRef]
- Kalogiannis, S. Cases of melanism in Dolichophis caspius (Gmelin, 1789) (Squamata: Colubridae) from Greece and a new distribution record. Parnass. Arch. 2021, 9, 19–22. [Google Scholar]
- Burbrink, F.T.; Lawson, R.; Slowinski, J.B. Mitochondrial DNA phylogeography of the North American rat snake (Elaphe obsoleta): A critique of the subspecies concept. Evolution 2000, 54, 2107–2114. [Google Scholar] [CrossRef]
- de Queiroz, A.; Lawson, R.; Lemos-Espinal, J.A. Phylogenetic relationships of North American garter snakes (Thamnophis) based on four mitochondrial genes: How much DNA sequence is enough? Mol. Phylogenet. Evol. 2002, 22, 315–329. [Google Scholar] [CrossRef]
- Lawson, R.; Slowinski, J.B.; Crother, B.I.; Burbrink, F.T. Phylogeny of the Colubroidea (Serpentes): New evidence from mitochondrial and nuclear genes. Mol. Phylogenet. Evol. 2005, 37, 581–601. [Google Scholar] [CrossRef]
- Vieites, D.R.; Min, M.S.; Wake, D.B. Rapid diversification and dispersal during periods of global warming by plethodontid salamanders. Proc. Nat. Acad. Sci. USA 2007, 104, 19903–19907. [Google Scholar] [CrossRef] [PubMed]
- Growth, J.G.; Barrowclough, G.F. Basal Divergences in Birds and the Phylogenetic Utility of the Nuclear RAG-1 Gene. Mol. Phylogenet. Evol. 1999, 12, 115–123. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Javorčík, A.; Strachinis, I.; Thanou, E.; Kornilios, P.; Avcı, A.; Üzüm, N.; Olgun, K.; Ilgaz, Ç.; Kumlutaş, Y.; Lymberakis, P.; et al. Phylogeography of Dolichophis Populations in the Aegean Region (Squamata: Colubridae) with Taxonomic Remarks. Diversity 2024, 16, 184. https://doi.org/10.3390/d16030184
Javorčík A, Strachinis I, Thanou E, Kornilios P, Avcı A, Üzüm N, Olgun K, Ilgaz Ç, Kumlutaş Y, Lymberakis P, et al. Phylogeography of Dolichophis Populations in the Aegean Region (Squamata: Colubridae) with Taxonomic Remarks. Diversity. 2024; 16(3):184. https://doi.org/10.3390/d16030184
Chicago/Turabian StyleJavorčík, Adam, Ilias Strachinis, Evanthia Thanou, Panagiotis Kornilios, Aziz Avcı, Nazan Üzüm, Kurtuluş Olgun, Çetin Ilgaz, Yusuf Kumlutaş, Petros Lymberakis, and et al. 2024. "Phylogeography of Dolichophis Populations in the Aegean Region (Squamata: Colubridae) with Taxonomic Remarks" Diversity 16, no. 3: 184. https://doi.org/10.3390/d16030184
APA StyleJavorčík, A., Strachinis, I., Thanou, E., Kornilios, P., Avcı, A., Üzüm, N., Olgun, K., Ilgaz, Ç., Kumlutaş, Y., Lymberakis, P., Nagy, Z. T., & Jablonski, D. (2024). Phylogeography of Dolichophis Populations in the Aegean Region (Squamata: Colubridae) with Taxonomic Remarks. Diversity, 16(3), 184. https://doi.org/10.3390/d16030184






