Advances in the Study of Orchidinae Subtribe (Orchidaceae) Species with 40,42-Chromosomes in the Mediterranean Region
Abstract
1. Introduction
2. Materials and Methods
2.1. Cytological Analysis
2.2. Chromosome Numbers and Karyotype Parameters
2.3. Nomenclature
3. Results
3.1. Genus Neotinea s.l.
3.2. Genus Orchis s.str.
3.3. Genus Dactylorhiza
3.4. Genus Gymnadenia
3.5. B-Chromosomes
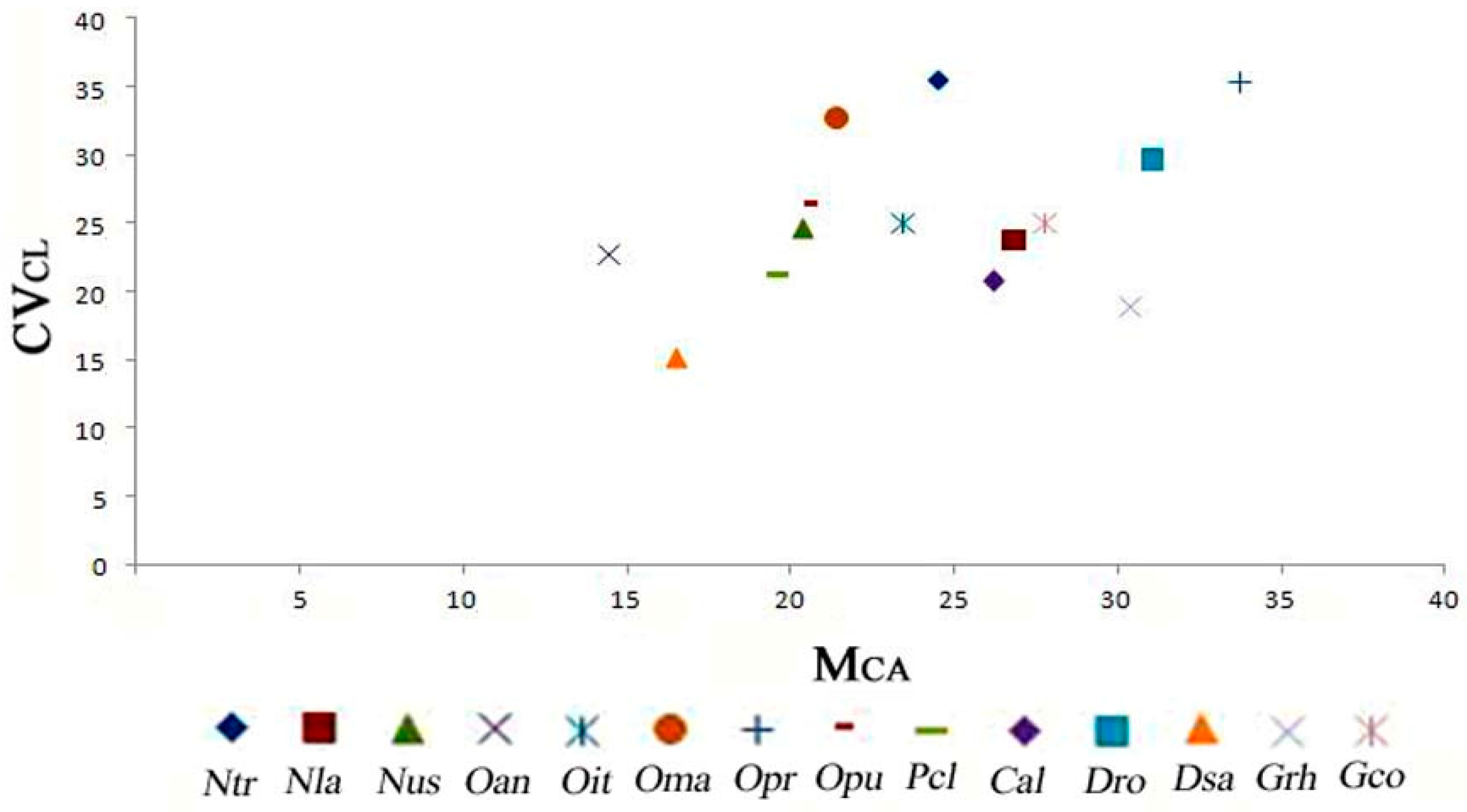
3.6. Diagram of the Morphometric Parameters
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pridgeon, A.M.; Bateman, R.M.; Cox, A.V.; Hapeman, J.R.; Chase, M.W. Phylogenetics of subtribe Orchidinae (Orchidoideae, Orchidaceae) based on nuclear ITS sequences. 1. Intergeneric relationships and polyphyly of Orchis sensu lato. Lindleyana 1997, 12, 89–109. [Google Scholar]
- Aceto, S.; Caputo, P.; Cozzolino, S.; Gaudio, L.; Moretti, A. Phylogeny and evolution of Orchis and allied genera based on ITS DNA variation: Morphological gaps and molecular continuity. Mol. Phylogenet. Evol. 1999, 13, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Bateman, R.M.; Hollingsworth, M.P.; Preston, J.; Yi-Bo, L.; Pridgeon, M.A.; Chase, W.M. Molecular phylogenetics and evolution of Orchidinae and selected Habenariinae (Orchidaceae). Bot. J. Linn. Soc. 2003, 142, 1–40. [Google Scholar] [CrossRef]
- Shipunov, A.B.; Fay, M.F.; Pillon, Y.; Bateman, R.M.; Chase, M.W. Dactylorhiza (Orchidaceae) in European Russia: Combined molecular and morphological analysis. Am. J. Bot. 2004, 91, 1419–1426. [Google Scholar] [CrossRef] [PubMed]
- Bateman, R.M. Evolutionary classification of European orchids: The crucial importance of maximising explicit evidence and minimising authoritarian speculation. J. Eur. Orchid. 2009, 41, 501–572. [Google Scholar]
- Gamarra, R.; Galan, P.; Herrera, I.; Ortunez, E. Seed micromorphology supports the splitting of Limnorchis from Platanthera (Orchidaceae). Nord. J. Bot. 2008, 26, 61–65. [Google Scholar] [CrossRef]
- Sramko, G.; Molnar, A.V.; Hawkins, J.A.; Bateman, R.M. Molecular phylogeny and evolutionary history of the Eurosiatic orchid genus Himantoglossum s.l. (Orchidaceae). Ann. Bot. 2014, 114, 1609–1626. [Google Scholar] [CrossRef] [PubMed]
- Chase, M.W.; Cameron, K.M.; Freudestein, J.V.; Pridgeon, A.M.; Salazar, G.; van der Berg, C.; Schuiteman, A. An updated classification of Orchidaceae. Bot. J. Linn. Soc. 2015, 177, 151–174. [Google Scholar] [CrossRef]
- Efimov, P.G. A revision of Platanthera (Orchidaceae; Orchidoideae; Orchideae) in Asia. Phytotaxa 2016, 254, 1–233. [Google Scholar] [CrossRef]
- Jin, X.H.; Schuitemann, A.; Chase, M.W.; Li, J.W.; Chung, S.W.; Hsu, T.C.; Jin, X.H. Phylogenetics of subtribe Orchidinae s.l. (Orchidaceae; Orchidoideae) based on seven markers (plastid matK, psaB, rbcL, trnL-F, trnH-psba, and nuclear nrITS, Xdh): Implications for generic delimitation. BMC Plant Biol. 2017, 17, 222. [Google Scholar] [CrossRef]
- Bateman, R.M.; Murphy, A.R.M.; Hollingsworth, P.M.; Hart, M.L.; Denholm, I.; Rudall, P.J. Molecular and morphological phylogenetics of the digitate-tubered clade within subtribe Orchidinae s.s. (Orchidaceae: Orchideae). Kew Bull. 2018, 73, 54. [Google Scholar] [CrossRef]
- Trávníček, P.; Chumová, Z.; Záveská, E.; Hanzlíčková, J.; Kupková, L.; Kučera, J.; Gbúrová Štubňová, E.; Rejlová, L.; Mandáková, T.; Ponert, J. Integrative Study of Genotypic and Phenotypic Diversity in the Eurasian Orchid Genus Neotinea. Front. Plant Sci. 2021, 12, 734240. [Google Scholar] [CrossRef] [PubMed]
- D’Emerico, S.; Bianco, P.; Medagli, P.; Ruggiero, L. Karyological studies of some taxa of the genera Himantoglossum, Orchis, Serapias and Spiranthes (Orchidaceae) from Apulia (Italy). Caryologia 1990, 43, 267–276. [Google Scholar] [CrossRef]
- Bianco, P.; D’Emerico, S.; Medagli, P.; Ruggiero, L. Polyploidy and aneuploidy in Ophrys, Orchis and Anacamptis (Orchidaceae). Plant Syst. Evol. 1991, 178, 235–245. [Google Scholar] [CrossRef]
- D’Emerico, S.; Pignone, D.; Bianco, P. Karyomorphological analyses and heterochromatin characteristic disclose phyletic relationships among 2n = 32 and 2n = 36 species of Orchis (Orchidaceae). Plant Syst. Evol. 1996, 200, 111–124. [Google Scholar] [CrossRef]
- D’Emerico, S.; Galasso, I.; Pignone, D.; Scrugli, A. Localization of rDNA loci by Fluorescent In situ Hybridization in some wild orchids from Italy (Orchidaceae). Caryologia 2001, 54, 31–36. [Google Scholar] [CrossRef]
- Bernardos, S.; Tyteca, D.; Amich, F. Cytotaxonomic study of some taxa of the subtribe Orchidinae (Orchidoideae, Orchidaceae) from the Iberian Peninsula. Isr. J. Plant Sci. 2004, 52, 161–170. [Google Scholar] [CrossRef]
- D’Emerico, S. Cytogenetic diversity in Orchis s.l. and allied genera (Orchidinae, Orchidaceae). In Plant Genome; Sharma, A.K., Sharma, A., Eds.; Part B: Phanerogams (Higher Groups); Science Publishers, Inc.: Enfield, NH, USA, 2005; Volume 1, pp. 61–87. [Google Scholar]
- Bellusci, F.; Aquaro, G. Contribution to the cytotaxonomical knowledge of four species of Serapias L. (Orchidaceae). Caryologia 2008, 61, 294–299. [Google Scholar]
- Šegota, V.; Hršak, V.; Vuković, N.; Alegro, A.; Besendorfer, V.; Sedlar, Z.; Bogdanović, S.; Poljak, I. Disentangling the kinship of Serapias todaroi Tin. (Orchidaceae) along the eastern Adriatic using chromosome count and morphometry. Flora 2018, 249, 9–15. [Google Scholar] [CrossRef]
- Turco, A.; Albano, A.; Medagli, P.; Pulvirenti, S.; D’Emerico, S. New cytological data in Ophrys sect. Pseudophrys Godfery and comparative karyomorphological studies in Ophrys L. (Orchidaceae). Plant Biosyst. 2018, 152, 901–910. [Google Scholar] [CrossRef]
- Hedrén, M.; Lorenz, R.; Teppner, H.; Dolinar, B.; Giotta, C.; Griebl, N.; Hansson, S.; Heidtke, U.; Klein, E.; Perazza, G.; et al. Evolution and systematics of polyploidy Nigritella (Orchidaceae). Nord. J. Bot. 2018, 36, 1–32. [Google Scholar] [CrossRef]
- Turco, A.; Albano, A.; Medagli, P.; Wagensommer, R.P.; D’Emerico, S. Comparative chromosome studies in species of subtribe Orchidinae (Orchidaceae). Comp. Cytogen. 2021, 15, 507–525. [Google Scholar] [CrossRef] [PubMed]
- Turco, A.; Albano, A.; Medagli, P.; Wagensommer, R.P.; D’Emerico, S. Comparative Cytogenetic of the 36-Chromosomes Genera of Orchidinae Subtribe (Orchidaceae) in the Mediterranean Region: A Summary and New Data. Plants 2023, 12, 2798. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Xiang, D.; Xia, K.; Sun, B.; Khurshid, H.; Esh, A.M.H.; Zhang, H. Characterization of Repetitive DNA in Saccharum officinarum and Saccharum spontaneum by Genome Sequencing and Cytological Assays. Front. Plant Sci. 2022, 13, 814620. [Google Scholar] [CrossRef] [PubMed]
- Guerra, M. Cytotaxonomy: The end of childhood. Plant Biosyst. 2012, 146, 703–710. [Google Scholar] [CrossRef]
- Guerra, M. Patterns of heterochromatin distribution in plant chromosomes. Genet. Mol. Biol. 2000, 23, 1029–1041. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S.; Schwarzacher, T. Organisation of the plant genome in chromosomes. Plant J. 2011, 66, 18–33. [Google Scholar] [CrossRef]
- Sharma, S.K.; Mukai, Y. Chromosome research in orchids: Current status and future prospects with special emphasis from molecular and epigenetic perspective. Nucleus 2015, 58, 173–184. [Google Scholar] [CrossRef]
- Abbott, R.J.; Ireland, H.E.; Rogers, H.J. Population decline despite high genetic diversity in the new allopolyploid species Senecio cambrensis (Asteraceae). Mol. Ecol. 2007, 16, 1023–1033. [Google Scholar] [CrossRef]
- Eilam, T.; Anikster, E.Y.; Millet, J.; Anisterski, M.; Feldman, M. Genome Size in Diploids, Allopolyploids, and Autopolyploids of Mediterranean Triticeae. J. Bot. 2010, 2010, 341380. [Google Scholar] [CrossRef]
- Burns, R.; Mandáková, T.; Gunis, J.; Soto-Jiménez, L.M.; Liu, C.; Lysak, M.A.; Novikova, P.Y.; Nordborg, M. Gradual evolution of allopolyploidy in Arabidopsis suecica. Nat. Ecol. Evol. 2021, 5, 1367–1381. [Google Scholar] [CrossRef] [PubMed]
- Trávníček, P.; Jersáková, J.; Kubátová, B.; Krejčíková, J.; Bateman, R.M.; Lučanová, M.; Krajníková, E.; Těšitelová, T.; Štípková, Z.; Amardeilh, J.P.; et al. Minority cytotypes in European populations of the Gymnadenia conopsea complex (Orchidaceae) greatly increase intraspecific and intrapopulation diversity. Ann. Bot. 2012, 110, 977–986. [Google Scholar] [CrossRef] [PubMed]
- Pavarese, G.; Tranchida Lombardo, V.; Galesi, R.; D’Emerico, S.; Casotti, R.; Cristaudo, A.; Cozzolino, S. When polyploidy and hybridization produce a fuzzy taxon: The complex origin of the insular neoendemic Neotinea commutata (Orchidaceae). Bot. J. Linn. Soc. 2013, 173, 707–720. [Google Scholar] [CrossRef]
- Gennaio, R.; Pellegrino, G. Serapias ausoniae (Orchidaceae; Orchideae): A new species from southern Italy confirmed by morphological, cytological and molecular analyses. Phytotaxa 2021, 516, 159–168. [Google Scholar] [CrossRef]
- Cozzolino, S.; Aceto, S.; Caputo, P.; Gaudio, L.; Nazzaro, R. Phylogenetic relationships in Orchis and some related genera: An approach using chloroplast DNA. Nord. J. Bot. 1997, 18, 79–87. [Google Scholar] [CrossRef]
- Bateman, R.M.; Pridgeon, A.M.; Chase, M.W. Phylogenetics of subtribe Orchidinae (Orchidoideae, Orchidaceae) based on nuclear ITS sequences. 2. Infrageneric relationships and reclassification to achieve monophyly of Orchis sensu stricto. Lindlejana 1997, 12, 113–141. [Google Scholar]
- Aceto, S.; Caputo, P.; Gaudio, L.; Nazzaro, R.; Cozzolino, S. Molecular approach to the identification and characterization of natural hybrids between Orchis pauciflora Ten. And 130 Orchis quadripunctata Cyr. ex Ten. (Orchidaceae). Bot. Helv. 2000, 110, 31–39. [Google Scholar]
- Pellegrino, G.; Cozzolino, S.; D’Emerico, S.; Grünanger, P. The taxonomic position of the controversial taxon Orchis clandestina (Orchidaceae): Karyomorphological and molecular analyses. Bot. Helv. 2000, 110, 101–107. [Google Scholar]
- D’Emerico, S.; Cozzolino, S.; Pellegrino, G.; Pignone, D.; Scrugli, A. Heterochromatin distribution in selected taxa of the 42-chromosomes Orchis s. l. (Orchidaceae). Caryologia 2002, 55, 55–62. [Google Scholar] [CrossRef]
- Cozzolino, S.; D’Emerico, S.; Widmer, A. Evidence for reproductive isolate selection in Mediterranean orchids: Karyotype differences compensate for the lack of pollinator specificity. Proc. R. Soc. Lond. B 2004, 271 (Suppl. S5), 259–262. [Google Scholar] [CrossRef]
- Pellegrino, G.; D’Emerico, S.; Musacchio, A.; Scrugli, A.; Cozzolino, S. Confirmation of hybridization among sympatric insular populations of Orchis mascula and O. provincialis. Plant Syst. Evol. 2005, 251, 131142. [Google Scholar] [CrossRef]
- Scopece, G.; Cozzolino, S.; Bateman, R.M. Just what is a genus? Comparing levels of postzygotic isolation to test alternative taxonomic hypotheses in Orchidaceae subtribe Orchidinae. Taxon 2010, 59, 1754–1764. [Google Scholar] [CrossRef]
- Kadri, T.; Fay, M.F.; Bateman, R.M. Little genetic differentiation across Europe between early-flowering and late-flowering populations of the rapidly declining orchid Neotinea ustulata. Biol. J. Linn. Soc. 2006, 87, 13–25. [Google Scholar]
- Hürkan, K.; Taşkin, K.M. Internal Transcribed Spacer (ITS) Fails Barcoding of the Genus Neotinea Rchb.f. (Orchidaceae). J. Agric. Sci. (Tarim Bilim. Derg.) 2021, 27, 69–75. [Google Scholar] [CrossRef]
- Battaglia, E. A simplified Feulgen method using cold hydrolysis. Caryologia 1957, 9, 372–373. [Google Scholar] [CrossRef]
- Perrino, R.; Pignone, D. Contribution to the taxonomy of Vicia species belonging to the section Faba. Kulturpflanze 1981, 29, 311–319. [Google Scholar] [CrossRef]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Paszko, B. A critical review and a new proposal of karyotype asymmetry indices. Plant Syst. Evol. 2006, 258, 39–48. [Google Scholar] [CrossRef]
- Peruzzi, L.; Eroğlu, H.E. Karyotype asymmetry: Again, how to measure and what to measure? Comp. Cytogen. 2013, 7, 1–9. [Google Scholar] [CrossRef]
- Delforge, P. Orchidés d’Europe, d’Afrique du Nord et do Proche-Orient, 4th ed.; Delachaux et Niestle: Paris, France, 2016. [Google Scholar]
- Plants of the World Online (POWO). Facilitated by the Royal Botanic Gardens, Kew 2023. Available online: http://www.plantsoftheworldonline.org/.3.2 (accessed on 20 July 2023).
- D’Emerico, S.; Bianco, P.; Medagli, P. Karyological studies on Orchidaceae. Tribe Ophrydeae, subtribe Serapiadinae. Caryologia 1992, 45, 301–311. [Google Scholar] [CrossRef]
- Mazzola, P. Cytogeographic aspects of Orchis commutata Tod. (Orchidaceae). Webbia 1984, 38, 773–779. [Google Scholar] [CrossRef]
- Wood, J. Orchis L. Distribution. In Genera Orchidacearum 2: Orchidoideae; Pridgeon, A.M., Cribb, P.J., Chase, M.C., Rasmussen, F.N., Eds.; Oxford University Press: Oxford, UK, 2001; Part 1; p. 333. [Google Scholar]
- D’Emerico, S.; Pignone, D.; Scrugli, A. Giemsa C-banded karyotypes in Serapias L. (Orchidaceae). Bot. J. Linn. Soc. 2000, 133, 485–492. [Google Scholar] [CrossRef]
- Schweizer, D.; Nagl, W. Heterochromatin diversity in Cymbidium, and its relationship to differential DNA replication. Exp. Cell Res. 1976, 98, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Linares, C.; Vega, C.; Ferrer, E.; Fominaya, A. Identification of C-banded chromosomes in meiosis and the analysis of nucleolar activity in Avena byzantina C. Koch cv ‘Kanota’. Theor. Appl. Genet. 1992, 83, 650–654. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, R.; Sasaki, T.; Ishikawa, R.; Osabe, K.; Kawanabe, T.; Dennis, E.S. Molecular Mechanisms of Epigenetic Variation in Plants. Int. J. Mol. Sci. 2012, 13, 9900–9922. [Google Scholar] [CrossRef]
- Seraj, R.G.M.; Tohidfar, M.; Piri, H. Plant Epigenetics: Mechanisms and Applications. J. Epigenetics 2019, 1, 24–34. [Google Scholar]
- Zhang, H.; Lang, Z.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Zhu, Q.; Cai, Z.; Tang, Q.; Jin, W. Repetitive sequence analysis and karyotyping reveal different genome evolution and speciation of diploid and tetraploid Tripsacum dactyloides. Crop J. 2016, 4, 247–255. [Google Scholar] [CrossRef]
- Li, S.F.; Zhang, G.J.; Yuan, J.H.; Deng, C.L.; Gao, W.J. Repetitive sequences and epigenetic modification: Inseparable partners play important roles in the evolution of plant sex chromosomes. Planta 2016, 243, 1083–1095. [Google Scholar] [CrossRef]
- Ayres-Alves, T.; Cardoso, A.L.; Nagamachi, C.Y.; de Sousa, L.M.; Pieczarka, J.C.; Noronha, R.C.R. Karyotypic evolution and chromosomal organization of repetitive DNA sequences in species of Panaque, Panaqolus, and Scobinancistrus (Siluriformes and Loricariidae) from the Amazon Basin. Zebrafish 2017, 14, 251–260. [Google Scholar] [CrossRef]
- Tang, S.J. New evidence for the theory of chromosome organization by repetitive elements (CORE). Genes 2017, 8, 81. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Pan, Y.J.; Zhao, X.Q.; Dwivedi, D.; Zhu, L.H.; Ali, J.; Fu, B.Y.; Li, Z.K. Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). J. Exper. Bot. 2011, 62, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Surdonja, K.; Eggert, K.; Hajirezaei, M.R.; Harshavardhan, V.; Seiler, C.; von Wirén, N.; Sreenivasulu, N.; Kuhlmann, M. Increase of DNA methylation at the HvCKX2.1 promoter by terminal drought stress in barley. Epigenomes 2017, 1, 9. [Google Scholar] [CrossRef]
- Forestan, C.; Farinati, S.; Rouster, J.; Lassagne, H.; Lauria, M.; Dal Ferro, N.; Varotto, S. Control of maize vegetative and reproductive development, fertility, and rRNAs silencing by histone deacetylase 108. Genetics 2018, 208, 1443–1466. [Google Scholar] [CrossRef] [PubMed]
- Begcy, K.; Dresselhaus, T. Epigenetic responses to abiotic stresses during reproductive development in cereals. Plant Reprod. 2018, 31, 343–355. [Google Scholar] [CrossRef]
- D’Emerico, S.; Grünanger, P. Giemsa C-banding in some Gymnadenia species and in Chamorchis alpina from the Dolomites (Italy). J. Eur. Orch. 2001, 33, 405–414. [Google Scholar]
- De Soó, R. Genus Dactylorhiza. In Flora Europaea; Tutin, T.G., Heywood, V.H., Burges, N.A., Moore, D.M., Valentine, D.H., Walters, S.M., Webb, D.A., Eds.; Cambridge University Press: Cambridge, UK, 1980; Volume 5. [Google Scholar]
- Moore, R.G. Flora Europaea. Check-List Chromosome Index; Cambridge University Press: Cambridge, UK, 1982. [Google Scholar]
- Scrugli, A. Numeri cromosomici per la flora italiana. Inform. Bot. Ital. 1980, 12, 149–153. [Google Scholar]
- Vöth, W.; Greilhuber, J. Zur Karyosystematik von Dactylorhiza maculata s.l. und ihrer Verbreitung, insbesondere in Niederösterreich. Linz. Biol. Beitr. 1980, 12, 415–468. [Google Scholar]
- D’Emerico, S.; Cozzolino, S.; Pellegrino, G.; Pignone, D.; Scrugli, A. Karyotype structure, supernumerary chromosomes and heterochromatin distribution suggest a pathway of karyotype evolution in Dactylorhiza (Orchidaceae). Bot. J. Linn. Soc. 2002, 138, 85–91. [Google Scholar] [CrossRef]
- Baumann, H.; D’Emerico, S.; Lorenz, R.; Pulvirenti, S. Supernumerary chromosomes and speciation processes in Dactylorhiza urvilleana subsp. phoenissa (Orchidaceae) from Lebanon. J. Eur. Orch. 2012, 44, 811–824. [Google Scholar]
- Teppner, H.; Klein, E. Nigritella rhellicani spec. nova und N. nigra (L.) Rchb. F. s. str. (Orchidaceae–Orchideae). Phyton 1990, 31, 5–26. [Google Scholar]
- Teppner, H.; Ster, T. Nigritella buschmanniae spec. nova (Orchidaceae–Orchideae) und eine Biographie für Frau Adolfine Buschmann. Phyton 1996, 36, 277–294. [Google Scholar]
- Teppner, H.; Klein, E. Etiam atque etiam—Nigritella versus Gymnadenia: Neukombinationen und Gymnadenia dolomitensis spec. nova (Orchidaceae-Orchideae). Phyton 1998, 38, 220–224. [Google Scholar]
- Teppner, H. Adventitions embryony in Nigritella (Orchidaceae). Folia Geobot. Phytot. 1996, 31, 323–331. [Google Scholar] [CrossRef]
- Cerbah, M.; Coulaud, J.; Siljak-Yakovlev, S. rDNA organization and evolutionary relationships in the genus Hypochaeris (Asteraceae). J. Hered. 1998, 89, 312–318. [Google Scholar] [CrossRef]
- Jiang, J. Fluorescence in situ hybridization in plants: Recent developments and future applications. Chromosome Res 2019, 27, 153–165. [Google Scholar] [CrossRef]







| Taxon | Provenance | No. Plants | 2n | No. of Plants with B-Chromosome |
|---|---|---|---|---|
| Neotinea commutata (Tod.) R.M. Bateman | Italy | 8 | 84 | |
| N. lactea (Poir.) R.M. Bateman, Pridgeon & M.W. Chase | Italy | 12 | 42 42+1B | 1 |
| N. maculata (Desf.) Stearn | Italy | 4 | 42 | |
| N. tridentata (Scop.) R.M. Bateman, Pridgeon & M.W. Chase | Italy | 15 | 42 42+1B | 1 |
| N. ustulata (L.) R.M. Bateman, Pridgeon & M.W. Chase | Italy | 9 | 42 42+1B | 1 |
| Orchis anthropophora (L.) All. | Italy | 5 | 42 | |
| O. italica Poir. | Italy | 12 | 42 42+1B | 1 |
| O. mascula (L.) L. | Italy | 10 | 42 42+1B | 2 |
| O. patens Desf. | Italy | 2 | 84 | |
| O. pauciflora Ten. | Italy | 5 | 42 | |
| O. provincialis Balb. ex Lam. & DC. | Italy | 10 | 42 | |
| O. purpurea Huds. | Italy | 5 | 42 | |
| Platanthera chlorantha (Custer) Rchb. | Italy | 5 | 42 | |
| Dactylorhiza romana (Sebast.) Soó | Italy | 15 | 40 40+1B, 40+2B, 40+3B | 3 |
| D. phoenissa (B. Baumann & H. Baumann) P. Delforge | Lebanon | 3 | 80 80+4B | 1 |
| D. saccifera (Brogniart) Soó | Italy | 5 | 40 | |
| D. sambucina (L.) Soó | Italy | 10 | 40 | |
| Gymnadenia austriaca (Teppner & E. Klein) P. Delforge | Italy | 3 | 80 | |
| G. conopsea (L.) R.Br. | Italy | 2 | 40 | |
| G. rhellicani Teppner & E. Klein | Italy | 2 | 40 |
| Taxon | Code | Formula | THL | MCA | CVCL | CVCI |
|---|---|---|---|---|---|---|
| Neotinea lactea (Poir.) R.M. Bateman, Pridgeon & M.W. Chase | Nla | 16m+26sm | 61.72 | 26.82 | 23.73 | 14.91 |
| N. tridentata (Scop.) R.M. Bateman, Pridgeon & M.W. Chase | Ntr | 24m+18sm | 39.57 | 24.52 | 35.49 | 16.46 |
| N. ustulata (L.) R.M. Bateman, Pridgeon & M.W. Chase | Nus | 28m+14sm | 42.89 | 20.38 | 24.63 | 14.38 |
| Orchis anthropophora (L.) All. | Oan | 36m+6sm | 32.33 | 14.47 | 22.65 | 11.88 |
| O. italica Poir. | Oit | 28m+14sm | 40.43 | 23.43 | 25.01 | 15.44 |
| O. mascula (L.) L. | Oma | 28m+14sm | 46.60 | 21.41 | 32.67 | 15.40 |
| O. provincialis Balb. ex Lam. & DC. | Opr | 10m+24sm+8st | 44.43 | 33.75 | 35.27 | 25.53 |
| O. purpurea Huds. | Opu | 34m+8sm | 51.72 | 20.47 | 26.36 | 12.29 |
| Platanthera chlorantha (Custer) Rchb. | Pch | 30m+12sm | 58.23 | 19.59 | 21.25 | 11.75 |
| Chamorchis alpina (L.) Rich. | Cal | 24m+18sm | 46.48 | 26.22 | 20.73 | 18.30 |
| Dactylorhiza romana (Sebast.) Soó | Dro | 14m+20sm+6st | 44.50 | 31.08 | 29.65 | 20.22 |
| D. saccifera (Brogniart) Soó | Dsa | 36m+4sm | 46.78 | 16.50 | 15.18 | 07.69 |
| Gymnadenia conopsea (L.) R.Br. | Gco | 22m+12sm+6st | 44.93 | 27.77 | 24.96 | 25.41 |
| G. rhellicani Teppner & E. Klein | Grh | 14m+26sm | 35.23 | 30.37 | 18.90 | 19.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turco, A.; Wagensommer, R.P.; Medagli, P.; Albano, A.; D’Emerico, S. Advances in the Study of Orchidinae Subtribe (Orchidaceae) Species with 40,42-Chromosomes in the Mediterranean Region. Diversity 2024, 16, 41. https://doi.org/10.3390/d16010041
Turco A, Wagensommer RP, Medagli P, Albano A, D’Emerico S. Advances in the Study of Orchidinae Subtribe (Orchidaceae) Species with 40,42-Chromosomes in the Mediterranean Region. Diversity. 2024; 16(1):41. https://doi.org/10.3390/d16010041
Chicago/Turabian StyleTurco, Alessio, Robert Philipp Wagensommer, Pietro Medagli, Antonella Albano, and Saverio D’Emerico. 2024. "Advances in the Study of Orchidinae Subtribe (Orchidaceae) Species with 40,42-Chromosomes in the Mediterranean Region" Diversity 16, no. 1: 41. https://doi.org/10.3390/d16010041
APA StyleTurco, A., Wagensommer, R. P., Medagli, P., Albano, A., & D’Emerico, S. (2024). Advances in the Study of Orchidinae Subtribe (Orchidaceae) Species with 40,42-Chromosomes in the Mediterranean Region. Diversity, 16(1), 41. https://doi.org/10.3390/d16010041









