Zoonotic Pathogens Isolated from an Introduced Population of Red Swamp Crayfish (Procambarus clarkii) in Tenerife (Canary Islands, Spain)
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
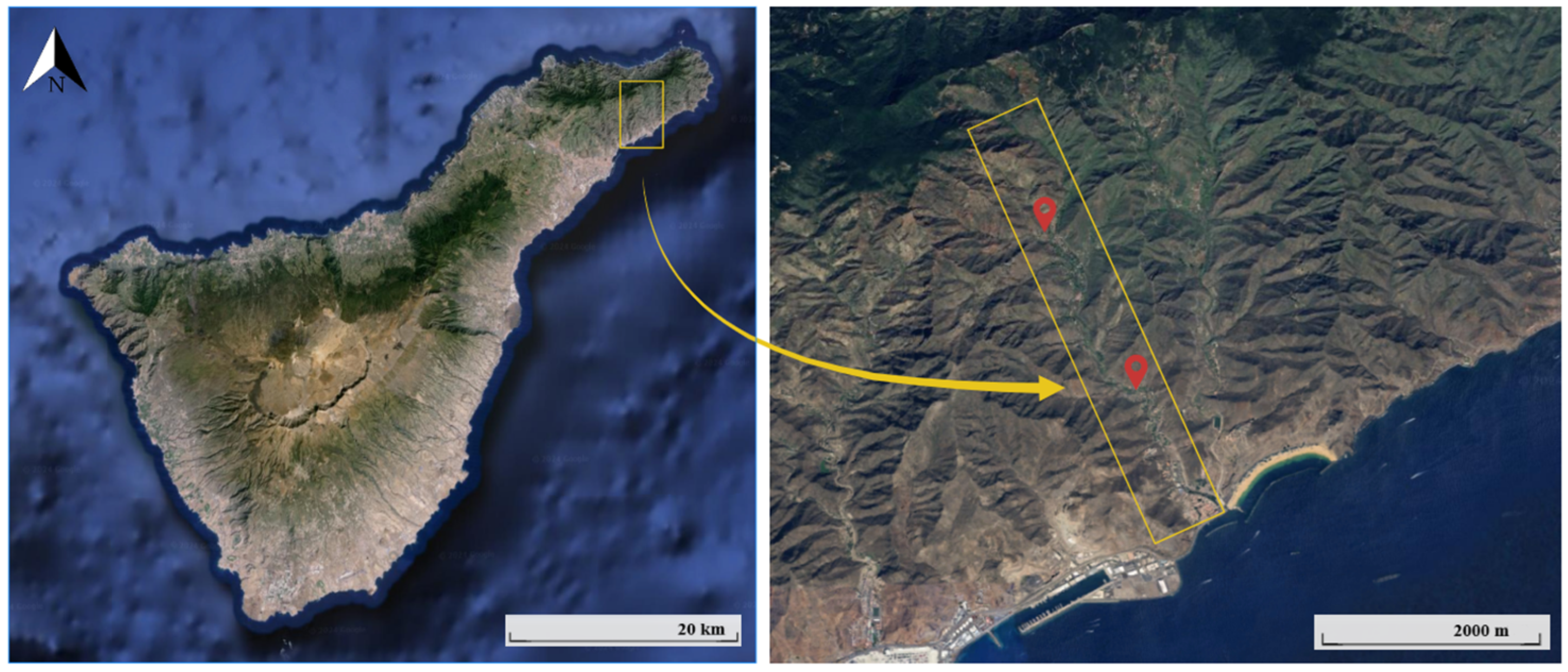
2.2. Sample Preparation
2.3. Detection and Identification of Pathogenic Bacteria
2.3.1. Detection of Pathogenic Bacteria Using the Biofire FilmArray™ System (BioMérieux)
2.3.2. Detection of Pathogenic Bacteria Using Culture–PCR
2.3.3. Molecular Identification of Isolates
DNA Extraction
PCR Assays
2.4. Statistical Analysis
3. Results
3.1. Assays Conducted with the Biofire FilmArray™ System
3.2. Culturing-PCR Assays
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Claire, W.H.; Wroiten, J.W. First Record of the Crayfish, Procambarus clarkii, from Idaho, USA (Decapoda, Cambaridae). Crustaceana 1978, 35, 317–319. [Google Scholar]
- Hobbs, H.H., Jr. Crayfish distribution, adaptive radiation and evolution. In Feshwater Crayfish: Biology Management and Exploitation; Croom Helm: London, UK, 1988; pp. 52–82. [Google Scholar]
- Gherardi, F. Crayfish Invading Europe: The Case Study of Procambarus clarkii. Mar. Freshw. Behav. Physiol. 2006, 39, 175–191. [Google Scholar] [CrossRef]
- Dörr, A.J.M.; Scoparo, M.; Cardinali, I.; La Porta, G.; Caldaroni, B.; Magara, G.; Pallottini, M.; Selvaggi, R.; Cenci-Goga, B.; Goretti, E.; et al. Population Ecology and Genetic Diversity of the Invasive Alien Species Procambarus clarkii in Lake Trasimeno (Italy). Biology 2021, 10, 1059. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, C.F.; Bécares, E.; Fernández-Aláez, M. Shift from clear to turbid pase in Lake Chozas (NW Spain) due to the introduction of American red swamp crayfish (Procambarus clarkii). Hydrobiologia 2003, 506, 421–426. [Google Scholar] [CrossRef]
- Dörr, A.J.; Rodolfi, M.; Scalici, M.; Elia, A.C.; Garzoli, L.; Picco, A.M. Phoma glomerata, a Potential New Threat to Italian Inland Waters. J. Nat. Conserv. 2011, 19, 370–373. [Google Scholar] [CrossRef]
- Dörr, A.J.; Elia, A.C.; Rodolfi, M.; Garzoli, L.; Picco, A.M.; D’Amen, M.; Scalici, M.A. Model of Co-Occurrence: Segregation and Aggregation Patterns in the Mycoflora of the Crayfish Procambarus clarkii in Lake Trasimeno (Central Italy). J. Limnol. 2012, 71, 135–143. [Google Scholar] [CrossRef][Green Version]
- Isaacs, J.C.; Lavergne, D.R. Louisiana Commercial Crawfish Harvester’s Survey Report. La. Department of Wildlife and Fisheries, Socioeconomic Research and Development Section. 2010. Available online: https://www.lsuagcenter.com/NR/rdonlyres/CF07A02D-B4FA-4304-93A7-64809619B6FC/80317/CrawfishHarvestersReport2010.pdf (accessed on 11 April 2021).
- El-Kholie, E.M.; Khader, S.A.; Abdelreheem, M.A.T. Chemical, physical, microbiological and quality attributes studies on River Nile crayfish. Afr. J. Biotechnol. 2012, 11, 11262–11270. [Google Scholar] [CrossRef]
- Wang, W.; Gu, W.; Ding, Z.; Ren, Y.; Chen, J.; Hou, Y. A novel Spiroplasma pathogen causing systemic infection in the crayfish Procambarus clarkii Crustacea: Decapod, in China. FEMS Microbiol. Lett. 2005, 249, 131–137. [Google Scholar] [CrossRef]
- Xiao, X.; Han, D.; Zhu, X.; Yang, Y.; Xie, S.; Huang, Y. Effect of dietary cornstarch levels on growth performance, enzyme activity and hepatopancreas histology of juvenile red swamp crayfish, Procambarus clarkii (Girard). Aquaculture 2014, 426, 112–119. [Google Scholar] [CrossRef]
- Gutiérrez Yurrita, P.J. El Papel Ecológico del Cangrejo Rojo (“Procambarus clarkii”), en los Ecosistemas Acuáticos del Parque Nacional de Doñana una Perspectiva Ecofisiológica y Bioenértica. Doctoral Dissertation, Universidad Autónoma de Madrid, Madrid, Spain, 1997. [Google Scholar]
- Geiger, W.; Alcorlo, P.; Batanás, A.; Montes, C. Impact of an introduced Crustacean on the trophic webs of Mediterranean wetlands. Biol. Invasions 2005, 7, 49–73. [Google Scholar]
- Gil-Sánchez, J.M.; Alba-Tercedor, J. Ecology of the native and introduced crayfishes Austropotamobius pallipes and Procambarus clarkii in southern Spain and implications for conservation of the native species. Biol. Conserv. 2002, 105, 75–80. [Google Scholar] [CrossRef]
- Ramos, A.; Pereira, T. Un novo Astacidae para a fauna portuguesa: Procambarus clarkii (Girard, 1852). Bol. Inst. Nac. Investig. Pescas Lisb. 1981, 6, 37–47. [Google Scholar]
- Correira, A.M.; Costa, A.C. Introduction of Red Swamp Crayfish Procambarus clarkii (Crustacea, Decapoda) in São Miguel, Azores, Portugal. Archipiélago 12ª. 1994, pp. 67–73. Available online: https://repositorio.uac.pt/bitstream/10400.3/2124/1/LMSpp67-73_CORREIA_COSTA-N12A.pdf (accessed on 2 June 2021).
- Herrera-Arteaga, G.A.; Barquín Díez, J.; de los Santos Gómez, A. Colonización de la isla de Tenerife (Islas Canarias) por el cangrejo rojo americano Procambarus clarkii Girard (1852) (Decapoda, Cambaridae). Rev. Acad. Canar. Cienc. 2006, 4, 81–88. [Google Scholar]
- Mingorance, M.C.; Gómez, J.I. Una actividad de educación ambiental basada en observaciones del cangrejo de río americano (Procambrus clarkii) en el Barranco del Cercado (Tenerife, Islas Canarias). Ecosistemas 2002, 11, 1–5. Available online: https://www.academia.edu/121093318/Una_actividad_de_educaci%C3%B3n_ambiental_basada_en_observaciones_del_cangrejo_de_r%C3%ADo_americano_Procambarus_clarkii_en_el_Barranco_del (accessed on 2 June 2021).
- Verdú, J.R.; Numa, C.; Galante, E. Atlas y Libro Rojo de los Invertebrados Amenazados de España (Especies Vulnerables) Volumen II. Dirección General de Medio Natural y Política Forestal; Ministerio de Medio Ambiente, Medio Rural y Marino: Madrid, Spain, 2011; 595p. [Google Scholar]
- Dong, X.; Li, Z.; Wang, X.; Zhou, M.; Lin, L.; Zhou, Y.; Li, J. Characteristics of Vibrio parahaemolyticus isolates obtained from crayfish (Procambarus clarkii) in freshwater. Int. J. Food Microbiol. 2016, 238, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Palillo, J.A.; Mollenkopf, D.; Marsh, A.E.; Wittum, T.E.; James, J.P.B.; Reichley, S.R.; Ghosh, S.; Palillo, M.B.; Malbrue, R. Detection of Zoonotic Bacteria and Paragonimus kellicotti in Red Swamp Crayfish (Procambarus clarkii) and the Assessment of Traditional Crayfish Boils. J. Food Prot. 2022, 85, 1388–1396. [Google Scholar] [CrossRef]
- Elmossalami, M.K.; Emara, M.T. Safety and quality of freshwater crayfish Procambarus clarkii in the river Nile. Food/Nahrung 1999, 43, 126–128. [Google Scholar] [CrossRef]
- Saad El-Deen, A.G. Isolation and identification of enterobacteriaceae from freshwater grayfish (Procambarus clarkii). Assiut Vet. Med. J. 2009, 55, 1–11. [Google Scholar] [CrossRef]
- Kia, G.S.N.; Mathias, S.; Esonu, D.O.; Benjamin, E. Ocurrence of Salmonella and Shigella on dried crayfish (Procambarus clarkii) sold in Zaria and Kaduna central market, Kaduna State, Nigeria. Niger. Vet. J. 2020, 41, 32–40. [Google Scholar] [CrossRef]
- Anda, P.; Segura del Pozo, J.; Díaz García, J.M.; Escudero, R.; García Peña, J.; López Velasco, M.C. Waterborne outbreak of Turalemia associated with Crayfish Fishing. Emerg. Infect. Dis. 2001, 7, 575–582. [Google Scholar] [CrossRef][Green Version]
- Ordax, J. Turalemia posiblemente transmitida por cangrejos. Gac. Sanit. 2003, 17, 164–165. [Google Scholar] [CrossRef] [PubMed]
- Ikerd, J.L.; Burnett, K.G.; Burnett, L.E. Effects of salinity on the accumulation of hemocyte aggregates and bacteria in the gills of Callinectes sapidus, the Atlantic blue crab, injected with Vibrio campbellii. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2015, 183, 97106. [Google Scholar] [CrossRef] [PubMed]
- Cuellar-Gempeler, C.; Munguia, P. Habitat filters mediate successional trajectories in bacterial communities associated with the striped shore crab. Oecologia 2019, 191, 957–970. [Google Scholar] [CrossRef]
- Ma, R.; Wang, Y.; Zhao, S.; Ma, Q.; Yin, M.; Li, X.; Fang, W. Bacterial Flora in the gill tissues and intestinal tracts of male and female chinese mitten crabs (Eriocheir sinensis) with different diets in a mud pond. Curr. Microbiol. 2021, 78, 2291–2297. [Google Scholar] [CrossRef]
- Kawamura, K.; Kanelso, M. Microbial quality of human wastes and treatment plant effluent. Water Sci Technol. 1986, 18, 257–265. [Google Scholar] [CrossRef]
- Lowman, F.G.; Rice, T.R.; Richards, F.A. Radioactivity in the Marine Environment; National Academy of Sciences: Washington, DC, USA, 1971; pp. 161–199. [Google Scholar] [CrossRef]
- Tretsven, W.I. Bacteriological survey of filleting processes in the Greater Northwest. III. Bacterial and physical effects of pughing fish incorrectly. J. Food Prot. 1964, 27, 13–17. [Google Scholar] [CrossRef]
- Amer, H.A.; Sedik, M.F.; Khalafalla, F.A.; El-Ghany Awad, H. Results of chemical análisis of prawn muscle as influenced by sex variations. Food/Nahrung 1991, 35, 133–138. [Google Scholar] [CrossRef]
- López, C.; Clemente, S.; Almeida, C.; Brito, A.; Hernández, M. A genetic approach to the origin of Millepora sp. in the Eastern Atlantic. Coral Reefs. 2015, 34, 631–638. [Google Scholar] [CrossRef]
- Guimarães de Freitas, C.; Patrícia Santana, A.; Helena Caldeira da Silva, P.; Salvador Picão Gonçalves, V.; Ferreira Barros, M.A.; Gonçalves Torres, F.A.; Sayori Murata, L.; Perecmanis, S. PCR multiplex for detection of Salmonella Enteritidis, Typhi and Typhimurium and occurrence in poultry meat. Int. J. Food Mirobiol. 2010, 139, 15–22. [Google Scholar] [CrossRef]
- Pérez-Roth, E.; Claverie-Martín, F.; Villar, J.; Méndez-Álvarez, S. Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J. Clin. Microbiol. 2001, 39, 4037–4041. [Google Scholar] [CrossRef]
- Schets, F.M.; List, C.; Kadar, M.; Ruiter, H.; Medema, G.J. Suppression of Plesiomonas shigelloides Growth in the Direct Plating Method for the Enumeration of Escherichia coli in Water. RIVM Report No. 289202020. 1998. Bilthoven, The Netherlands: RIVM. Available online: https://www.rivm.nl/publicaties/suppression-of-plesiomonas-shigelloides-growth-in-direct-plating-method-for-enumeration#abstract_en (accessed on 2 June 2021).
- Monteil, H.; Harf-Monteil, C. Les infections à Aeromonas. Presse Méd. 1997, 26, 1790–1798. [Google Scholar] [PubMed]
- Medema, G.; Schets, C. Occurrence of Plesiomonas shigelloides in surface water: Relationship with faecal pollution and trophic state. Int. J. Hyg. Environ. Health 1993, 194, 398–404. [Google Scholar]
- Schubert, R.H.W.; Beichert, R. The influence of treated sewage effluents on the numbers of Plesiomonas shigelloides isolated from river waters. Hyg. Med. 1993, 18, 57–59. [Google Scholar]
- Gonzalez-Rey, C.; Svenson, S.B.; Eriksson, L.M.; Ciznar, I.; Krovacek, K. Unexpected finding of the “tropical” bacterial pathogen Plesiomonas shigelloides from lake water north of the Polar Circle. Polar Biol. 2003, 26, 495–499. [Google Scholar] [CrossRef]
- Bravo, L.; Ramírez, M.; Cabrera, R.; Cabrera, L.; Rodríguez, A.; Hernández, R.; Castañeda, N.; Fernández, A. Serotypes and antibiotypes of Plesiomonas shigelloides in Cuba. Rev. Panam. Infectol. 2004, 6, 23–27. [Google Scholar]
- Marshall, D.L.; Kim, J.J.; Donnelly, S.P. Antimicrobial susceptibility and plasmid-mediated streptomycin resistance of Plesiomonas shigelloides isolated from blue crab. J. Appl. Bacteriol. 1996, 81, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Ingham, S. Growth of Aeromonas hydrophila and Plesiomonas shigelloides on cooked crayfish tails during cold storage under air, vacuum, and a modified atmosphere. J. Food Prot. 1990, 53, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhang, Q.; Zhang, T.; Chen, J.; Wang, S.; Hao, J.; Lin, Y.; Li, A. Association of the microbiota dysbiosis in the hepatopáncreas of farmed crayfish (Procambarus clarkii) with disease outbreaks. Aquaculture 2021, 536, 736492. [Google Scholar] [CrossRef]
- Wilson, D.K.; Cleary, T.G. Escherichia coli, Aeromonas y Plesiomonas. In Nelson. Tratado de Pediatría, 15th ed.; Behrman, R.E., Kliegman, R.M., Jenson, H.B., Eds.; Editorial Ciencias Médicas: La Habana, Cuba, 1998; pp. 95–99. [Google Scholar]
- Brooks, G.F.; Carrol, K.C.; Butel, J.S.; Morse, S.; Mitzner, T.A. Microbiología Médica de Jawetz, Melnick & Adelberg, 25th ed.; MAcGraw Hill, Ed.; El Manual Moderno: Ciudad de México, México, 2010. [Google Scholar]
- Bravo, L.; Cabrera, R.; Ramírez, M.; Llop, A.; Fernández, A.; García, B. Plesiomonas shigelloides, a Vibrionaceae to be account. Rev. Cuba 2000, 52, 10–14. [Google Scholar]
- Guevara, J.; Tafur, C. Bilicultivos de colecistectomizados en el Hospital Nacional “Edgardo Rabagliati Martin” del IPSS. Perú. Diagnóstico 1998, 17, 116–120. [Google Scholar]
- Young, A.Z.; Neujahr, D.; Estok, L. Case report. Epididymo-orchitis and bacterimia caused by Plesiomonas shigelloides in an HIV infected patient. AIDS Read. 2001, 11, 617–619. [Google Scholar] [PubMed]
- Abbott, S.L. Klebsiella, Enterobacter, Citrobacter, Serratia, Plesiomonas, and other Enterobacteriaceae. In Manual of Clinical Microbiology, 8th ed.; Murray, P.R., Baron, E.J., Jorgensen, J.H., Pfaller, M.A., Yolken, R.H., Eds.; mBio: Washington, DC, USA, 2003; Volume 44, pp. 684–700. [Google Scholar]
- Miller, M.L.; Koburger, J.A. Plesiomonas shigelloides: An Opportunistic Food and Waterborne Pathogen. J. Food Prot. 1985, 48, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Hernández, P.; Rodríguez de García, R. Prevalence of Plesiomonas shigelloides in surface water. Arch. Latinoam. Nutr. 1997, 47, 47–49. [Google Scholar] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Wong, S.S.Y.; Yuen, K. Two cases of continuous ambulatory peritoneal dialysis-associated peritonitis due to Plesiomonas shigelloides. J. Clin. Microbiol. 2003, 42, 933–935. [Google Scholar] [CrossRef][Green Version]
- Barkate, J.A. Incidence and Growth of Some Pathogens in Freshwater Crayfish (Procambarus clarkii, Girard). Ph.D. Thesis, Louisiana State University and Agricultural & Mechanical College, Baton Rouge, LA, USA, 1967. [Google Scholar]
- CDC. Typhoid Fever. 2018. Available online: https://www.cdc.gov/typhoid-fever/ (accessed on 6 May 2024).
- Dorrestein, G.M. Diagnostic approaches and management of diseases in captive passerines. In Seminars in Avian and Exotic Pet Medicine; WB Saunders: Philadelphia, PA, USA, 2003; Volume 12, pp. 11–20. [Google Scholar]
- Madadgar, O.; Zahraei Salehi, T.; Ghafari, M.M.; Ashrafi Tamai, I.; Madani, S.A.; Yahyareyat, R. Study of an unusual paratyphoid epornitic in canaries (Serinus canaria). Avian Pathol. 2009, 38, 437–441. [Google Scholar] [CrossRef]
- Spinu, M.; Gurzau, A.E.; Niculae, M.; Brudasca, G.F.; Sandru, C.D.; Kireb, C.; Pall, E. Reciprocal relationships in antibiotic resistance of Salmonella spp. carried by wild birds and fish in the danube delta. Int. J. Infect. Dis. 2019, 79, 43. [Google Scholar] [CrossRef]
- Fit, N.I.; Mureşan, C.; Chirilă, F.; Răpuntean, S.; Criste, A.; Nadăş, G. Bacterial isolates in Asio otus and Strix aluco birds of prey. Bull. Univ. Agric. Sci. Vet. Med. Cluj-Napoca Vet. Med. 2015, 72, 424–429. [Google Scholar] [CrossRef]
- Kirkpatrick, C.E.; Colvin, B.A. Salmonella spp. in nestling common barn-owls (Tyto alba) from southwestern New Jersey. J. Wild. Dis. 1986, 22, 340–343. [Google Scholar] [CrossRef]
- Vasconcelos, R.H.; De Castro Teixeira, R.S.; Goes da Silva, I.N.; de Souza Lopes, E.; Cardoso Maciel, W. Feral pigeons (Columbia livia) as potential reservoirs of Salmonella sp. and Escherichia coli. Arq. Inst. Biológico 2018, 85, e0412017. [Google Scholar] [CrossRef]
- Lapuz, R.; Tani, H.; Sasai, K.; Shirota, K.; Katoh, H.; Baba, E. An epidemiological analysis of Salmonella enteritidis contamination in a rat-infested chicken layer farm, an egg processing facility, and liquid egg samples by pulsed-field gel electrophoresis. J. Vet. Med. Sci. 2007, 69, 649–652. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lapuz, R.; Tani, H.; Sasai, K.; Shirota, K.; Katoh, H.; Baba, E. The role of roof rats (Rattus rattus) in the spread of Salmonella enteritidis and S. infantis contamination in layer farms in eastern Japan. Epidemiol. Infect. 2008, 136, 1235–1243. [Google Scholar] [CrossRef] [PubMed]
- Meerburg, B.G.; Kijlstra, A. Role of rodents in transmisión of Salmonella and Campylobacter. J. Sci. Food Agric. 2007, 87, 2774–2781. [Google Scholar] [CrossRef]
- Clarkson, L.S.; Tobin-D’Angelo, M.; Shuler, C.; Hanna, S.; Benson, J.; Voetsch, A.C. Sporadic Salmonella entérica serotype Javiana infections in Georgia and Tennesse: A hypothesis-generating study. Epidemiol. Infcect. 2009, 138, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.R.; Neil, K.P.; Barton Behravesh, C.; Sortir, M.J.; Angulo, F.J. Recent multistate outbreaks of human Salmonella infections acquired from turtles: A continuing public health challenge. Clin. Infect. Dis. 2010, 50, 554–559. [Google Scholar] [CrossRef]
- González-Lama, Z.; Rodríguez Melián, R.; Lupiola Gómez, P.A. Salmonelas en lagartos de la Isla de Tenerife. Rev. Canar. Cienc. Vet. 2005, 2, 27–31. [Google Scholar]
- Fitter, S.; Heuzenroeder, M.; Thomas, C.J. A combined PCR and selective enrichment method for rapid detection of Listeria monocytogenes. J. Appl. Bacteriol. 1992, 73, 53–59. [Google Scholar] [CrossRef]
- Dróżdż, M.; Malaszczuk, M.; Paluch, E.; Pawlak, A. Zoonotic potential and prevalence of Salmonella serovars isolated from pets. Infect. Ecol. Epidemiol. 2021, 11, 1975530. [Google Scholar] [CrossRef]
- Acha, P.; Szyfres, B. Zoonosis y Enfermedades Transmisibles Comunes al Hombre y a los Animales, 2nd ed.; Publicación Científica y Técnica No. 580; OPAS: Washington, DC, USA, 1986; 420p. [Google Scholar]
- RENAVE, 2022a. Informe Epidemiológico Sobre la Situación de la Salmonelosis en España. Año 2022. Centro Nacional de Epidemiología. Instituto de Salud Carlos III. 7 pp. Available online: https://cne.isciii.es/documents/d/cne/vigilancia_salmonelosis_2022-pdf (accessed on 9 January 2024).
- Foster, N.; Tang, Y.; Berchieri, A.; Geng, S.; Jiao, X.; Barrow, P. Revisiting persistent Salmonella infection and the carrier state: What do we know? Pathogens 2021, 10, 1299. [Google Scholar] [CrossRef]
- González-Escobedo, G.; Marshall, J.M.; Gunn, J.S. Chronic and acute infection of the gall bladder by Salmonella Typhi: Understanding the carrier state. Nat. Rev. Microbiol. 2011, 9, 9–14. [Google Scholar] [CrossRef]
- Huang, Z.; Li, Y.; Cai, C.; Dong, N. Isolation, molecular characterization and antimicrobial resistance of selected culturable bacteria from crayfish (Procambarus clarkii). Front. Microbiol. 2022, 13, 911777. [Google Scholar] [CrossRef] [PubMed]
- Kambire, O.; Adingra, A.A.; Yao, K.M.; Koffi-Nevry, R. Prevalence of virulence genes associated with diarrheagenic pathotypes of Escherichia coli isolates from water, sediment, fish and crab in Aby Lagoon, Côte d’Ivorie. Int. J. Microbiol. 2017, 2017, 9532170. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Graniel, J.E. Escherichia coli enteropatógena (EPEC): Una causa frecuente de diarrea infantil. Salud Tabasco 2003, 9, 188–193. [Google Scholar]
- Mare, A.D.; Ciurea, C.N.; Man, A.; Tudor, B.; Moldovan, V.; Decean, L.; Toma, F. Enteropathogenic Escherichia coli—A summary of the literature. Gastroenterol. Insights 2021, 12, 28–40. [Google Scholar] [CrossRef]
- Trabulsi, L.R.; Keller, R.; Gomes, T.A.T. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 2002, 8, 508. [Google Scholar] [CrossRef]
- Deborah Chen, H.; Frankel, G. Enteropathogenic Escherichia coli: Unravelling pathogenesis. FEMS Microbiol. Rev. 2005, 29, 83–98. [Google Scholar] [CrossRef]
- Yang, K.; Pagaling, E.; Yan, T. Estimating the prevalence of potential enteropathogenic Escherichia coli and intimin gene diversity in a human community by monitoring sanitary sewage. Appl. Environ. Microbiol. 2014, 80, 119–127. [Google Scholar] [CrossRef]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef] [PubMed]
- Ronin, I.; Katsowich, N.; Rosenshine, I.; Balaban, N.Q. A long-term epigenetic memory switch controls bacterial virulence bimodality. eLife 2017, 6, e19599. [Google Scholar] [CrossRef]
- Hernandes, R.T.; Elias, W.P.; Vieira, M.A.M.; Gomes, T.A.T. An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiol. Lett. 2009, 297, 137–149. [Google Scholar] [CrossRef]
- Swennes, A.G.; Buckley, E.M.; Parry, N.M.A.; Madden, C.M.; García, A.; Morgan, P.B.; Astrofsky, K.M.; Fox, J.G. Enzootic enteropathogenic Escherichia coli Infection in laboratory rabbits. J. Clin. Microbiol. 2012, 50, 2353–2358. [Google Scholar] [CrossRef] [PubMed]
- Sekse, C.; Sunde, M.; Lindstedt, B.A.; Hopp, P.; Bruheim, T.; Cudjoe, K.S.; Kvitle, B.; Urdahl, A.M. Potentially human-pathogenic Escherichia coli O26 in Norwegian sheep flocks. Appl. Environ. Microbiol. 2011, 77, 4949–4958. [Google Scholar] [CrossRef]
- Moura, R.A.; Sircili, M.P.; Leomil, L.; Matté, M.H.; Trabulsi, L.R.; Elias, W.P.; Irino, K.; Pestana de Castro, A.F. Clonal relationship among atypical enteropathogenic Escherichia coli strains isolated from different animal species and humans. Appl. Environ. Microbiol. 2009, 75, 7399–7408. [Google Scholar] [CrossRef]
- Sanches, L.A.; da Silva, M.; Friciello Teixeira, R.H.; Vieira Cunha, M.P.; Xavier de Oliveria, M.G.; Midolli Vieira, M.A.; Tardelli Gomes, T.A.; Knobl, T. Captive wild birds as reservoirs of enteropathogenic E. coli (EPEC) and Shiga-toxin producing E. coli (STEC). Braz. J. Microbiol. 2017, 48, 760–763. [Google Scholar] [CrossRef]
- Gomes, T.A.T.; Elias, W.P.; Scaletsky, I.C.A.; Guth, B.E.C.; Rodrigues, J.F.; Piazza, R.M.F. Diarrheagenic Escherichia coli. Braz. J. Microbiol. 2016, 47, 3–30. [Google Scholar] [CrossRef] [PubMed]
- Escher, M.; Scavia, G.; Morabito, S.; Tozzoli, R.; Maugliani, A.; Cantoni, S. A severe foodborne outbreak of diarrhoea linked to a canteen in Italy caused by enteroinvasive Escherichia coli, an uncommon agent. Epidemiol. Infect. 2014, 142, 2559–2566. [Google Scholar] [CrossRef]
- Michelacci, V.; Prosseda, G.; Maugliani, A.; Tozzoli, R.; Sanchez, S.; Herrera-León, S. Characterization of an emergent clone of enteroinvasive Escherichia coli circulating in Europe. Clin. Microbiol. Infect. 2016, 22, 287.e11–287.e19. [Google Scholar] [CrossRef] [PubMed]
- Newitt, S.; MacGregor, V.; Robbins, V.; Bayliss, L.; Chattaway, M.A.; Dallman, T. Two linked enteroinvasive Escherichia coli outbreaks, Nottingham, UK, June 2014. Emerg. Infect. Dis. 2016, 22, 1178–1184. [Google Scholar] [CrossRef]
- Pasqua, M.; Michelacci, V.; Di Martino, M.L.; Tozzoli, R.; Grossi, M.; Colonnam, B.; Morabitom, S.; Prosseda, G. The intriguing evolutionary journey of enteroinvasive E. coli (EIEC) toward pathogenicity. Front. Microbiol. 2017, 8, 312178. [Google Scholar] [CrossRef]
- Huang, S.W.; Hsu, B.M.; Su, Y.J.; Ji, D.D.; Lin, W.C.; Chen, J.L.; Shih, F.C.; Kao, P.M.; Chiu, Y.C. Ocurrence of diarreheagenic Escherichia coli genes in raw water of water treatment plant. Environ. Sci. Pollut. Res. 2012, 19, 2776–2783. [Google Scholar] [CrossRef]
- Franz, E.; Veenman, C.; Van Hoek, A.H.A.M.; de Roda Husman, A.; Blaak, H. Pathogenic Escherichia coli producing extended spectrum β-lactamases isolates from Surface water and wastewater. Sci. Rep. 2015, 5, 14372. [Google Scholar] [CrossRef] [PubMed]
- Liu, D. Diarrhoeagenic Escherichia coli. In Molecular Medical Microbiology; Academic Press: Cambridge, MA, USA, 2015; pp. 1133–1146. [Google Scholar] [CrossRef]
- RENAVE, 2022b. Informe Epidemiológico Sobre la Situación de la Campilobacteriosis en España. Año 2022. Centro Nacional de Epidemiología. Instituto de Salud Carlos III. 9pp. Available online: https://cne.isciii.es/documents/d/cne/informe_campylobacter_2022_final-pdf (accessed on 30 January 2024).
- Humphrey, T.; O’Brien, S.; Madsen, M. Campylobacters as zoonotic pathogens: A food production perspective. Int. J. Food Microbiol. 2007, 117, 237–257. [Google Scholar] [CrossRef]
- Sippy, R.; Sandoval-Green, C.M.J.; Sahin, O.; Plummer, P.; Sue Fairbanks, W.; Zhang, Q.; Blanchong, J.A. Ocurrence and molecular analysis in wildlife on livestock farms. Vet. Microbiol. 2012, 157, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Mishu Allos, B.; Lastovica, A.J. Campylobacter Infections. In Tropical Infectious Diseases: Principles, Pathogens and Practice, 3rd ed.; Saunders Elsevier: Boston, MA, USA, 2011; Volume 19, pp. 145–149. [Google Scholar] [CrossRef]
- RCVE 2022. Red Canaria de Vigilancia Epidemiológica. Informe 2022. Servicio Canario de la Salud. Gobierno de Canarias. Available online: https://www3.gobiernodecanarias.org/sanidad/scs/content/a74fe267-1c22-11ee-bd63-17442ee4d369/InformeRCVE2022.pdf (accessed on 30 January 2024).
- RENAVE, 2022c. Informe Epidemiológico Sobre la Situación de la Yersiniosis en España. Año 2022. Centro Nacional de Epidemiología. Instituto de Salud Carlos III. 6 pp. Available online: https://cne.isciii.es/documents/d/cne/informe_yersinia_2022_final-pdf (accessed on 30 January 2024).
- Krauss, H.; Weber, A.; Appel, M.; Enders, B.; Isenberg, H.D.; Schiefer, H.G.; Slenczka, W.; Von Graevenitz, A.; Zahner, H. Zoonoses Infectious Diseases Transmissible from Animals to Humans, 3rd ed.; ASM Press: Washington, DC, USA, 2003. [Google Scholar]
- Murray, P.R.; Baron, E.J.; Jorgensen, J.H.; Landry, M.L.; Pfaller, M.A. Manual of Clinical Microbiology, 9th ed.; ASM Press: Washington, DC, USA, 2007. [Google Scholar]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union one health 2019 zoonoses report. EFSA J. 2021, 19, e06406. [Google Scholar] [CrossRef]
- Bucher, M.; Meyer, C.; Grötzbach, B.; Wacheck, S.; Stolle, A.; Fredriksson-Ahomaa, M. Epidemiological data on pathogenic Yersinia enterocolitica in southern Germany during 2000–2006. Foodborne Pathog. Dis. 2008, 5, 273–280. [Google Scholar] [CrossRef]
- Rouffaer, L.O.; Baert, K.; Van den Abeele, A.M.; Cox, I.; Vanantwerpen, G.; De Zutter, L.; Strubbe, D.; Vranckx, K.; Lens, L.; Haesebrouck, F.; et al. Low prevalence of human enteropathogenic Yersinia spp. in brown rats (Rattus norvegicus) in Flanders. PLoS ONE 2017, 12, e0175648. [Google Scholar] [CrossRef]
- Platt-Samoraj, A.; Klaudia Konczyk-Kmiecik, K.; Bakuła, T. Occurrence and genetic correlations of Yersinia spp. isolated. from commensal rodents in Northeastern Poland. Pathogens 2021, 10, 1247. [Google Scholar] [CrossRef] [PubMed]
- Actis, L.A.; Tolmasky, M.E.; Crosa, J.H. Vibriosis. In Fish Diseases and Disorders Viral, Bacterial and Fungal Infections, 2nd ed.; Woo, P.K., Bruno, D.W., Eds.; CAB International: London, UK, 2011; pp. 570–605. [Google Scholar]
- Thune, R.L.; Hawke, J.P.; Siebeling, R.J. Vibriosis in the red swamp crawfish. J. Aquat. Anim. Health 1991, 3, 188–191. [Google Scholar] [CrossRef]
- Zhou, J.; Zhao, H.; Huang, Z.; Ye, X.; Zhang, L.; Li, Q.; Zhao, Z.; Su, X.; Liu, G.; Du, J. Differential transcriptomic análisis of crayfish (Procambarus clarkii) from a rice coculture system challenged by Vibrio parahaemoyticus. CBP Genom. Proteom. 2020, 36, 100741. [Google Scholar] [CrossRef]
- Austin, B.; Austin, D.A. Diseases of farmed and wild fish. In Bacterial Fish Pathogens, 5th ed.; Springer Praxis: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Austin, B. Vibrios as causal agents of zoonoses. Vet. Microbiol. 2009, 140, 310–317. [Google Scholar] [CrossRef]
- Teng, L.; Zou, G.; Zhou, Y.; Li, J.; Song, Z.; Dong, X.; Ma, Z.; Zheng, Z.; Chen, H.; Li, J. Phage controlling method against novel freshwater-derived Vibrio parahaemolyticus in ready-to-eat crayfish (Procambarus clarkii). Food Res. Int. 2022, 162, 111986. [Google Scholar] [CrossRef] [PubMed]
- Sack, R.B. A search for canine carriers of Vibrio. J. Infect. Dis. 1973, 127, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-Rodríguez, E.; Martín-Carrillo, N.; Valladares, B.; Foronda, P. Study of zoonotic enteric pathogens of Atelerix algirus in Tenerife, Canary Islands, Spain. Front. Vet. Sci. 2020, 7, 579602. [Google Scholar] [CrossRef]
- Sanyal, S.C.; Singh, S.J.; Tiwari, P.C.; Sen, I.C.; Marwah, S.M.; Hazarika, U.R.; Singh, H.; Shimada, T.; Sakazaki, R. Role of household animals in maintenance of cholerae infection in a community. J. Infect. Dis. 1974, 130, 575–579. [Google Scholar] [CrossRef]
- Rodríguez, C.; Taminiau, B.; Van Broeck, J.; Delmée, M.; Daube, G. Clostridium difficile in food and animals: A comprehensive review. Adv. Microbiol. Infect. Dis. Public Health 2016, 4, 65–92. [Google Scholar]
- Simango, C. Prevalence of Clostridium difficile in the environment in a rural community in Zimbabwe. Trans. R. Soc. Trop. Med. Hyg. 2006, 100, 1146–1150. [Google Scholar] [CrossRef]
- Zidaric, V.; Beigot, S.; Lapajne, S.; Rupnik, M. The occurrence and high diversity of Clostridium difficile genotypes in rivers. Anaerobe 2010, 16, 371–375. [Google Scholar] [CrossRef]
- Romano, V.; Pasquale, V.; Krovacek, K.; Mauri, F.; Demarta, A.; Dumontet, S. Toxigenic Clostridium difficile PCR ribotypes from wastewater treatment plants in Southern Switzerland. Appl. Environ. Microbiol. 2012, 78, 6643–6646. [Google Scholar] [CrossRef]
- Viau, E.; Peccia, J. Survey of wastewater indicators and human pathogen genomes in biosolids produced by class A and class B stabilization treatments. Appl. Environ. Microbiol. 2009, 75, 164–174. [Google Scholar] [CrossRef]
- Bazaid, F. Distribution and Sources of Clostridium difficile Present in Water Sources. Doctoral Dissertation, University of Guelph, Guelph, ON, Canada, 2012. [Google Scholar]
- Krijger, I.M.; Meerburg, B.G.; Harmanus, C.; Burt, S.A. Clostridium difficile in wild rodents and insectivores in Nertherlands. Lett. Appl. Microbiol. 2019, 69, 35–40. [Google Scholar] [CrossRef]
- Hernández, M.; Ramos, M.; Lecuona, M. P032: Clostridium difficile infection in Tenerife Canary Island, Spain. Antimicrob. Resist. Infect. Control 2013, 2, P32. [Google Scholar] [CrossRef][Green Version]
- Aroca-Ferri, M.; Tosco-Núñez, T.; Peñate-Bolaños, M.; Molina-Cabrilla, J.; Ojeda-Vargas, M. Primer brote de Clostridioides difficile ribotipo 027 en Canarias. Rev. Esp. Quimioter. 2023, 36, 317. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.; Tabashsum, Z.; Alvarado-Martínez, Z.; Scriba, A.; Sellers, G.; Kapadia, S.; Canagarajah, C.; Biswas, D. Ecological distribution of Staphylococcus in integrated farmas withn Washington DC-Maryland. J. Food Saf. 2024, 44, e13123. [Google Scholar] [CrossRef]
- Dong, X.; Wang, X.; Chen, X.; Yan, Z.; Cheng, J.; Gao, L.; Liu, Y.; Li, J. Genetic diversity and virulence potential of Staphylococcus aureus isolated from crayfish (Procambarus clarkii). Curr. Microbiol. 2017, 74, 28–33. [Google Scholar] [CrossRef]
- Shao, L.; Qiu, X.; Li, J.; Chen, J.; Wang, R.; Han, Q.; Yang, P. Isolation and identification of intestinal cellulolytic bacteria from red swamp crayfish (Procambarus clarkii). Isr. J. Aquac. Bamidgeh 2024, 76, 1–7. [Google Scholar] [CrossRef]
- Solliman, W.; Abbas, W.T.; Ibrahim, T.B.; Kenawy, A.M.; Elgendy, M.Y. Disease causing organisms in Procambarus clarkii and Gambusia affinis with emphasis on their role in biomonitoring of aquatic pollution. Egypt J. Vet. Sci. 2016, 47, 63–81. [Google Scholar] [CrossRef][Green Version]
- Sousa, M.; Silva, N.; Igrejas, G.; Silva, F.; Sargo, R.; Alegria, N.; Benito, D.; Gómez, P.; Lozano, C.; Gómez-Sanz, E.; et al. Antimicrobial resistance determinants in Staphylococcus spp. recovered from birds of prey in Portugal. Vet. Microbiol. 2014, 171, 436–440. [Google Scholar] [CrossRef]
- Hunter, P.R.; Thompson, R.A. The zoonotic transmission of Giardia and Cryptosporidium. Int. J. Parasitol. 2005, 35, 1181–1190. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Zahedi, A.; Paparini, A. Cryptosporidium in humans and animals—A one health approach to prophylaxis. Parasite Immunol. 2016, 38, 535–547. [Google Scholar] [CrossRef]
- Rosado-García, F.M.; Guerrero-Flórez, M.; Karanis, G.; Hinojosa, M.D.C.; Karanis, P. Water-borne protozoa parasites: The Latin American perspective. Int. J. Hyg. Environ. Health 2017, 220, 783–798. [Google Scholar] [CrossRef]
- Khan, A.; Shaik, J.S.; Grigg, M.E. Genomics and molecular epidemiology of Cryptosporidium species. Acta Trop. 2018, 184, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Hijjawi, N.; Xiao, L. Foodborne cryptosporidiosis. Int. J. Parasitol. 2018, 48, 1–12. [Google Scholar] [CrossRef]
- Karanis, P. Cryptosporidium: Waterborne and foodborne transmission and worldwide outbreaks. In Recent Advances in Environmental Science from the Euro-Mediterranean and Surrounding Regions, Proceedings of the Euro-Mediterranean Conference for Environmental Integration (EMCEI-1), Sousse, Tunisia, 20–25 November 2017; Springer International Publishing: Heidelberg, Germany, 2018; pp. 41–44. [Google Scholar] [CrossRef]
- Abreu-Acosta, N.; Foronda-Rodríguez, P.; López, M.; Valladares, B. Occurrence of Cryptosporidium hominis in pigeons (Columbia livia). Acta Parasitol. 2009, 54, 1–5. [Google Scholar] [CrossRef]
- Feliu, C.; López, M.; Gómez, M.S.; Torres, J.; Sánchez, S.; Miquel, J.; Abreu-Acosta, N.; Segovia, J.M.; Martín-Alonso, A.; Montoliu, I.; et al. Parasite fauna of rodents (Murinae) from El Hierro (Canary Islands, Spain): A multidisciplinary approach. Acta Parasitol. 2012, 57, 171–178. [Google Scholar] [CrossRef]
- García-Livia, K.; Martín-Alonso, A.; Foronda, P. Diversity of Cryptosporidium spp. in wild rodents from the Canary Islands, Spain. Parasit. Vectors 2020, 13, 445. [Google Scholar] [CrossRef] [PubMed]
- Baz-González, E.; Martín-Carrillo, N.; García-Livia, K.; Foronda, P. Rabbits (Oryctolagus cuniculus) from Tenerife, Canary Islands, Spain. Vet. Sci. 2022, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Abreu-Acosta, N.; Quispe, M.A.; Foronda-Rodríguez, P.; Alcoba-Florez, J.; Lorenzo-Morales, J.; Ortega-Rivas, A.; Valladares, B. Cryptosporidium in patients with diarrhoea, on Tenerife, Canary Islands, Spain. Ann. Trop. Med. Parasitol. 2007, 101, 539–545. [Google Scholar] [CrossRef]
- RENAVE, 2022e. Informe Epidemiológico Sobre la Situación de la Criptosporidiosis en España. Años 2019 y 2020. Centro Nacional de Epidemiología. Instituto de Salud Carlos III. 8 pp. Available online: https://cne.isciii.es/documents/d/cne/informe_criptosporidium_2022-pdf (accessed on 30 January 2024).
- Raef, A.M.; Mohamed, A.A.; Mohamed, M.E.M.; Abd El-Maksoud, S.A. Further studies on the role of crayfish “Procambarus clarki” in transmisión of some zoonotic parasites in East Delta. In The Third International Scientific Conference; Faculty of Veterinary Medicine Mansoura University: Mansoura, Egypt, 2003; pp. 29–30. [Google Scholar]
- Amro, M.; Magda, A.; Amer, O.; Merwad, A. Studies on the role of shellfish as a source for transmitting some parasites of zoonotic importance. Mansoura Vet. Med. J. 2005, 7, 1–25. [Google Scholar] [CrossRef]
- Graczyk, T.; McOliver, C.; Silbergeld, E.K.; Tamang, L.; Roberts, J.D. Risk of handling as a route of esposure to infectious waterborne Cryptosporidium parvum oocysts via Atlantic Blue Crabs (Callinectes sapidus). Appl. Environ. Microbiol. 2007, 73, 4069–4070. [Google Scholar] [CrossRef][Green Version]
- Santin, M. Cryptosporidium and Giardia in ruminants. Vet. Clin. N. Am. Food A 2020, 36, 223–238. [Google Scholar] [CrossRef]
- Dixon, B.R. Giardia duodenalis in humans and animals—Transmission and disease. FEMS Microbiol. Rev. 2021, 135, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Álvarez, A.; Martín-Alonso, A.; Abreu-Acosta, N.; Feliu, C.; Hugot, J.P.; Valladares, B.; Foronda, F. Identification of a novel assemblage G subgenotype and a zoonotic assemblage B in rodent isolates of Giardia duodenalis in the Canary Islands, Spain. Parasitology 2014, 141, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Ruíz, A.; Foronda, P.; González, J.F.; Guedes, A.; Abreu-Acosta, N.; Molina, J.M.; Valladares, B. Ocurrence and genotype characterization of Giardia duodenalis in goat kids from the Canary Islands, Spain. Vet. Parasitol. 2008, 154, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Abreu-Acosta, N.; Martín-Delgado, M.; Ortega-Rivas, A.; del Castillo Remiro, A.; Aguiar González, E.; Valladares Hernández, B. Presencia de Giardia lamblia y Cryptosporidium spp. en aguas residuales depuradas reutilizadas para riego agrícola en la isla de Tenerife, España. Efectos del transporte a larga distancia sobre la calidad del agua reutilizada. Rev. Salud Ambient. 2002, 2, 2–7. [Google Scholar]
- Abreu-Acosta, N.; Vera, L. Ocurrence and removal of parasites, enteric bacteria and faecal contamination indicators in wastewater natural reclamation systems in Tenerife-Canary Islands, Spain. Ecol. Eng. 2011, 37, 496–503. [Google Scholar] [CrossRef]
- RENAVE, 2022f. Informe Epidemiológico Sobre la Situación de la Giardiasis en España. Años 2019 y 2020. Centro Nacional de Epidemiología. Instituto Carlos III. 8 pp. Available online: https://cne.isciii.es/documents/d/cne/informe_giardia_2022_final-pdf (accessed on 1 February 2024).
- Graczyk, T.K.; Farley, C.A.; Fayer, R.; Lewis, E.J.; Trout, J.M. Detection of Cryptosporidium oocysts and Giardia cysts in the tissues of Eastern Oysters (Crassostrea virginica) carrying principal oyster infectious diseases. J. Parasitol. 1998, 84, 1039–1042. [Google Scholar] [CrossRef]
- Gómez-Couso, H.; Méndez-Hermida, F.; Castro-Hermida, J.A.; Ares-Mazás, E. Giardia in shellfish-farming areas: Detection in mussels, river water and waste Waters. Vet. Parasitol. 2005, 133, 13–18. [Google Scholar] [CrossRef]
| Bacterial Strain |
|---|
| Salmonella enterica serovar Typhimurium ATCC® 14028 |
| Salmonella enterica serovar Enteritidis ATCC® 13076 |
| Salmonella enterica serovar Typhi strain ATCC® 19430 |
| Staphylococcus aureus ATCC® 653 |
| Staphylococcus epidermidis ATCC® 12228 |
| Staphylococcus saprophyticus ATCC® 15305 |
| Staphylococcus aureus derived from ATCC® BAA-1708™ (Methicillin and Mupirocin resistant) |
| Staphylococcus haemolyticus ATCC® 29970 |
| Staphylococcus lugdunensis ATCC® 49576 |
| Staphylococcus hominis ATCC® 19536 |
| Bacteria | Target Gene | Primers | Sequence (5′-3′) | Size (bp) |
|---|---|---|---|---|
| Salmonella spp. | ompC | OMPCF | ATC GCT GAC TTA TGC AAT CG | 204 |
| OMPCR | CGG GTT GCG TTA TAG GTC TG | |||
| S. enterica ser. Enteritidis | Sdf1 | ENTF | TGT GTT TTA TCT GAT GCA AGA GG | 304 |
| ENTR | TGA ACT ACG TTC GTT CTTCTG G | |||
| S. enterica ser. Typhi | ViaB | ViaBF | CAC GCA CCA TCA TTT CAC CG | 738 |
| ViaBR | AAC AGG CTG TAG CGA TTT AGG | |||
| S. enterica ser. Typhimurium | Spy | TyphF | TTG TTC ACT TTT TAC CCC TGA A | 401 |
| TyphR | CCC TGA CAG CCG TTA GAT ATT |
| Specie (Locus) | Primer | Sequence (5′-3′) | Size (pb) |
|---|---|---|---|
| S. lugdunensis (fbI) | fbIF | AAA TCT CCA AGT TGA CCA AAC ATA C | 550 |
| fbIR | GAT TGC GCT GAA AGA ATT GC | ||
| Mupirocin resistance (ileS2) | ileS2F | TAT ATT ATG CGA TGG AAG GTT GG | 456 |
| ileS2R | AAT AAA ATC AGC TGG AAA GTG TTG | ||
| S. saprophyticus (sap) | sapF | AAC GGG CGT CTC GAT AGA AAA | 380 |
| sapR | AAC GGG CGT CCA CAA AAT CA | ||
| S. aureus (nuc) | nucF | TCG CTT GCT ATG ATT GTG G | 359 |
| nucR | GCC AAT GTT CTA CCA TAG C | ||
| Methicillin resistance (mecA) | mecA1 | GTA GAA ATG ACT GAA CGT CCG ATA A | 310 |
| mecA2 | CCA ATT CCA CAT TGT TTC GGT CTA A | ||
| S. haemolyticus (mvaA) | mvaA1 | GGT CGC TTA GTC GGA ACA AT | 271 |
| mvaA2 | CAC GAG CAA TCT CAT CAC CT | ||
| S. epidermidis (sep) | sepF | CAG TTA TAC GGT ATG AGA GC | 219 |
| sepR | CTG TAG AGT GAC AGT TTG GT | ||
| S. hominis (hom) | homF | TAC AGG GCC ATT TAA AGA CG | 177 |
| homR | GTT TCT GGT GTA TCA ACA CC |
| Bacteria | + (Prevalence) [95% CI] (n = 22) |
|---|---|
| Plesiomonas shigelloides | 9 (40.90%) [20.71, 63.64] |
| Enteroinvasive E. coli | 7 (31.81%) [13.86, 54.87] |
| Enteropathogenic E. coli | 2 (9.09%) [1.12, 29.16] |
| Salmonella spp. | 4 (18.18%) [5.19, 40.28] |
| Bacteria | + (Prevalence) [95% CI] (n = 13) |
|---|---|
| Salmonella ser. Enteritidis | 2 (15.38%) [1.92, 45.45] |
| Salmonella ser. Typhimurium | 2 (15.38%) [1.92, 45.45] |
| Salmonella ser. Typhi | 1 (7.69%) [0.19, 36.03] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abreu-Acosta, N.; Martín-Carrillo, N.; Foronda, P. Zoonotic Pathogens Isolated from an Introduced Population of Red Swamp Crayfish (Procambarus clarkii) in Tenerife (Canary Islands, Spain). Diversity 2024, 16, 643. https://doi.org/10.3390/d16100643
Abreu-Acosta N, Martín-Carrillo N, Foronda P. Zoonotic Pathogens Isolated from an Introduced Population of Red Swamp Crayfish (Procambarus clarkii) in Tenerife (Canary Islands, Spain). Diversity. 2024; 16(10):643. https://doi.org/10.3390/d16100643
Chicago/Turabian StyleAbreu-Acosta, Néstor, Natalia Martín-Carrillo, and Pilar Foronda. 2024. "Zoonotic Pathogens Isolated from an Introduced Population of Red Swamp Crayfish (Procambarus clarkii) in Tenerife (Canary Islands, Spain)" Diversity 16, no. 10: 643. https://doi.org/10.3390/d16100643
APA StyleAbreu-Acosta, N., Martín-Carrillo, N., & Foronda, P. (2024). Zoonotic Pathogens Isolated from an Introduced Population of Red Swamp Crayfish (Procambarus clarkii) in Tenerife (Canary Islands, Spain). Diversity, 16(10), 643. https://doi.org/10.3390/d16100643







