Morphological and Molecular Characterization of Discolaimus haridwarensis sp. n. (Nematoda: Dorylaimida: Qudsianematidae) from India
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, Extraction, and Morphological Identification
2.2. DNA Extraction, PCR and Sequencing
2.3. Phylogenetic Analyses
3. Results
3.1. Systematics
- Phylum: Nematoda Potts, 1932
- Class: Enoplea Inglis, 1983
- Order: Dorylaimida Pearse, 1942
- Family: Qudsianematidae Jairajpuri, 1965
- Genus: Discolaimus Cobb, 1913
- Species: haridwarensis sp. n.
- Isolates/strains: HN13
3.1.1. Discolaimus haridwarensis sp. n.
- urn:lsid:zoobank.org:pub:679768CE-BC71-4CE1-B561-39B8D645C52F
3.1.2. Material Examined
3.1.3. Description
- Male. Not found.
3.1.4. Measurements
| Holotype | Paratype | |
|---|---|---|
| Character a | ♀ | 9 ♀♀ |
| L | 2.28 | 2.17 ± 0.89 (2.11–2.32) |
| a | 35.9 | 38.6 ± 3.8 (32.8–42.5) |
| b | 5.2 | 4.8 ± 0.3 (4.3–5.2) |
| c | 59.3 | 59.7 ± 5.8 (53.2–68.3) |
| c′ | 1.0 | 1.1 ± 0.1 (1.0–1.3) |
| V | 48.3 | 51.4 ± 2.2 (48.1–55.2) |
| G1 | 6.2 | 5.9 ± 0.7 (5.2–6.8) |
| G2 | 6.4 | 5.9 ± 0.6 (5.1–6.5) |
| Lip region diameter | 29.5 | 28.4 ± 1.1 (27.0–30.0) |
| Odontostyle length | 23.0 | 21.8 ± 1.2 (20.0–23.0) |
| Odontophore length | 36.5 | 36.1 ± 2.5 (34.0–38.0) |
| Neck length | 437 | 460.3 ± 20.9 (436–487) |
| Pharyngeal expansion | 279 | 271.4 ± 5.9 (266–280) |
| Diameter at neck base | 62.5 | 54.3 ± 5.4 (48.5–62.5) |
| at midbody | 63.0 | 58.5 ± 7.5 (49.0–66.0) |
| at anus | 35.0 | 31.7 ± 2.2 (28.0–33.5) |
| Prerectum | 26.5 | 32.7 ± 5.4 (26.5–38.5) |
| Rectum | 33.5 | 32.2 ± 1.2 (30.5–33.5) |
| Tail | 38.5 | 37.2 ± 2.4 (34.0–40.0) |
3.1.5. Type Habitat and Locality
3.1.6. Type Material
3.1.7. Etymology
3.1.8. Diagnosis
3.1.9. Relationships
3.2. Molecular Characterization
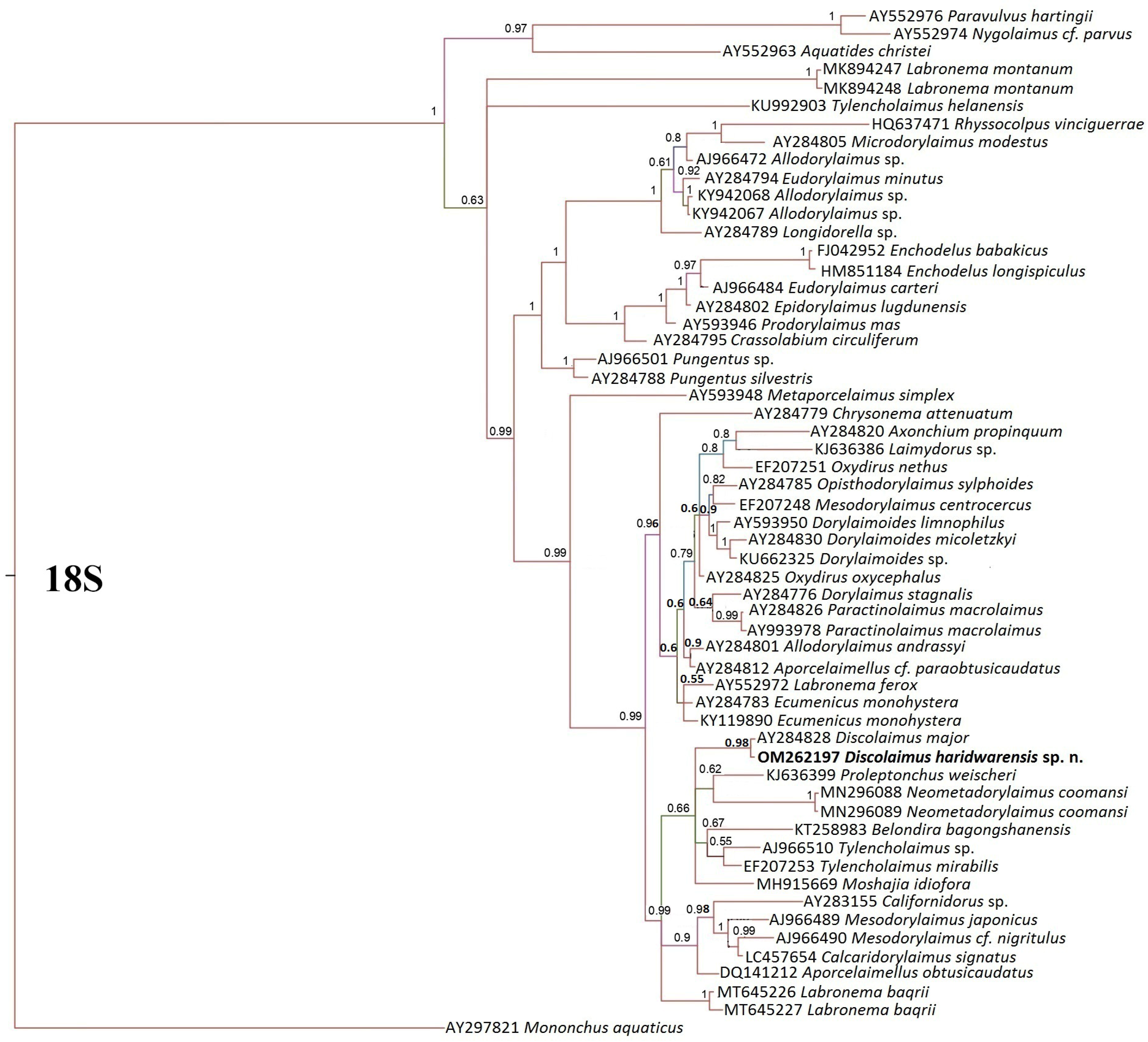
3.3. Phylogenetic Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cobb, N.A. New nematode genera found inhabiting freshwater and non-brackish soils. J. Wash. Acad. Sci. 1913, 3, 432–444. [Google Scholar]
- Andrássy, I. Free-Living Nematodes of Hungary (Nematoda errantia). III. Pedozoologica Hungarica 5; Hungarian Natural History Museum and Systematic Research Group of the Hungarian Academy of Sciences: Budapest, Hungary, 2009; 608p. [Google Scholar]
- Peña-Santiago, R. Dorylaimida Mundi (Nematoda): Checklist of Genera and Species, with Their Records; Monographic Papers on Nematology 7; University of Jaén: Jaén, Spain, 2021. [Google Scholar]
- Wu, W.; Yan, L.; Xu, C.L.; Wang, K.; Jin, S.Y.; Xie, H. A new species of the genus Discolaimus Cobb, 1913 (Nematoda: Dorylaimida: Qudsianematidae) from Qinghai, China. Zootaxa 2016, 4088, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Erum, I.; Nasira, K.; Sagir, H.; Raza, S.; Firoza, K. Description of Discolaimus miniodontii n. sp. and Laevides hunderansis n. sp. with notes on Discolaimoides spatilabium (Nematoda: Dorylaimida) from District Ghizer, Gilgit-Baltistan, Pakistan. Pakistan J Nematol. 2020, 38, 1–13. [Google Scholar]
- Sharma, V. Diversity of Terrestrial nematodes in Uttarakhand. J. Exp. Zool. India 2011, 14, 575–582. [Google Scholar]
- Sharma, V. A preliminary study of nematodes in Govind Wildlife Sanctuary, Uttarkashi, Uttarakhand, India. Ann. For. Res. 2013, 21, 92–94. [Google Scholar]
- Gantait, V.V.; Bhattacharya, T.; Chatterjee, A. A new species of the genus Discolaimus Cobb, 1913 (Qudsianematidae: Dorylaimida) from West Bengal, India with revised key to the genus. Proc. Zool. Soc. 2009, 62, 67–73. [Google Scholar] [CrossRef]
- Gantait, V.V.; Chatterjee, A.; Bhattacharya, T. Trophic groups of nematodes associated with banana plantation in Paschim Medinipur district of West Bengal, India. Trop. Ecol. 2011, 52, 331–335. [Google Scholar]
- Sen, D.; Chatterjee, A.; Manna, B. Two New and Two Known Species of Dorylaimoidea (Nematoda) from West Bengal, India with a Key to the Species of the Genus Indodorylaimus Ali and Prabha, 1974. Rec. Zool. Surv. India 2012, 112, 11–20. [Google Scholar] [CrossRef]
- Khan, E.; Laha, S.K. Seven new dorylaimid nematodes from Indian Agricultural Research Institute Farm, Delhi, India. Indian J. Nematol. 1982, 12, 232–249. [Google Scholar]
- Siddiqi, M.R. Studies on Discolaimus spp. (Nematoda: Dorylaimidae) from India. Z. Zool. Syst. Evol. 1964, 2, 174–184. [Google Scholar] [CrossRef]
- Lal, A.; Khan, E. Nematodes associated with forest trees in nurseries and reserved natural forests of northern India. Indian J. For. 1988, 11, 276–282. [Google Scholar]
- Bilgrami, A.L.; Kulshreshtha, R.; Pervez, R. Community analysis of predaceous and freeliving nematodes of Aligarh district. Indian J. Nematol. 1998, 27, 104–110. [Google Scholar]
- Pervez, R. Prevalence of plant parasitic and predatory nematodes associated with different crops of Badaun districts (Uttar Pradesh). Curr. Nematol. 2004, 15, 43–46. [Google Scholar]
- Baqri, Q.H.; Bohra, P. Studies on Plant and Soil Nematodes Associated with Crops of Economic Importance in Gujarat; Records of the Zoological Survey of India, Occasional Paper; Zoological Survey of India: Kolkata, Indian, 2005; No. 240; 48p. [Google Scholar]
- Bohra, P.; Baqri, Q.H. Biodiversity of plant and soil nematodes from Andaman and Car Nicobar Islands. Rec. Zool. Surv. India 2005, 105, 43–49. [Google Scholar] [CrossRef]
- Bohra, P.; Baqri, Q.H.; Dwivedi, A.K. Diversity of phytophagous nematodes associated with economically important crops in Gujarat, India. Indian J. Nematol. 2005, 35, 130–133. [Google Scholar]
- Pervez, R.; Ali, S.S. Biodiversity of the predatory nematodes associated with pulse crops from Jhansi district of Uttar Pradesh. Curr. Nematol. 2008, 19, 57–61. [Google Scholar]
- Khan, Z.A.; Ahmad, W.A.; Jairajpuri, M.S. Description of two new species of Dorylaimoidea from India. Fund. Appl. Nematol. 1994, 17, 147–151. [Google Scholar]
- Rahaman, P.F.; Ahmad, I. Community analysis of predatory nematode species from Aligarh soils, India. Nematol. Mediterr. 1995, 23, 57–60. [Google Scholar]
- Sharma, V. Ten new records of free-living nematodes from Uttarakhand, India. Indian J. For. 2018, 41, 39–42. [Google Scholar]
- Ahmad, W.; Jairajpuri, M.S. Some new and known species of Dorylaimoidea. Nematologica 1982, 28, 39–61. [Google Scholar] [CrossRef]
- Mohilal, N.; Dhanachand, C. Investigations of soil nematodes from Manipur, species of the family Qudsianematidae. Uttar Pradesh J. Zool. 2001, 21, 41–45. [Google Scholar]
- Baniyamuddin, M.; Ahmad, W. Some new and known species of Dorylaimoidea (Dorylaimida: Nematoda) from natural forest of Arunachal Pradesh, India. J. Nematode Morphol. Syst. 2007, 10, 75–95. [Google Scholar]
- Cobb, N.A. Estimating the nematode population of the soil Bureau of Plant Industry. U. S. Dep. Agric. Agric. Technol. Circ. 1918, 1, 19–24. [Google Scholar]
- Goodey, J.B. Laboratory Methods for Work with Plant and Soil Nematodes; Technical Bulletin No. 2; Ministry of Agriculture, Fisheries and Food: London, UK, 1957; 47p. [Google Scholar]
- Southey, J.F. Laboratory Methods for Work with Plant and Soil Nematodes; Reference Book 402; Ministery of Agriculture, Fisheries and Food: London, UK, 1985. [Google Scholar]
- Courtney, W.D.; Polley, D.; Miller, V.L. TAF, an improved fixative in nematode technique. Plant Dis. Rep. 1955, 39, 570–571. [Google Scholar]
- Seinhorst, J.W. A rapid method for the transfer of nematodes from fixative to anhydrous glycerin. Nematologica 1959, 4, 67–69. [Google Scholar] [CrossRef]
- Sharma, H.; Chaubey, A.K. Molecular and phenotypic characterization of Hemicriconemoides rosae (Rathour et al., 2003) from mustard rhizosphere in India. J. Basic Appl. Zool. 2023, 84, 16. [Google Scholar] [CrossRef]
- De Man, J.G. Die einheimischen, frei in der reinen Erde und imsüssen Wasser lebenden Nematoden. Tijdschr. Ned. Dierkd. Ver. 1880, 5, 1–104. [Google Scholar]
- Floyd, R.M.; Rogers, A.D.; Lambshead, P.J.D.; Smith, C.R. Nematode-specific PCR primers for the 18S small subunit rRNA gene. Mol. Ecol. Notes 2005, 5, 611–612. [Google Scholar] [CrossRef]
- De Ley, P.; Félix, M.A.; Frisse, L.M.; Nadler, S.A.; Sternberg, P.W.; Thomas, W.K. Molecular and morphological characterisation of two reproductively isolated species with mirror-image anatomy (Nematoda: Cephalobidae). Nematology 1999, 1, 591–612. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment and analysis program for Windows 95/98.NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Loof, P.A.A.; Coomans, A. On the development and location of the oesophageal gland nuclei in the Dorylaimina. Zesz. Probl. Postepow. Nauk Roln. 1970, 92, 79–161. [Google Scholar]
- Thorne, G. A monograph of the nematodes of the superfamily Dorylaimoidea. Capita Zool. 1939, 8, 1–261. [Google Scholar]
- Peña-Santiago, R.; Torres, B.; Liébanas, G.; Abolafia, J. Nematodes of the order Dorylaimida from Andalucía Oriental, Spain. The genus Discolaimus Cobb, 1913. II. Two previously known species, with comments on their taxonomy. Russ. J. Nematol. 2002, 10, 79–88. [Google Scholar]
- Siddiqi, M.R. Ten new species of Discolaimus Cobb, 1913 (Nematoda: Dorylaimida). Int. J. Nematol. 2005, 15, 215–229. [Google Scholar]
- Sauer, M.R.; Annells, C.M. Species of Discolaimus (Nematoda: Dorylaimoidea) from Australia. Nematologica 1985, 31, 121–133. [Google Scholar] [CrossRef]
- Lordello, L.G.E. On the morphology of Proleptonchus aestivus n. gen., n. sp. and Dorylaimus lourdesae n. sp., two new soil nematodes from Brazil. Proc. Helminthol. Soc. Wash. 1955, 22, 71–75.
- Tulaganov, A.T. Plant parasitic and soil nematodes in Uzbekistan. Izdat. Akad. Nauk. Uzbeksk. SSR Tashkent. 1949; 3–227. (In Russian) [Google Scholar]
- Andrássy, I. The scientific results of the Hungarian Soil Zoological Expedition to the Brazzaville-Congo, 31. Nematodes from Groundwater. Ann. Univ. Sci. Budapestinensis Rolando Eötvös Nomin. (Sect. Biol.) 1968, 9–10, 3–26. [Google Scholar]
- Andrássy, I. The superfamily Dorylaimoidea (Nematoda)—A review. Family Qudsianematidae, II. Opusc. Zool. 1991, 24, 3–55. [Google Scholar]
- Heyns, J. A report on South African nematodes of the genera Labronema Thorne, Discolaimus Cobb, Discolaimoides n. gen., and Discolaimium Thorne (Nemata: Dorylaimoidea). Proc. Helminthol. Soc. Wash. 1963, 30, 1–6. [Google Scholar]
- Jairajpuri, M.S.; Ahmad, W. Dorylaimida: Free-Living, Predacious and Plant-Parasitic Nematodes; Brill: Leiden, The Netherlands, 1992. [Google Scholar]
- De Man, J.G. Onderzoekingen over vrij in de aarde levende Nematoden. Tijdschr. Ned. Dierkd. Ver. 1876, 2, 78–196. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, H.; Chaubey, A.K.; Álvarez-Ortega, S. Morphological and Molecular Characterization of Discolaimus haridwarensis sp. n. (Nematoda: Dorylaimida: Qudsianematidae) from India. Diversity 2024, 16, 598. https://doi.org/10.3390/d16100598
Sharma H, Chaubey AK, Álvarez-Ortega S. Morphological and Molecular Characterization of Discolaimus haridwarensis sp. n. (Nematoda: Dorylaimida: Qudsianematidae) from India. Diversity. 2024; 16(10):598. https://doi.org/10.3390/d16100598
Chicago/Turabian StyleSharma, Himani, Ashok Kumar Chaubey, and Sergio Álvarez-Ortega. 2024. "Morphological and Molecular Characterization of Discolaimus haridwarensis sp. n. (Nematoda: Dorylaimida: Qudsianematidae) from India" Diversity 16, no. 10: 598. https://doi.org/10.3390/d16100598
APA StyleSharma, H., Chaubey, A. K., & Álvarez-Ortega, S. (2024). Morphological and Molecular Characterization of Discolaimus haridwarensis sp. n. (Nematoda: Dorylaimida: Qudsianematidae) from India. Diversity, 16(10), 598. https://doi.org/10.3390/d16100598







