Revealing the Diversity of Thin Filamentous Cyanobacteria, with the Discovery of a Novel Species, Pegethrix qiandaoensis sp. nov. (Oculatellaceae, Oculatellales), in a Freshwater Lake in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Collection and Cultivation
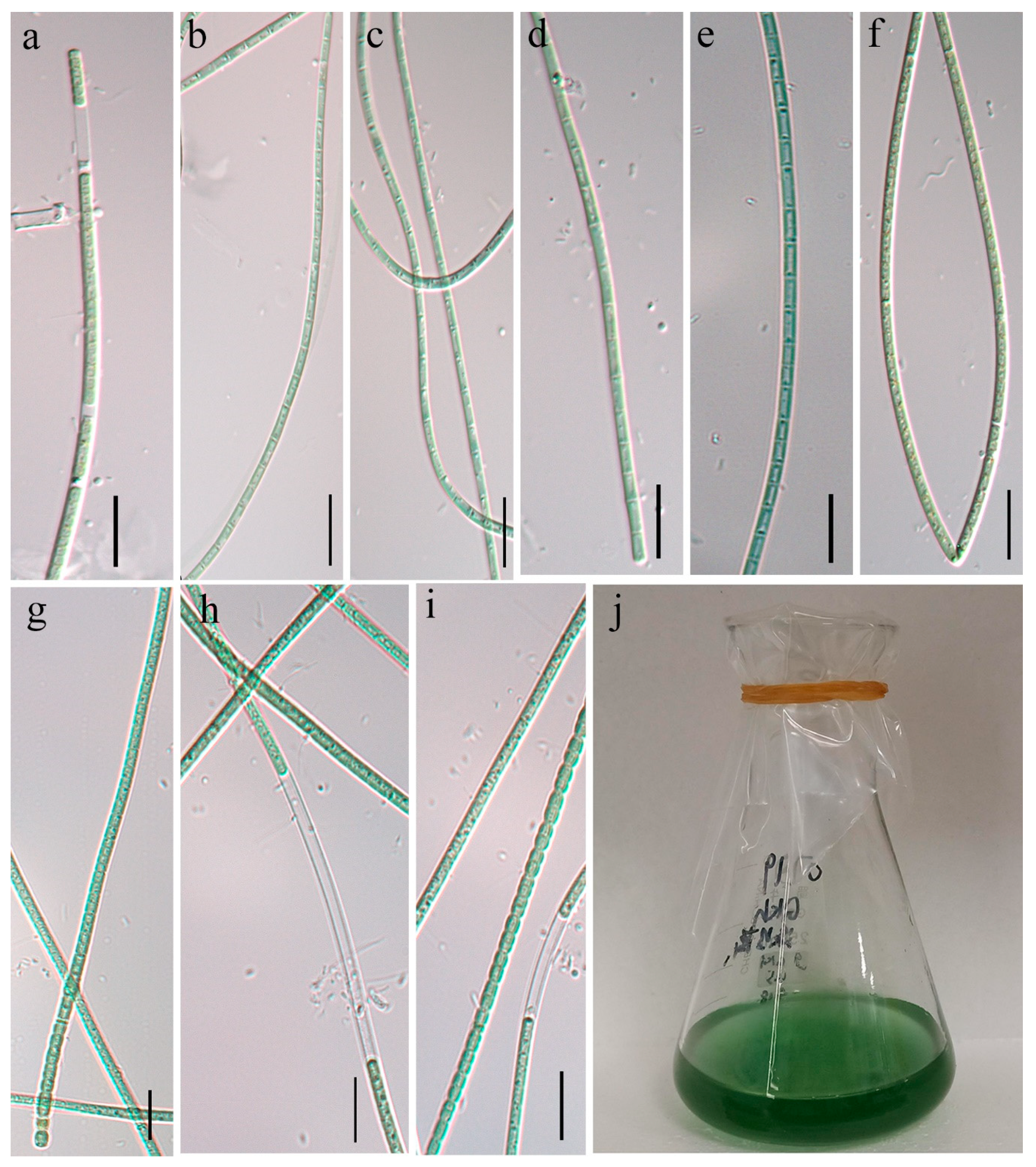
2.2. Morphological Characterization
2.3. DNA Extraction, PCR Amplification, and Sequencing
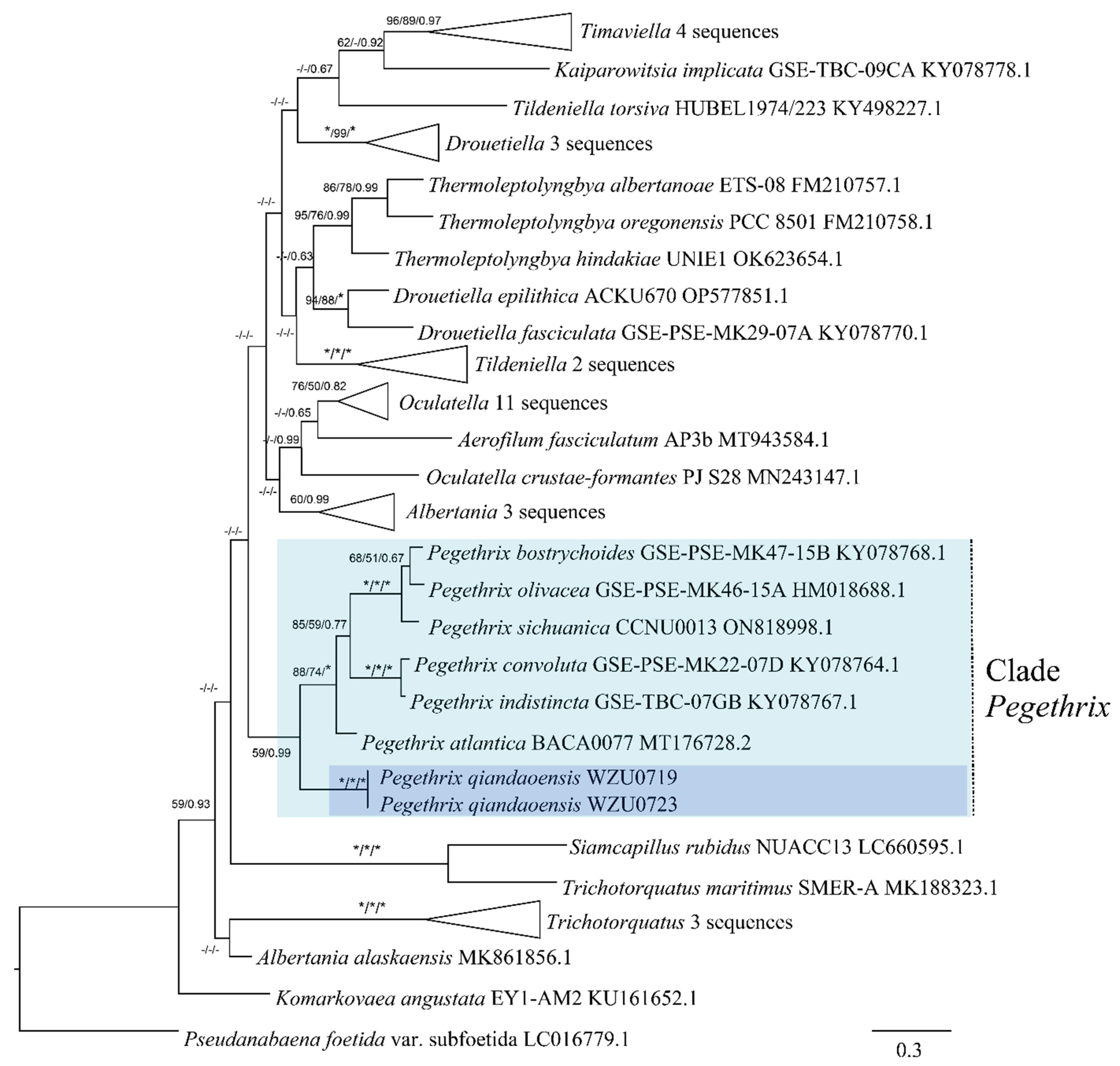
2.4. Phylogenetic Analysis
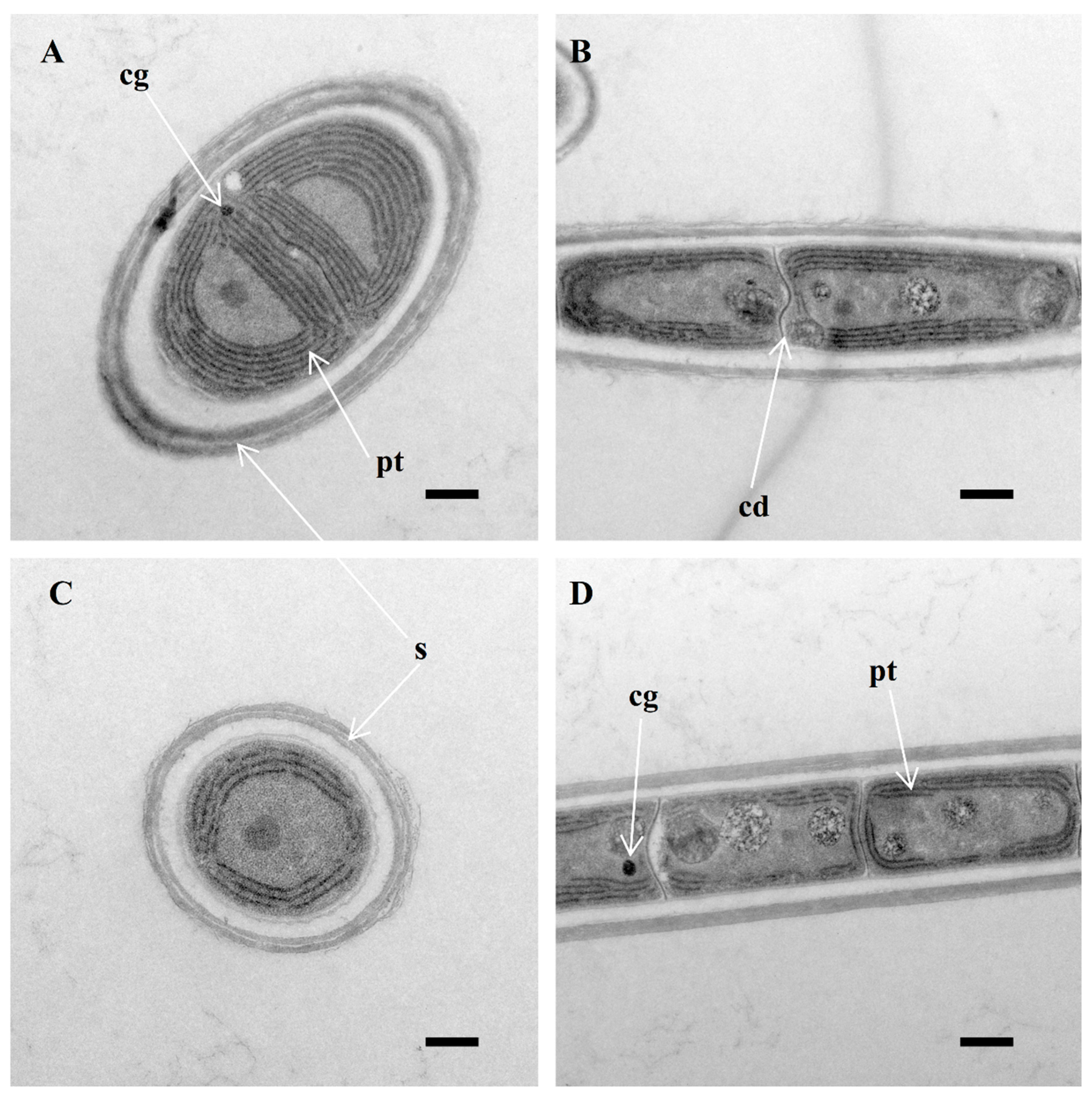
2.5. Morphological Ultrastructural Characterization
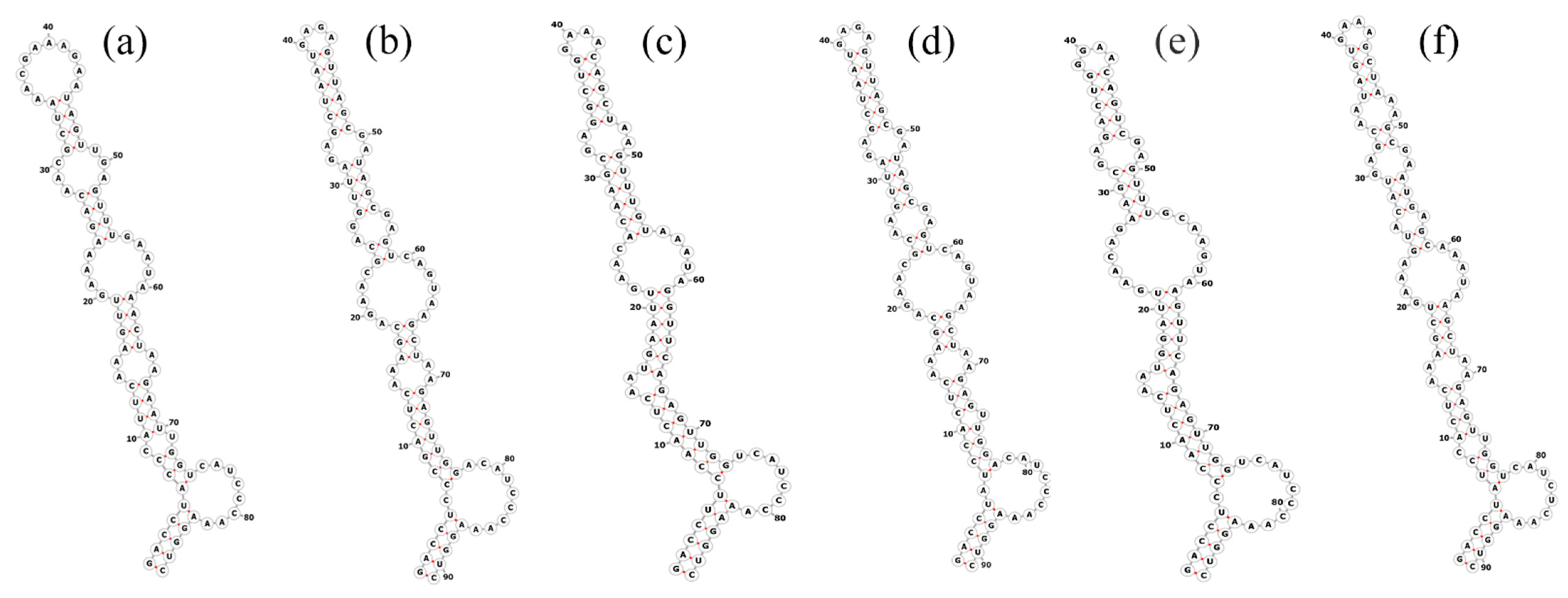
2.6. Analyses of 16S–23S Internal Transcribed Spacer (ITS)
3. Results
3.1. Morphological Description
3.2. Molecular and Phylogenetic Analyses
3.3. Molecular Analyses on the ITS between 16S–23S rRNA Genes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whitton, B.A. The Ecology of Cyanobacteria II: Their Diversity in Time and Space; Springer: Dordrecht, The Netherlands, 2012; 669p. [Google Scholar]
- Paerl, H.W.; Huisman, J. Climate—Blooms like it hot. Science 2008, 320, 57–58. [Google Scholar] [CrossRef]
- Anagnostidis, K.; Komárek, J. Modern approach to the classification system of the Cyanophytes 3: Oscillatoriales. Algol. Stud. 1988, 50, 327–472. [Google Scholar]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphyletic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Kaštovský, J.; Gomez, E.B.; Hladil, J.; Johansen, J.R. Cyanocohniella calida gen. et sp. nov. (Cyanobacteria: Aphanizomenonaceae) a new cyanobacterium from the thermal springs from Karlovy Vary, Czech Republic. Phytotaxa 2014, 181, 279–292. [Google Scholar] [CrossRef]
- Casamatta, D.A.; Johansen, J.R.; Vis, M.L.; Broadwater, S.T. Molecular and morphological characterization of ten polar and near-polar strains within the Oscillatoriales (Cyanobacteria). J. Phycol. 2005, 41, 421–438. [Google Scholar] [CrossRef]
- Hašler, P.; Dvořák, P.; Poulíčková, A.; Casamatta, D.A. A novel genus Ammassolinea gen. nov. (Cyanobacteria) isolated from sub–tropical epipelic habitats. Fottea 2014, 14, 241–248. [Google Scholar] [CrossRef]
- Osorio-Santos, K.; Pietrasiak, N.; Bohunicka, M.; Miscoe, L.; Kovacik, L.; Martin, M.P.; Johansen, J.R. Seven new species of Oculatella (Pseudanabaenales, Cyanobacteria): Taxonomically recognizing cryptic diversification. Eur. J. Phycol. 2014, 49, 450–470. [Google Scholar] [CrossRef]
- Dvořák, P.; Poulíčková, A.; Hašler, P.; Belli, M.; Casamatta, D.A.; Papini, A. Species concepts and speciation factors in cyanobacteria, with connection to the problems of diversity and classification. Biodivers. Conserv. 2015, 24, 739–757. [Google Scholar] [CrossRef]
- Genuário, D.B.; Vaz, M.G.M.V.; Hentschke, G.S.; Sant’anna, C.L.; Fiore, M.F. Halotia gen. nov., a phylogenetically and physiologically coherent cyanobacterial genus isolated from marine coastal environments. Int. J. Syst. Evol. Microbiol. 2015, 15, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Hauerová, R.; Hauer, T.; Kaštovský, J.; Komárek, J.; Lepšová-Skácelová, O.; Mareš, J. Tenebriella gen. nov.—The dark twin of Oscillatoria. Mol. Phylogenet. Evol. 2021, 165, 107293. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.R.; Olsen, C.E.; Lowe, R.L.; Fučíková, K.; Casamatta, D.A. Leptolyngbya species from selected seep walls in the Great Smoky Mountains National Park. Algol. Stud. 2008, 126, 21–36. [Google Scholar] [CrossRef]
- Turicchia, S.; Ventura, S.; Komárková, J.; Komárek, J. Taxonomic evaluation of the cyanobacterial microflora from alkaline marshes of northern Belize. 2. Diversity of oscillatorialean genera. Nova Hedwig. 2009, 89, 165–200. [Google Scholar] [CrossRef]
- Komárek, J. Several problems of the polyphasic approach in the modern cyanobacterial system. Hydrobiologia 2018, 811, 7–17. [Google Scholar] [CrossRef]
- Strunecký, O.; Raabova, L.; Bernardova, A.; Ivanova, A.P.; Semanova, A.; Crossley, J.; Kaftan, D. Diversity of cyanobacteria at the Alaska North Slope with description of two new genera: Gibliniella and Shackletoniella. FEMS Microbiol. Ecol. 2019, 96, fiz189. [Google Scholar] [CrossRef]
- Strunecký, O.; Ivanova, A.P.; Mareš, J. An updated classification of cyanobacterial orders and families based on phylogenomic and polyphasic analysis. J. Phycol. 2022, 59, 12–51. [Google Scholar] [CrossRef]
- Mai, T.; Johansen, J.R.; Pietrasiak, N.; Bohunická, M.; Martin, M.P. Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. Phytotaxa 2018, 365, 1–59. [Google Scholar] [CrossRef]
- Shen, L.; Zhang, Z.; Huang, L.; Zhang, L.; Yu, G.; Chen, M.; Li, R.; Qiu, B. Chlorophyll f production in two new subaerial cyanobacteria of the family Oculatellaceae. J. Phycol. 2023, 59, 370–382. [Google Scholar] [CrossRef]
- Luz, R.; Cordeiro, R.; Kaštovský, J.; Johansen, J.R.; Dias, E.; Fonseca, A.; Urbatzka, R.; Vasconcelos, V.; Gonçalves, V. New terrestrial cyanobacteria from the Azores Islands: Description of Venetifunis gen. nov. and new species of Albertania, Kovacikia and Pegethrix. Phycologia 2023, 62, 483–498. [Google Scholar] [CrossRef]
- Stanier, R.Y.; Kunisawa, R.; Mandel, M.; Cohen–Bazire, G. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 1971, 35, 171–205. [Google Scholar] [CrossRef]
- Neilan, B.A.; Jacobs, D.; Goodman, A.E. Genetic diversity and phylogeny of toxic cyanobacteria determined by DNA polymorphisms within the phycocyanin locus. Appl. Environ. Microbiol. 1995, 61, 3875–3883. [Google Scholar] [CrossRef]
- Edwards, U.; Rogall, T.; Blöcker, H.; Emde, M.; Böttger, E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar] [CrossRef]
- Gkelis, S.; Rajaniemi, P.; Vardaka, E.; Moustaka–Gouni, M.; Lanaras, T.; Sivonen, K. Limnothrix redekei (Van Goor) Meffert (Cyanobacteria) strains from Lake Kastoria, Greece from a separate phylogenetic group. Microb. Ecol. 2005, 49, 176–182. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Page, R.D.M. TreeView: An application to display phylogenetic trees on personal computers. Bioinformatics 1996, 12, 357–358. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Mathews, D.H.; Disney, M.D.; Childs, J.L.; Schroeder, S.J.; Zuker, M.; Turner, D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Nucleic Acids Res. 2004, 101, 7287–7292. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Geng, R.; Shan, L.; Liu, Y.; Zhang, H.; Xiao, P.; Li, R. Taxonomic Discussion on Cyanobacterial Systematics at Family Level, with Special Regards to Phormidiaceae by Using the Strains of Chinese Newly Recorded Genera Ancylothrix and Potamolinea. Diversity 2022, 14, 301. [Google Scholar] [CrossRef]
- Cai, F.; Li, S.; Zhang, H.; Yu, G.; Li, R. Nodosilinea hunanesis sp. nov. (Prochlorotrichaceae, Synechococcales) from a Freshwater Pond in China Based on a Polyphasic Approach. Diversity 2022, 14, 364. [Google Scholar] [CrossRef]
- Perkerson, R.B.; Johansen, J.R.; Kovacik, L.; Brand, J.; Kastovsky, J.; Casamatta, D.A. A unique pseudanabaenalean (Cyanobacteria) genus Nodosilinea gen. nov. based on morphological and molecular data. J. Phycol. 2011, 47, 1397–1412. [Google Scholar] [CrossRef]
- Sciuto, K.; Moro, I. Detection of the new cosmopolitan genus Thermoleptolyngbya (Cyanobacteria, Leptolyngbyaceae) using the 16S rRNA gene and 16S–23S ITS region. Mol. Phylogenet. Evol. 2016, 105, 15–35. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Ebers, J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 2006, 4, 6–9. [Google Scholar]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.H. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Erwin, P.M.; Thacker, R.W. Cryptic diversity of the symbiotic cyanobacterium Synechococcus spongiarum among sponge hosts. Mol. Ecol. 2008, 17, 2937–2947. [Google Scholar] [CrossRef]
- Strunecký, O.; Bohunická, M.; Johansen, J.R.; Čapková, K.; Raabová, L.; Dvořák, P.; Komárek, J. A revision of the genus Geitlerinema and a description of the genus Anagnostidinema gen. nov. (Oscillatoriophycidae, Cyanobacteria). Fottea 2017, 17, 114–126. [Google Scholar] [CrossRef]
- Rasouli–Dogaheh, S.; Noroozi, M.; Khansha, J.; Amoozegar, M.A.; Rahimi, R.; Nikou, M.M.; Hauer, T. Khargia gen. nov., a new genus of simple trichal Cyanobacteria from the Persian Gulf. Fottea 2023, 23, 49–61. [Google Scholar] [CrossRef]



| Strain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. PP084669 Pegethrix qiandaoensis WZU 0719 | ||||||||||
| 2. OR815386 Pegethrix qiandaoensis WZU 0723 | 100.00% | |||||||||
| 3. OP410738.1 Pegethrix sichuanica CCNU0013 | 96.54% | 96.54% | ||||||||
| 4. MT176728.2 Pegethrix atlantica BACA0077 | 96.75% | 96.75% | 98.64% | |||||||
| 5. KY078768.1 Pegethrix bostrychoides GSE-PSE-MK47-15B | 97.38% | 97.38% | 98.01% | 98.22% | ||||||
| 6. KY078764.1 Pegethrix convoluta GSE-PSE-MK22-07D | 96.75% | 96.75% | 98.53% | 98.32% | 98.43% | |||||
| 7. KY078767.1 Pegethrix indistincta GSE-TBC-07GB | 96.86% | 96.86% | 98.64% | 98.43% | 98.53% | 99.90% | ||||
| 8. HM018688.1 Pegethrix olivacea GSE-PSE-MK46-15A | 97.17% | 97.17% | 98.22% | 98.01% | 98.74% | 98.64% | 98.74% | |||
| 9. NR172705.1 Trichotorquatus maritimus SMER-A | 91.51% | 91.51% | 90.46% | 90.46% | 90.04% | 90.67% | 90.78% | 90.46% | ||
| 10. HM018690.1 Drouetiella lurida Lukesova 1986/6 | 93.50% | 93.50% | 93.61% | 93.19% | 93.19% | 93.40% | 93.50% | 92.98% | 91.51% | |
| 11. HQ917692.1 Oculatella subterranea SP1402/Zammit 2007/5 | 92.14% | 92.14% | 92.45% | 92.24% | 92.24% | 92.98% | 92.87% | 92.35% | 91.72% | 92.24% |
| Strain | Complete ITS | D1-D1′ Helix | D2 | tRNA Ile | V2 Spacer | tRNA Ala | Box-B Helix | Box-A Helix | D4 | V3 Helix |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. Pegethrix qiandaoensis WZU 0719 | 649 | 87 | 12 | 74 | 60 | 73 | 50 | 12 | 7 | 126 |
| 2. Pegethrix qiandaoensis WZU 0723 | 649 | 87 | 12 | 74 | 60 | 73 | 50 | 12 | 7 | 126 |
| 3. KY078768.1 Pegethrix bostrychoides GSE-PSE-MK47-15B | 614 | 87 | 12 | 74 | 36 | 73 | 36 | 12 | 7 | 94 |
| 4. KY078764.1 Pegethrix convoluta GSE-PSE-MK22-07D | 627 | 91 | 12 | 74 | 14 | 73 | 36 | 12 | 7 | 108 |
| 5. KY078767.1 Pegethrix indistincta GSE-TBC-07GB | 627 | 91 | 12 | 74 | 14 | 73 | 36 | 12 | 7 | 108 |
| 6. HM018688.1 Pegethrix olivacea GSE-PSE-MK46-15A | 614 | 87 | 12 | 74 | 36 | 73 | 36 | 12 | 7 | 94 |
| 7. OP410738.1 Pegethrix sichuanica CCNU0013 | 559 | 87 | 12 | 74 | 15 | 73 | 38 | 12 | 7 | 95 |
| 8. MT176728.2 Pegethrix atlantica BACA0077 | 630 | 91 | 12 | 74 | 29 | 73 | 33 | 12 | 7 | 96 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, K.; Cheng, Y.; Geng, R.; Xiao, P.; Zhang, H.; Wu, Z.; Cai, F.; Li, R. Revealing the Diversity of Thin Filamentous Cyanobacteria, with the Discovery of a Novel Species, Pegethrix qiandaoensis sp. nov. (Oculatellaceae, Oculatellales), in a Freshwater Lake in China. Diversity 2024, 16, 161. https://doi.org/10.3390/d16030161
Gao K, Cheng Y, Geng R, Xiao P, Zhang H, Wu Z, Cai F, Li R. Revealing the Diversity of Thin Filamentous Cyanobacteria, with the Discovery of a Novel Species, Pegethrix qiandaoensis sp. nov. (Oculatellaceae, Oculatellales), in a Freshwater Lake in China. Diversity. 2024; 16(3):161. https://doi.org/10.3390/d16030161
Chicago/Turabian StyleGao, Kaihui, Yao Cheng, Rouzhen Geng, Peng Xiao, He Zhang, Zhixu Wu, Fangfang Cai, and Renhui Li. 2024. "Revealing the Diversity of Thin Filamentous Cyanobacteria, with the Discovery of a Novel Species, Pegethrix qiandaoensis sp. nov. (Oculatellaceae, Oculatellales), in a Freshwater Lake in China" Diversity 16, no. 3: 161. https://doi.org/10.3390/d16030161
APA StyleGao, K., Cheng, Y., Geng, R., Xiao, P., Zhang, H., Wu, Z., Cai, F., & Li, R. (2024). Revealing the Diversity of Thin Filamentous Cyanobacteria, with the Discovery of a Novel Species, Pegethrix qiandaoensis sp. nov. (Oculatellaceae, Oculatellales), in a Freshwater Lake in China. Diversity, 16(3), 161. https://doi.org/10.3390/d16030161








