Diversity and Composition of Posidonia oceanica-Associated Bacterial and Fungal Communities: Effect of Boat-Induced Mechanical Stress in the Villefranche-sur-Mer Bay (France)
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Sampling Activities
2.2. DNA Metabarcoding
2.3. Bioinformatics and Statistical Analyses
3. Results
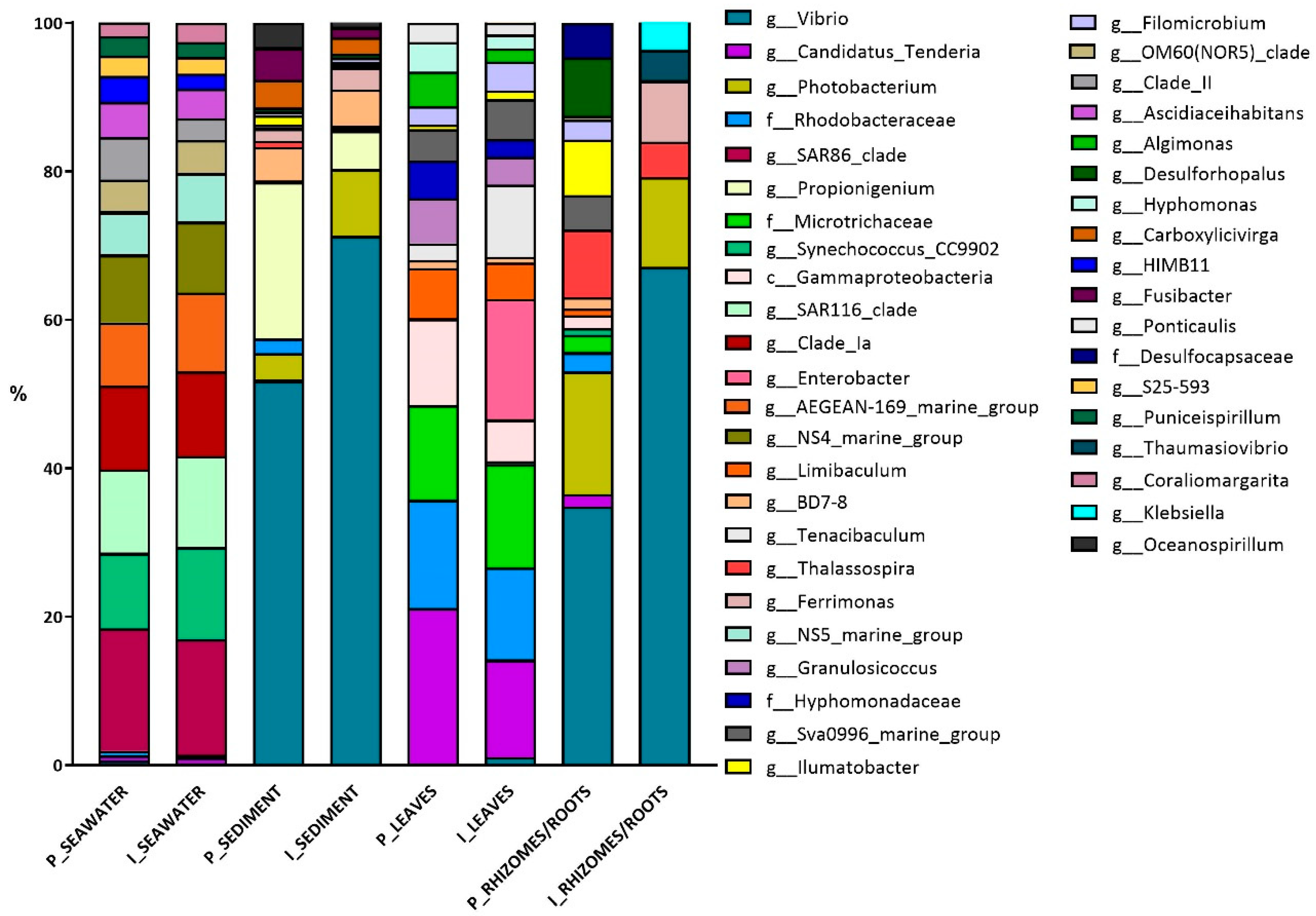
3.1. Bacterial Diversity
Bacterial Colonizers and FAPROTAX Analysis
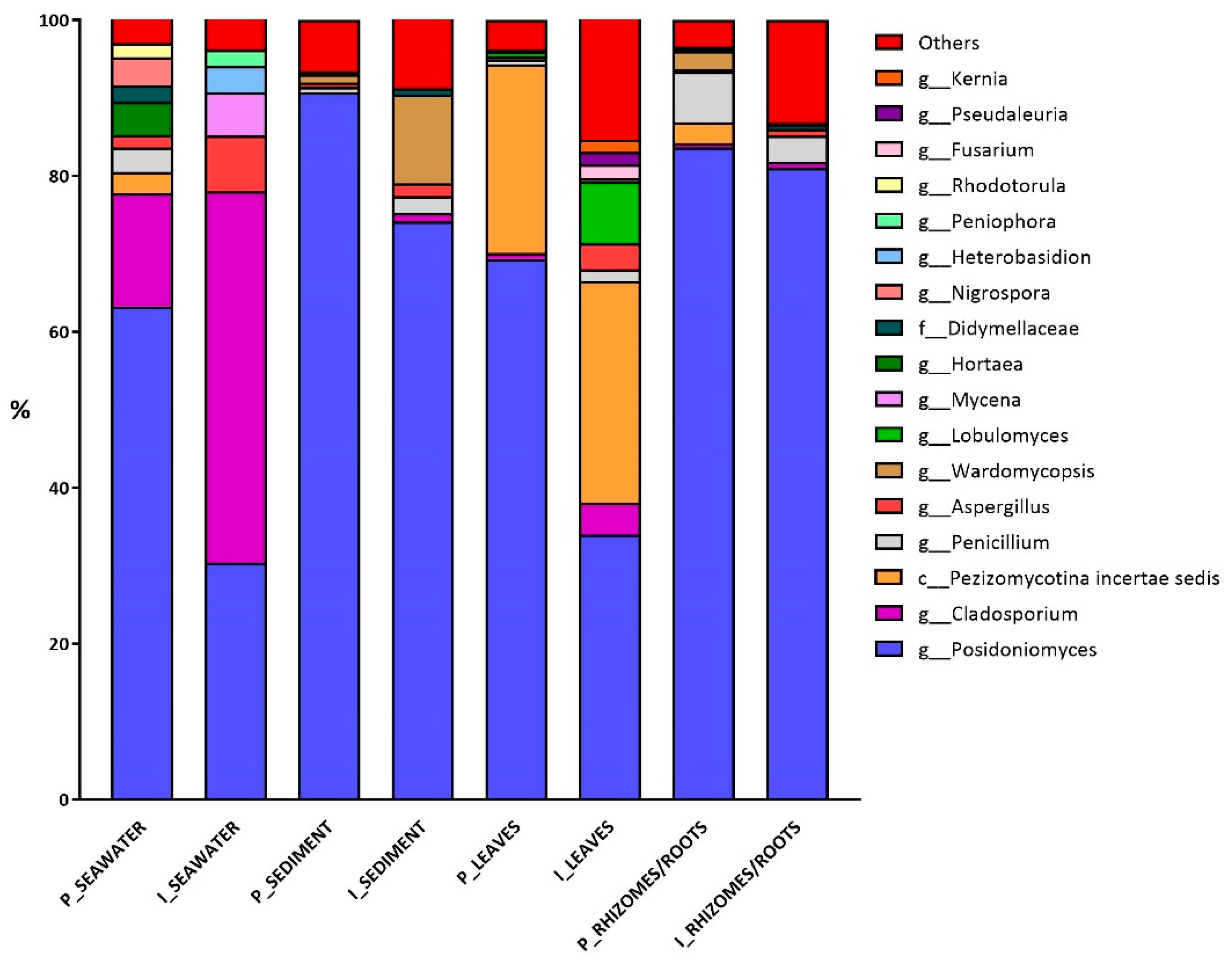
3.2. Fungal Diversity
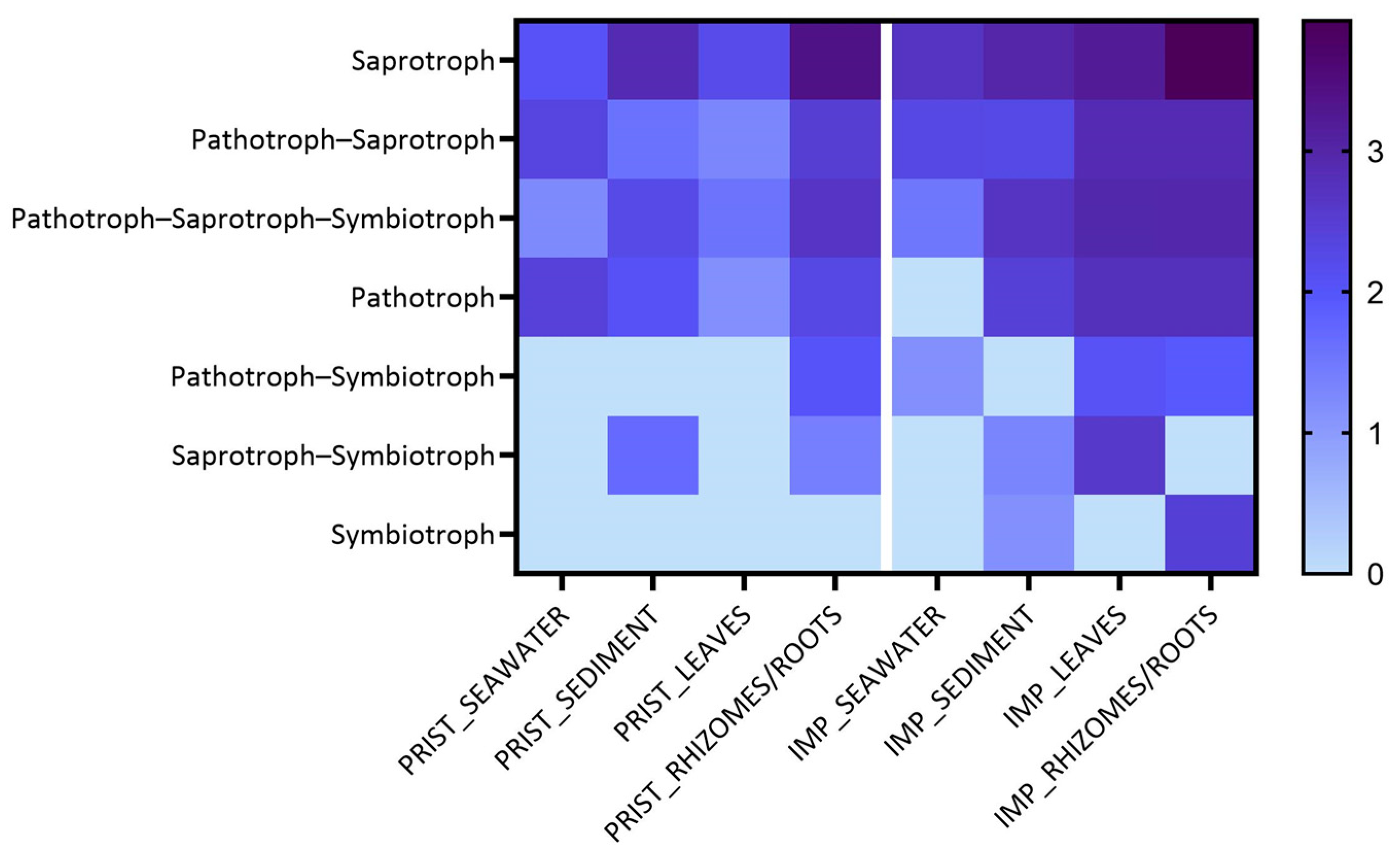
Fungal Colonizers and FUNGuild Analysis
4. Discussion
4.1. Bacterial Community
4.2. Fungal Community
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnaud-Haond, S.; Duarte, C.M.; Diaz-Almela, E.; Marba, N.; Sintes, T.; Serrao, E.A. Implications of extreme life span in clonal organisms: Millenary clones in meadows of the threatened seagrass Posidonia oceanica. PLoS ONE 2012, 7, e30454. [Google Scholar] [CrossRef] [PubMed]
- Boudouresque, C.F. Marine biodiversity in the Mediterranean: Status of species, populations and communities. Trav. Sci. PNPC 2004, 20, 97–146. [Google Scholar]
- Pergent-Martini, C.; Pergent, G.; Monnier, B.; Boudouresque, C.F.; Mori, C.; Valette-Sansevin, A. Contribution of Posidonia oceanica meadows in the context of climate change mitigation in the Mediterranean Sea. Mar. Environ. Res. 2021, 165, 105236. [Google Scholar] [CrossRef] [PubMed]
- Apostolaki, E.T.; Caviglia, L.; Santinelli, V.; Cundy, A.B.; Tramati, C.D.; Mazzola, A.; Vizzini, S. The importance of dead seagrass (Posidonia oceanica) matte as a biogeochemical sink. Front. Mar. Sci. 2022, 9, 861998. [Google Scholar] [CrossRef]
- Boudouresque, C.F.; Pergent, G.; Pergent-Martini, C.; Ruitton, S.; Thibaut, T.; Verlaque, M. The necromass of the Posidonia oceanica seagrass meadow: Fate, role, ecosystem services and vulnerability. Hydrobiologia 2016, 781, 25–42. [Google Scholar] [CrossRef]
- Monnier, B.; Pergent, G.; Valette-Sansevin, A.; Boudouresque, C.F.; Mateo, M.A.; Pergent-Martini, C. The Posidonia oceanica matte: A unique coastal carbon sink for climate change mitigation and implications for management. Vie Milieu/Life Environ. 2020, 70, 17–24. [Google Scholar]
- Council Directive 92/43/EEC on the Conservation of Natural Habitats and of Wild Fauna and Flora; Publications Office of the European Union: Luxemburg, 1992; Volume 206, pp. 7–50.
- Boudouresque, C.F.; Bernard, G.; Pergent, G.; Shili, A.; Verlaque, M. Regression of Mediterranean seagrasses caused by natural processes and anthropogenic disturbances and stress: A critical review. Bot. Mar. 2009, 52, 395–418. [Google Scholar] [CrossRef]
- Montefalcone, M.; Chiantore, M.; Lanzone, A.; Morri, C.; Albertelli, G.; Bianchi, C.N. BACI design reveals the decline of the seagrass Posidonia oceanica induced by anchoring. Mar. Pollut. Bull. 2008, 56, 1637–1645. [Google Scholar] [CrossRef]
- Francour, P.; Ganteaume, A.; Poulain, M. Effects of boat anchoring in Posidonia oceanica seagrass beds in the Port-Cros National Park (north-western Mediterranean Sea). Aquat. Conserv. Mar. Freshw. 1999, 9, 391–400. [Google Scholar] [CrossRef]
- Ardizzone, G.; Belluscio, A.; Maiorano, L. Long-term change in the structure of a Posidonia oceanica landscape and its reference for a monitoring plan. Mar. Ecol. 2006, 27, 299–309. [Google Scholar] [CrossRef]
- Ceccherelli, G.; Campo, D.; Milazzo, M. Short-term response of the slow growing seagrass Posidonia oceanica to simulated anchor impact. Mar. Environ. Res. 2007, 63, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Bourque, A.S.; Kenworthy, W.J.; Fourqurean, J.W. Impacts of physical disturbance on ecosystem structure in subtropical seagrass meadows. Mar. Ecol. Prog. Ser. 2015, 540, 27–41. [Google Scholar] [CrossRef]
- Rosenberg, E.; Koren, O.; Reshef, L.; Efrony, R.; Zilber-Rosenberg, I. The hologenome theory disregards the coral holobiont: Reply from Rosenberg et al. Nat. Rev. Microbiol. 2007, 5, 826. [Google Scholar] [CrossRef]
- Gilbert, S.F.; Rosenberg, E.; Zilber-Rosenberg, I. The holobiont with its hologenome is a level of selection in evolution. In Landscapes of Collectivity in the Life Sciences; MIT Press: Cambridge, MA, USA, 2017; pp. 305–324. [Google Scholar]
- Ganley, R.J.; Brunsfeld, S.J.; Newcombe, G. A community of unknown, endophytic fungi in western white pine. Proc. Natl. Acad. Sci. USA 2004, 101, 10107–10112. [Google Scholar] [CrossRef]
- Panno, L.; Bruno, M.; Voyron, S.; Anastasi, A.; Gnavi, G.; Miserere, L.; Varese, G.C. Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia oceanica. New Biotech. 2013, 30, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Conte, C.; Rotini, A.; Manfra, L.; D’Andrea, M.M.; Winters, G.; Migliore, L. The seagrass holobiont: What we know and what we still need to disclose for its possible use as an ecological indicator. Water 2021, 13, 406. [Google Scholar] [CrossRef]
- Rotini, A.; Conte, C.; Winters, G.; Vasquez, M.I.; Migliore, L. Undisturbed Posidonia oceanica meadows maintain the epiphytic bacterial community in different environments. Environ. Sci. Pollut. Res. 2023, 30, 95464–95474. [Google Scholar] [CrossRef]
- Tarquinio, F.; Hyndes, G.A.; Laverock, B.; Koenders, A.; Säwström, C. The seagrass holobiont: Understanding seagrass-bacteria interactions and their role in seagrass ecosystem functioning. FEMS Microbiol. Lett. 2019, 366, fnz057. [Google Scholar] [CrossRef]
- Korlević, M.; Markovski, M.; Zhao, Z.; Herndl, G.J.; Najdek, M. Seasonal dynamics of epiphytic microbial communities on marine macrophyte surfaces. Front. Microbiol. 2021, 12, 671342. [Google Scholar] [CrossRef]
- Szitenberg, A.; Beca-Carretero, P.; Azcárate-García, T.; Yergaliyev, T.; Alexander-Shani, R.; Winters, G. Teasing apart the host-related, nutrient-related and temperature-related effects shaping the phenology and microbiome of the tropical seagrass Halophila stipulacea. Environ. Microbiol. 2022, 17, 1–17. [Google Scholar] [CrossRef]
- Agawin, N.S.; Ferriol, P.; Cryer, C.; Alcon, E.; Busquets, A.; Sintes, E.; Moyà, G. Significant nitrogen fixation activity associated with the phyllosphere of Mediterranean seagrass Posidonia oceanica: First report. Mar. Ecol. Prog. Ser. 2016, 551, 53–62. [Google Scholar] [CrossRef]
- Lucas-Elío, P.; Goodwin, L.; Woyke, T.; Pitluck, S.; Nolan, M.; Kyrpides, N.C.; Detter, J.C.; Copeland, A.; Lu, M.; Bruce, D.; et al. Complete genome sequence of Marinomonas posidonica type strain (IVIA-Po-181(T)). Stand. Genom. Sci. 2012, 7, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, E.; Marco-Noales, E.; Gómez, D.; Lucas-Elío, P.; Ordax, M.; Garcías-Bonet, N.; Duarte, C.M.; Sanchez-Amat, A. Taxonomic study of Marinomonas strains isolated from the seagrass Posidonia oceanica, with descriptions of Marinomonas balearica sp. nov. and Marinomonas pollencensis sp. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Rotini, A.; Conte, C.; Seveso, D.; Montano, S.; Galli, P.; Vai, M.; Migliore, L.; Mejia, A. Daily variation of the associated microbial community and the Hsp60 expression in the Maldivian seagrass Thalassia hemprichii. J. Sea Res. 2020, 156, 101835. [Google Scholar] [CrossRef]
- Sapp, J. The dynamics of symbiosis: An historical overview. Canad. J. Bot. 2004, 82, 1046–1056. [Google Scholar] [CrossRef]
- Brachmann, A.; Parniske, M. The most widespread symbiosis on earth. PLoS Biol. 2006, 4, e239. [Google Scholar] [CrossRef]
- Vohník, M.; Borovec, O.; Župan, I.; Vondrášek, D.; Petrtýl, M.; Sudová, R. Anatomically and morphologically unique dark septate endophytic association in the roots of the Mediterranean endemic seagrass Posidonia oceanica. Mycorrhiza 2015, 25, 663–672. [Google Scholar] [CrossRef]
- Ettinger, C.L.; Eisen, J.A. Characterization of the mycobiome of the seagrass, Zostera marina, reveals putative associations with marine chytrids. Front. Microbiol. 2019, 10, 491431. [Google Scholar] [CrossRef]
- Poli, A.; Varese, G.C.; Garzoli, L.; Prigione, V. Seagrasses, seaweeds and plant debris: An extraordinary reservoir of fungal diversity in the Mediterranean Sea. Fungal Ecol. 2022, 60, 101156. [Google Scholar] [CrossRef]
- Vohník, M.; Borovec, O.; Kolaříková, Z.; Sudová, R.; Réblová, M. Extensive sampling and high-throughput sequencing reveal Posidoniomyces atricolor gen. et sp. nov. (Aigialaceae, Pleosporales) as the dominant root mycobiont of the dominant Mediterranean seagrass Posidonia oceanica. MycoKeys 2019, 55, 59. [Google Scholar] [CrossRef]
- Vohník, M. Are lulworthioid fungi dark septate endophytes of the dominant Mediterranean seagrass Posidonia oceanica? Plant. Biol. 2022, 24, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Lumibao, C.Y.; Harris, G.; Birnbaum, C. Global Diversity and Distribution of Rhizosphere and Root-Associated Fungi in Coastal Wetlands: A Systematic Review. Estuaries Coasts 2024, 47, 905–916. [Google Scholar] [CrossRef]
- Aminot, A.; Kérouel, R. Hydrologie des Écosystèmes Marins. Paramètres et Analyses; Ifremer: Plouzané, France, 2004; 336p. [Google Scholar]
- Kirkwood, D.S. Stability of solutions of nutrient salts during storage. Mar. Chem. 1992, 38, 151–164. [Google Scholar] [CrossRef]
- Treguer, P.; Le Corre, P. Manuel d’Analyse des Sels Nutritifs dans l’Eau de Mer (Utilisation de l’Autoanalyseur II Technicon R); Lab. Océano. Chim. Univ. Bretagne Occidentale: Brest, France, 1974; 110p. [Google Scholar]
- Kadivar, H.; Stapleton, A.E. Ultraviolet radiation alters maize phyllosphere bacterial diversity. Microb. Ecol. 2003, 45, 353–361. [Google Scholar] [CrossRef]
- Mejia, A.Y.; Rotini, A.; Lacasella, F.; Bookman, R.; Thaller, M.C.; Shem-Tov, R.; Winters, G.; Migliore, L. Assessing the ecological status of seagrasses using morphology, biochemical descriptors and microbial community analyses. A study in Halophila stipulacea (Forsk.) Aschers meadows in the northern Red Sea. Ecol. Indic. 2016, 60, 1150–1163. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef]
- White, T.J.; Burns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics; Academic Press: New York, NY, USA, 1990. [Google Scholar]
- Heeger, F.; Wurzbacher, C.; Bourne, E.C.; Mazzoni, C.J.; Monaghan, M.T. Combining the 5.8 S and ITS2 to improve classification of fungi. Methods Ecol. Evol. 2019, 10, 1702–1711. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Caporaso, J.G. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Larsson, K.H. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Tedersoo, L.; Mikryukov, V.; Anslan, S.; Bahram, M.; Khalid, A.N.; Corrales, A.; Abarenkov, K. The Global Soil Mycobiome consortium dataset for boosting fungal diversity research. Fungal Divers. 2021, 111, 573–588. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environm Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Liang, J.; Liu, J.; Wang, X.; Sun, H.; Zhang, Y.; Ju, F.; Thompson, F.; Zhang, X.H. Genomic analysis reveals adaptation of Vibrio campbellii to the Hadal Ocean. Appl. Environ. Microbiol. 2022, 88, e00575-22. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.T.; Liesack, W.; Finster, K. Desulfovibrio zosterae sp. nov., a new sulfate reducer isolated from surface-sterilized roots of the seagrass Zostera marina. Int. J. Syst. Evol. Microbiol. 1999, 49, 859–865. [Google Scholar] [CrossRef]
- Ivanova, E.P.; Gorshkova, N.M.; Sawabe, T.; Zhukova, N.V.; Hayashi, K.; Kurilenko, V.V.; Christen, R. Sulfitobacter delicatus sp. nov. and Sulfitobacter dubius sp. nov., respectively from a starfish (Stellaster equestris) and seagrass (Zostera marina). Int. J. Syst. Evol. Microbiol. 2004, 54, 475–480. [Google Scholar] [CrossRef]
- Garcias-Bonet, N.; Arrieta, J.M.; Duarte, C.M.; Marbà, N. Nitrogen-fixing bacteria in Mediterranean seagrass (Posidonia oceanica) roots. Aquat. Bot. 2016, 131, 57–60. [Google Scholar] [CrossRef]
- Ravikumar, S.; Gnanadesigan, M.; Saravanan, A.; Monisha, N.; Brindha, V.; Muthumari, S. Antagonistic properties of seagrass associated Streptomyces sp. RAUACT-1: A source for anthraquinone rich compound. Asian Pac. J. Trop. Med. 2012, 5, 887–890. [Google Scholar] [CrossRef]
- Wu, H.; Chen, W.; Wang, G.H.; Dai, S.K.; Zhou, D.Y.; Zhao, H.Z.; Li, X. Culture-dependent diversity of Actinobacteria associated with seagrass (Thalassia hemprichii). Afr. J. Microbiol. Res. 2012, 6, 87–94. [Google Scholar]
- Rabbani, G.; Yan, B.C.; Lee, N.L.Y.; Ooi, J.L.S.; Lee, J.N.; Huang, D.; Wainwright, B.J. Spatial and structural factors shape seagrass-associated bacterial communities in Singapore and peninsular Malaysia. Front. Mar. Sci. 2021, 8, 659180. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Q.; Yao, Y.; Tan, F.; Jiang, L.; Shi, W.; Liu, J. Bacterial Communities in Zostera marina Seagrass Beds of Northern China. Water 2024, 16, 935. [Google Scholar] [CrossRef]
- Brown, C.; Seidler, R.J. Potential pathogens in the environment: Klebsiella pneumoniae, a taxonomic and ecological enigma. Appl. Microbiol. 1973, 25, 900–904. [Google Scholar] [CrossRef] [PubMed]
- Shade, A.; Peter, H.; Allison, S.D.; Baho, D.L.; Berga, M.; Bürgmann, H.; Handelsman, J. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 2012, 3, 417. [Google Scholar] [CrossRef] [PubMed]
- Tarquinio, F.; Attlan, O.; Vanderklift, M.A.; Berry, O.; Bissett, A. Distinct endophytic bacterial communities inhabiting seagrass seeds. Front. Microbiol. 2021, 12, 703014. [Google Scholar] [CrossRef]
- Hansen, J.W.; James, W.U.; Perry, C.J.; Dennison, W.C.; Lomstein, B.A. Effect of the seagrass Zostera capricorni on sediment microbial processes. Mar. Ecol. Prog. Ser. 2000, 199, 83–96. [Google Scholar] [CrossRef]
- Singh, B.K.; Quince, C.; Macdonald, C.A.; Khachane, A.; Thomas, N.; Al-Soud, W.A.; Campbell, C.D. Loss of microbial diversity in soils is coincident with reductions in some specialized functions. Environ. Microbiol. 2014, 16, 2408–2420. [Google Scholar] [CrossRef]
- Hornick, K.M.; Buschmann, A.H. Insights into the diversity and metabolic function of bacterial communities in sediments from Chilean salmon aquaculture sites. Ann. Microbiol. 2018, 68, 63–77. [Google Scholar] [CrossRef]
- Addy, H.D.; Piercey, M.M.; Currah, R.S. Microfungal endophytes in roots. Can. J. Bot. 2005, 83, 1–13. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Company, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Bamisile, B.S.; Dash, C.K.; Akutse, K.S.; Keppanan, R.; Wang, L. Fungal endophytes: Beyond herbivore management. Front. Microbiol. 2018, 9, 544–577. [Google Scholar] [CrossRef]
- Quesada Moraga, E. Entomopathogenic fungi as endophytes: Their broader contribution to IPM and crop production. Biocontrol. Sci. Technol. 2020, 30, 864–877. [Google Scholar] [CrossRef]
- Heilmann-Clausen, J.; Christensen, M. Fungal diversity on decaying beech logs–implications for sustainable forestry. Biodivers. Conserv. 2003, 12, 953–973. [Google Scholar] [CrossRef]
- Jönsson, M.T.; Edman, M.; Jonsson, B.G. Colonization and extinction patterns of wood-decaying fungi in a boreal old-growth Picea abies forest. J. Ecol. 2008, 96, 1065–1075. [Google Scholar] [CrossRef]
- Meyer, S.; Rusterholz, H.P.; Baur, B. Saproxylic insects and fungi in deciduous forests along a rural–urban gradient. Ecol. Evol. 2021, 11, 1634–1652. [Google Scholar] [CrossRef]
- Poli, A.; Bovio, E.; Ranieri, L.; Varese, G.C.; Prigione, V. Fungal Diversity in the Neptune Forest: Comparison of the Mycobiota of Posidonia oceanica, Flabellia petiolata, and Padina pavonica. Front. Microbiol. 2020, 11, 933. [Google Scholar] [CrossRef]
- Florio Furno, M.; Ferrero, D.; Poli, A.; Prigione, V.; Tuohy, M.; Oliva, M.; Pretti, C.; Varese, G.C. Chapter Fungi from the Sediments of the Harbour of Livorno as Potential Bioremediation Agents; Firenze University Press: Florence, Italy, 2022; pp. 665–675. [Google Scholar]
- Migliore, L.; Rotini, A.; Randazzo, D.; Albanese, N.N.; Giallongo, A. Phenols content and 2-D electrophoresis protein pattern: A promising tool to monitor Posidonia meadows health state. BMC Ecol. 2007, 7, 6. [Google Scholar] [CrossRef]
- Rotini, A.; Belmonte, A.; Barrote, I.; Micheli, C.; Peirano, A.; Santos, R.O.; Silva, J.; Migliore, L. Effectiveness and consistency of a suite of descriptors for assessing the ecological status of seagrass meadows (Posidonia oceanica L. Delile). Estuar. Coast. Shelf Sci. 2013, 130, 252–259. [Google Scholar] [CrossRef]
- Conte, C.; Apostolaki, E.T.; Vizzini, S.; Migliore, L. A tight interaction between the native seagrass Cymodocea nodosa and the exotic Halophila stipulacea in the Aegean Sea highlights seagrass holobiont variations. Plants 2023, 12, 350. [Google Scholar] [CrossRef] [PubMed]
- Lunghini, D.; Granito, V.M.; Di Lonardo, D.P.; Maggi, O.; Persiani, A.M. Fungal diversity of saprotrophic litter fungi in a Mediterranean maquis environment. Mycologia 2013, 105, 1499–1515. [Google Scholar] [CrossRef]
- Zalar, P.D.; De Hoog, G.S.; Schroers, H.J.; Crous, P.W.; Groenewald, J.Z.; Gunde-Cimerman, N. Phylogeny and ecology of the ubiquitous saprobe Cladosporium sphaerospermum, with descriptions of seven new species from hypersaline environments. Stud. Mycol. 2007, 58, 157–183. [Google Scholar] [CrossRef] [PubMed]
- Vohník, M.; Borovec, O.; Kolařík, M. Communities of cultivable root mycobionts of the seagrass Posidonia oceanica in the northwest Mediterranean Sea are dominated by a hitherto undescribed pleosporalean dark septate endophyte. Microb. Ecol. 2016, 71, 442–451. [Google Scholar] [CrossRef]
- Tedersoo, L.; Sánchez-Ramírez, S.; Koljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 2018, 90, 135–159. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Abarenkov, K. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [PubMed]
- Borovec, O.; Vohník, M. Ontogenetic transition from specialized root hairs to specific root-fungus symbiosis in the dominant Mediterranean seagrass Posidonia oceanica. Sci. Rep. 2018, 8, 10773. [Google Scholar] [CrossRef]
- Rotini, A.; Mejia, A.Y.; Costa, R.; Migliore, L.; Winters, G. Ecophysiological plasticity and bacteriome shift in the seagrass Halophila stipulacea along a depth gradient in the Northern Red Sea. Front. Plant Sci. 2017, 7, 2015. [Google Scholar] [CrossRef]
- Conte, C.; Rotini, A.; Winters, G.; Vasquez, M.I.; Piazza, G.; Kletou, D.; Migliore, L. Elective affinities or random choice within the seagrass holobiont? The case of the native Posidonia oceanica (L.) Delile and the exotic Halophila stipulacea (Forssk.) Asch. from the same site (Limassol, Cyprus). Aquat. Bot. 2021, 174, 103420. [Google Scholar] [CrossRef]
- Lau, J.A.; Lennon, J.T. Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc. Natl. Acad. Sci. USA 2012, 109, 14058–14062. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.B.G. Are there more marine fungi to be described? Bot. Mar. 2011, 54, 343–354. [Google Scholar] [CrossRef]
- Gladfelter, A.S.; James, T.Y.; Amend, A.S. Marine fungi. Curr. Biol. 2019, 29, R191–R195. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.B.G.; Pang, K.L.; Abdel-Wahab, M.A.; Scholz, B.; Hyde, K.D.; Boekhout, T.; Ebel, R.; Rateb, M.E.; Henderson, L.; Sakayaroj, J.; et al. An online resource for marine fungi. Fungal Divers. 2019, 96, 347–433. [Google Scholar] [CrossRef]


| Site Status * | Shannon’s Index (H’) | No. of Taxa | Total No. of Sequences | P vs. I (Kruskall–Wallis) | |
|---|---|---|---|---|---|
| Seawater | P | 3.28 ± 0.06 | 310 | 123,400 | NS |
| I | 3.20 ± 0.04 | 234 | 124,233 | ||
| Sediment | P | 3.12 ± 0.36 | 450 | 73,999 | NS |
| I | 3.05 ± 0.03 | 432 | 73,252 | ||
| Leaves | P | 3.18 ± 0.16 | 252 | 36,829 | NS |
| I | 2.57 ± 1.19 | 188 | 30,367 | ||
| Rhizomes–Roots | P | 2.86 ± 0.90 | 267 | 42,588 | p < 0.05 |
| I | 1.43 ± 0.38 | 60 | 29,528 |
| Site Status * | Shannon’s Index (H’) | No. of Taxa | Total No. of Sequences | P vs. I (Kruskall–Wallis) | |
|---|---|---|---|---|---|
| Seawater | P | 1.04 ± 0.59 | 20 | 5242 | NS |
| I | 0.89 ± 0.42 | 14 | 2234 | ||
| Sediment | P | 0.54 ± 0.07 | 43 | 16,475 | NS |
| I | 1.09 ± 0.49 | 58 | 16,527 | ||
| Leaves | P | 0.70 ± 0.19 | 20 | 7551 | p < 0.05 |
| I | 1.67 ± 0.62 | 46 | 23,577 | ||
| Rhizomes–Roots | P | 0.68 ± 0.18 | 41 | 35,753 | NS |
| I | 1.49 ± 1.47 | 66 | 36,254 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frasca, S.; Alabiso, A.; D’Andrea, M.M.; Cattaneo, R.; Migliore, L. Diversity and Composition of Posidonia oceanica-Associated Bacterial and Fungal Communities: Effect of Boat-Induced Mechanical Stress in the Villefranche-sur-Mer Bay (France). Diversity 2024, 16, 604. https://doi.org/10.3390/d16100604
Frasca S, Alabiso A, D’Andrea MM, Cattaneo R, Migliore L. Diversity and Composition of Posidonia oceanica-Associated Bacterial and Fungal Communities: Effect of Boat-Induced Mechanical Stress in the Villefranche-sur-Mer Bay (France). Diversity. 2024; 16(10):604. https://doi.org/10.3390/d16100604
Chicago/Turabian StyleFrasca, Sara, Annamaria Alabiso, Marco M. D’Andrea, Raffaela Cattaneo, and Luciana Migliore. 2024. "Diversity and Composition of Posidonia oceanica-Associated Bacterial and Fungal Communities: Effect of Boat-Induced Mechanical Stress in the Villefranche-sur-Mer Bay (France)" Diversity 16, no. 10: 604. https://doi.org/10.3390/d16100604
APA StyleFrasca, S., Alabiso, A., D’Andrea, M. M., Cattaneo, R., & Migliore, L. (2024). Diversity and Composition of Posidonia oceanica-Associated Bacterial and Fungal Communities: Effect of Boat-Induced Mechanical Stress in the Villefranche-sur-Mer Bay (France). Diversity, 16(10), 604. https://doi.org/10.3390/d16100604








