Exploring Genetic and Morphological Integrity across Ocean Basins: A Case Study of the Mesopelagic Shrimp Systellaspis debilis (Decapoda: Oplophoridae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Material
2.2. DNA Extraction, Amplification, and Sequencing
2.3. Sequence Alignment and Phylogenetic Analyses
2.4. Morphological Analysis
3. Results
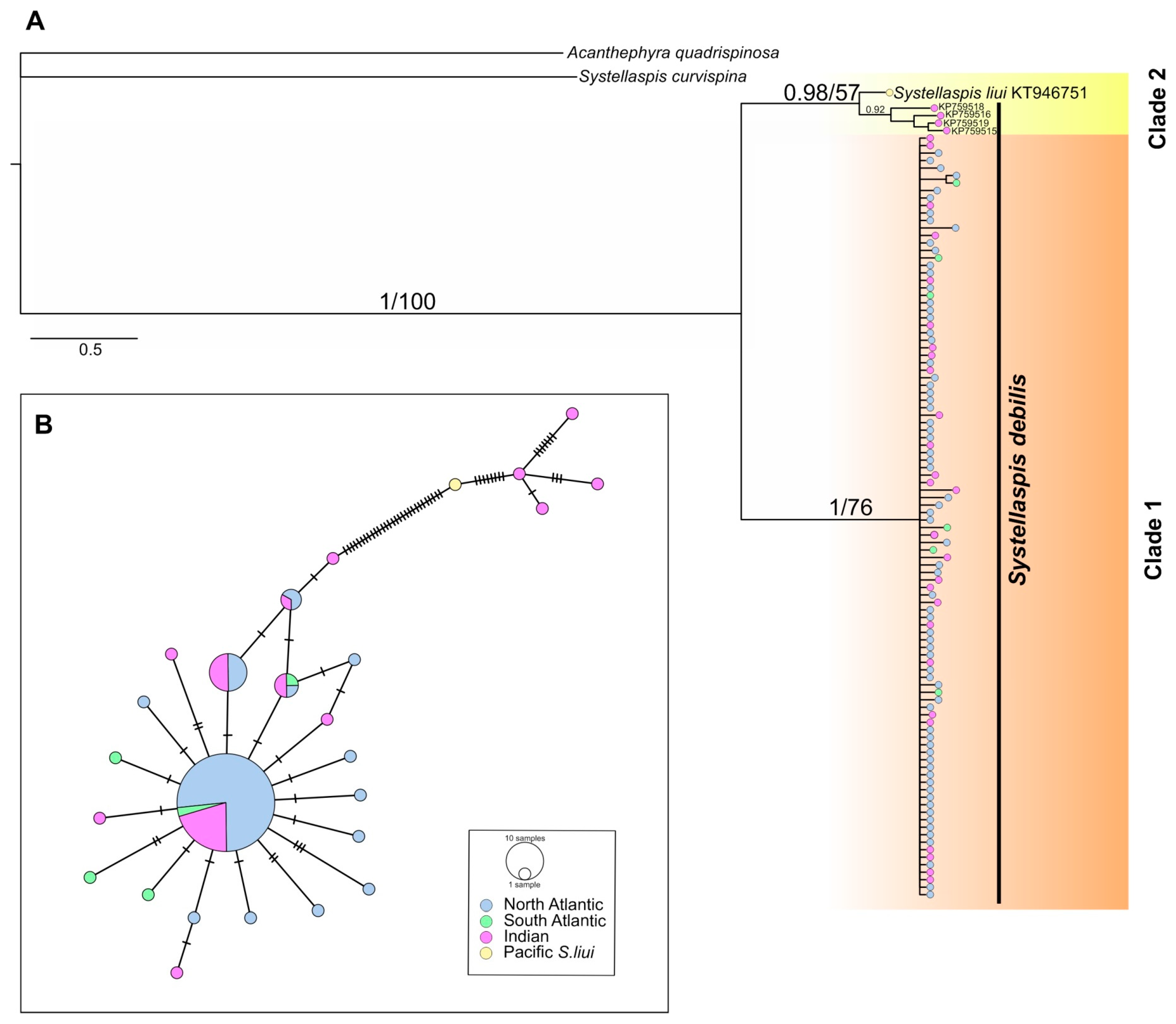
3.1. Genetic Variability and Spatial Structure
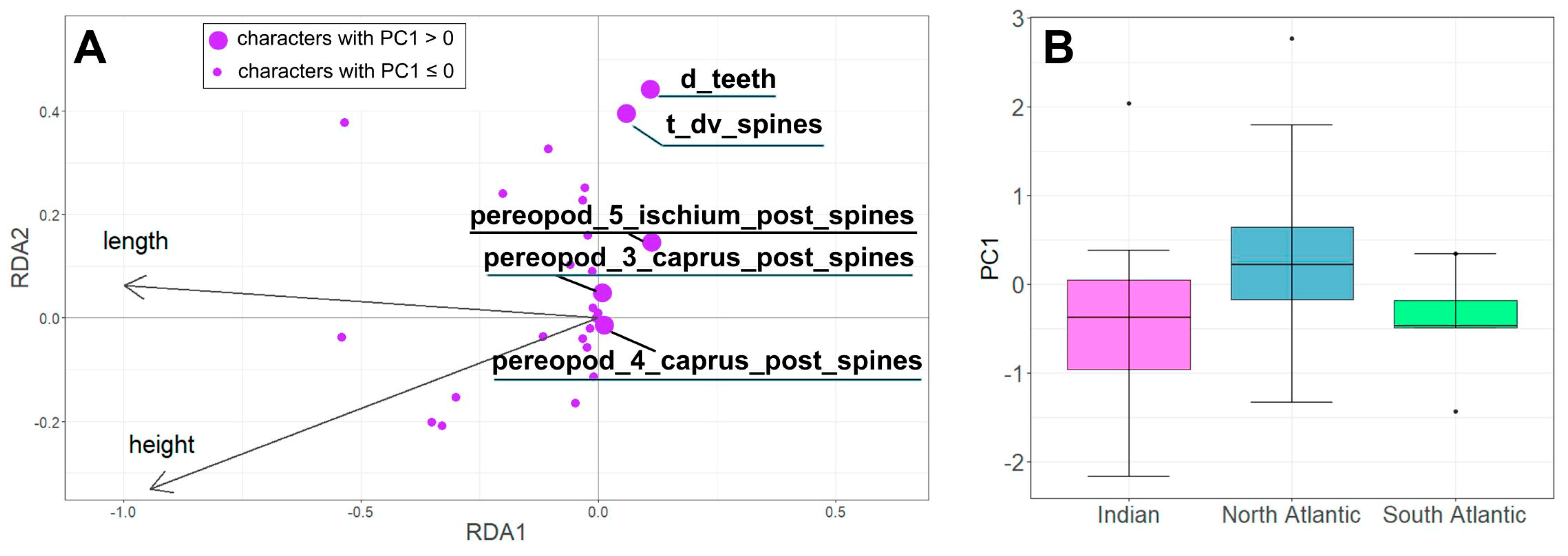
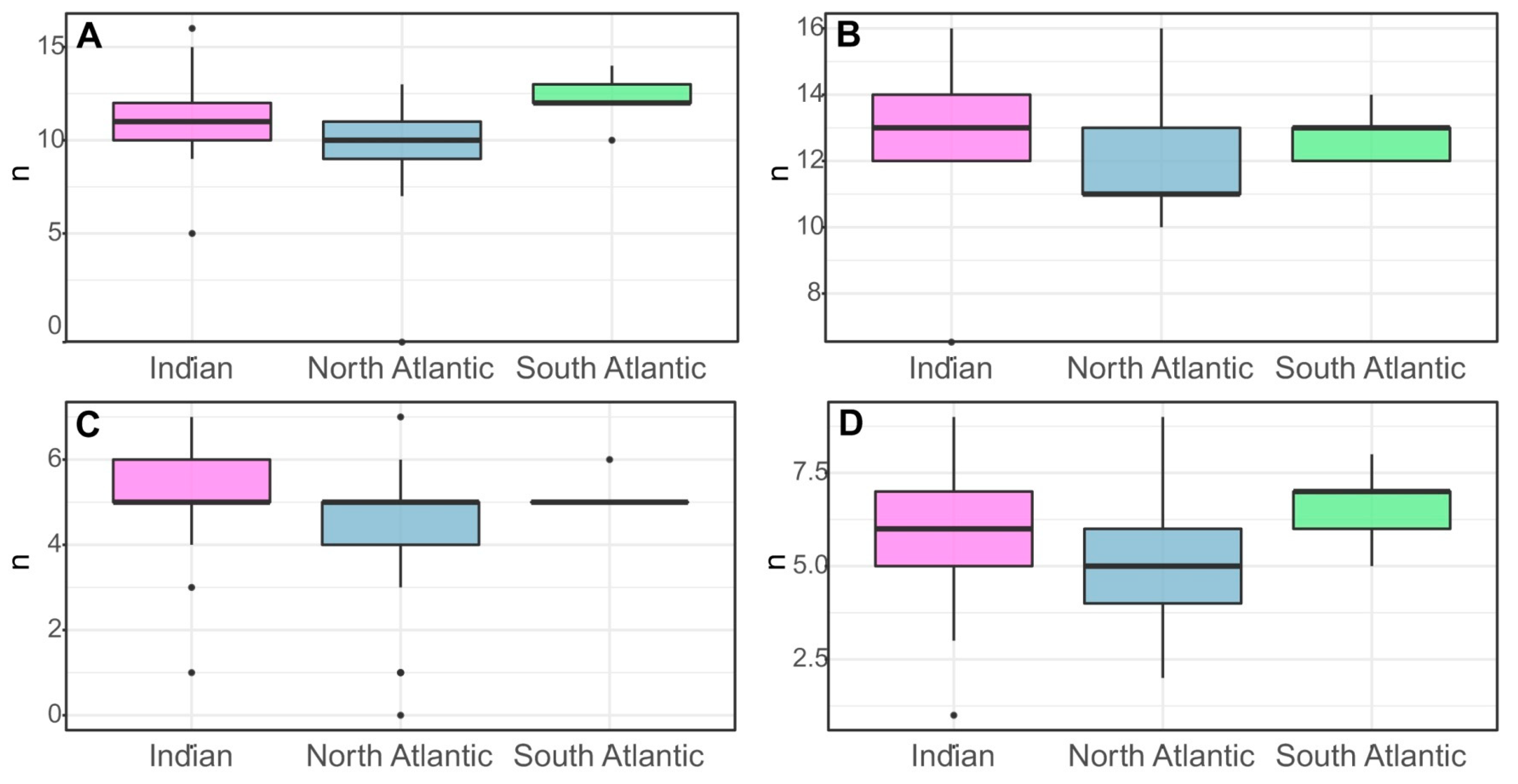
3.2. Morphological Variability
4. Discussion
4.1. Population Structure of Systellaspis debilis (Clade 1)
4.2. Morphological Variability of Systellaspis debilis (Clade 1)
4.3. The status of Systellaspis liui and Related Specimens (Clade 2)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Webb, T.J.; Berghe, E.V.; O’Dor, R. Biodiversity’s Big Wet Secret: The global distribution of marine biological records reveals chronic under-exploration of the deep pelagic ocean. PLoS ONE 2010, 5, 1–6. [Google Scholar] [CrossRef]
- Sutton, T.T.; Clark, M.R.; Dunn, D.C.; Halpin, P.N.; Rogers, A.D.; Guinotte, J.; Bograd, S.J.; Heino, M. A global biogeographic classification of the mesopelagic zone. Deep-Sea Res. I Oceanogr. Res. Pap. 2017, 126, 85–102. [Google Scholar] [CrossRef]
- Sarmiento, J.L.; Slater, R.; Barber, R.; Bopp, L.; Doney, S.C.; Hirst, A.C.; Kleypas, J.; Stouffer, R. Response of Ocean Ecosystems to Climate Warming. Glob. Biogeochem. Cycles 2004, 18, GB3003. [Google Scholar] [CrossRef]
- Mengerink, K.J.; Van Dover, C.L.; Ardron, J.; Baker, M.; Escobar-Briones, E.; Gjerde, K.; Koslow, J.A.; Levin, L.A. A call for deep-ocean stewardship. Science 2014, 344, 696–698. [Google Scholar] [CrossRef]
- Mackas, D.L.; Beaugrand, G. Comparisons of zooplankton time series. J. Mar. Syst. 2010, 79, 286–304. [Google Scholar] [CrossRef]
- Vereshchaka, A.L.; Mikaelyan, A.S.; Piontkovski, S.A.; Lunina, A.A. A mesoplankton biomass decline in the Central Atlantic coupled with an increase of surface temperature and an expansion of low-productive zones. Glob. Ecol. Biogeogr. 2023, 32, 1365–1376. [Google Scholar] [CrossRef]
- Palumbi, S.R. Genetic divergence, reproductive isolation, and marine speciation. Annu. Rev. Ecol. Evol. Syst. 1994, 25, 547–572. [Google Scholar] [CrossRef]
- Norton, E.L.; Goetze, E. Equatorial dispersal barriers and limited population connectivity among oceans in a planktonic copepod. Limnol. Oceanogr. 2013, 58, 1581–1596. [Google Scholar] [CrossRef]
- Kulagin, D.N.; Stupnikova, A.N.; Neretina, T.V.; Mugue, N.S. Spatial genetic heterogeneity of the cosmopolitan chaetognath Eukrohnia hamata (Möbius, 1875) revealed by mitochondrial DNA. Hydrobiologia 2014, 721, 197–207. [Google Scholar] [CrossRef]
- Deagle, B.E.; Faux, C.; Kawaguchi, S.; Meyer, B.; Jarman, S.N. Antarctic krill population genomics: Apparent panmixia, but genome complexity and large population size muddy the water. Mol. Ecol. 2015, 24, 4943–4959. [Google Scholar] [CrossRef]
- Timm, L.E.; Isma, L.M.; Johnston, M.W.; Bracken-Grissom, H.D. Comparative population genomics and biophysical modeling of shrimp migration in the Gulf of Mexico reveals current-mediated connectivity. Front. Mar. Sci. 2020, 7, 19. [Google Scholar] [CrossRef]
- Burridge, A.K.; Goetze, E.; Raes, N.; Huisman, J.; Peijnenburg, K.T.C.A. Global biogeography and evolution of cuvierina pteropods phylogenetics and phylogeography. BMC Evol. Biol. 2015, 15, 39. [Google Scholar] [CrossRef]
- Miyamoto, H.; Machida, R.J.; Nishida, S. “Complete mitochondrial genome sequences of the three pelagic chaetognaths Sagitta nagae, Sagitta decipiens and Sagitta enflata. Comp. Biochem. Physiol. Part D Genom. Proteom. 2010, 5, 65–72. [Google Scholar] [CrossRef]
- Yebra, L.; Bonnet, D.; Harris, R.P.; Lindeque, P.K.; Peijnenburg, K.T.C.A. Barriers in the Pelagic: Population structuring of Calanus helgolandicus and C. euxinus in European waters. Mar. Ecol. Prog. Ser. 2011, 428, 135–149. [Google Scholar] [CrossRef]
- Kulagin, D.N.; Lunina, A.A.; Simakova, U.V.; Vereshchaka, A.L. Progressing diversification and biogeography of the mesopelagic Nematoscelis (Crustacea: Euphausiacea) in the Atlantic”. ICES 2021, 78, 3457–3463. [Google Scholar] [CrossRef]
- Blanco-Bercial, L.; Álvarez-Marqués, F.; Bucklin, A. Comparative phylogeography and connectivity of sibling species of the marine copepod Clausocalanus (Calanoida). J. Exp. Mar. Biol. Ecol. 2011, 404, 108–115. [Google Scholar] [CrossRef]
- Churchill, C.K.C.; Valdés, Á.; Foighil, D.Ó. Molecular and morphological systematics of neustonic nudibranchs (Mollusca: Gastropoda: Glaucidae: Glaucus), with descriptions of three new cryptic species. Invertebr. Syst. 2014, 28, 174–195. [Google Scholar] [CrossRef]
- Churchill, C.K.C.; Valdés, Á.; Foighil, D.Ó. Afro-Eurasia and the Americas present barriers to gene flow for the cosmopolitan neustonic nudibranch Glaucus atlanticus. Mar. Biol. 2014, 161, 899–910. [Google Scholar] [CrossRef]
- Andrews, K.R.; Norton, E.L.; Fernandez-Silva, I.; Portner, E.; Goetze, E. Multilocus evidence for globally distributed cryptic species and distinct populations across ocean gyres in a mesopelagic copepod. Mol. Ecol. 2014, 23, 5462–5479. [Google Scholar] [CrossRef] [PubMed]
- Goetze, E.; Hüdepohl, P.T.; Chang, C.; Van Woudenberg, L.; Iacchei, M.; Peijnenburg, K.T.C.A. Ecological dispersal barrier across the Equatorial Atlantic in a migratory planktonic copepod. Prog. Oceanogr. 2017, 158, 203–212. [Google Scholar] [CrossRef]
- Kulagin, D.N.; Neretina, T.V. Genetic and morphological diversity of the cosmopolitan chaetognath Pseudosagitta maxima (Conant, 1896) in the Atlantic Ocean and its relationship with the congeneric species. ICES 2017, 74, 1875–1884. [Google Scholar] [CrossRef]
- Choo, L.Q.; Bal, T.M.P.; Goetze, E.; Peijnenburg, K.T.C.A. Oceanic dispersal barriers in a holoplanktonic gastropod. J. Evol. Biol. 2021, 34, 224–240. [Google Scholar] [CrossRef]
- Bucklin, A.; DiVito, K.R.; Smolina, I.; Choquet, M.; Questel, J.M.; Hoarau, G.; O’Neill, R.J. Population Genomics of Marine Zooplankton. In Population Genomics: Marine Organisms; Oleksiak, M., Rajora, O., Eds.; Springer: Cham, Switzerland, 2018; pp. 61–102. [Google Scholar] [CrossRef]
- Burukovsky, R.N.O. Biologii krevetki Systellaspis debilis (A. Milne Edwards, 1881) (Decapoda, Natantia, Oplophoridae). Bul. MOIP. Otd. boil 1992, 97, 60–70. (In Russian) [Google Scholar]
- Judkins, D.C. Geographical distribution of pelagic decapod shrimp in the Atlantic Ocean. Zootaxa 2014, 3895, 301–345. [Google Scholar] [CrossRef]
- Vereshchaka, A.; Abyzova, G.; Lunina, A.; Musaeva, E. The deep-sea zooplankton of the North, Central, and South Atlantic: Biomass, abundance, diversity. Deep Sea Res. Part II Top. Stud. Oceanogr. 2017, 137, 89–101. [Google Scholar] [CrossRef]
- Iwasaki, N.; Nemoto, T. Pelagics (Crustacea: Decapoda) from the Southern Ocean between 150° E and 115° E. Mem. Natl. Inst. Polar. Res. Ser. 1987, 38, 16–18. [Google Scholar]
- Lunina, A.A.; Kulagin, D.N.; Vereshchaka, A.L. Oplophoridae (Decapoda: Crustacea): Phylogeny, taxonomy and evolution studied by a combination of morphological and molecular methods. Zool. J. Linn. Soc. 2019, 186, 213–232. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenkoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Wormhoudt, A.V.; Adjeroud, M.; Rouzé, H.; Leray, M. Recent and old duplications in crustaceans “Internal Transcribed Spacer 1 ″: Structural and phylogenetic implications. Mol. Biol. Rep. 2019, 46, 5185–5195. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Ostell, J.; Pruitt, K.D.; Sayers, E.W. GenBank. Nucleic Acids Res. 2018, 46, D41–D47. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Sys. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Team, R.C. R: A language and environment for statistical computing. Suppl. Inf. Ref. S 2021, 1, 371–378. [Google Scholar]
- Legendre, P.; Legendre, L. Numerical Ecology; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; Wagner, H. Vegan: Community Ecology Package. Ordination Methods, Diversity Analysis and Other Functions for Community and Vegetation Ecologists; R Package Version. 2.5-7. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 25 April 2023).
- Charif, D.; Lobry, J.R. SeqinR 1.0-2: A contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In Structural Approaches to Sequence Evolution; Springer: Berlin/Heidelberg, Germany, 2007; pp. 207–232. [Google Scholar]
- Wickham, H.; Chang, W.; Wickham, M.H. Package ‘ggplot2′. Create Elegant data Visualisations Using the Grammar of Graphics; Version 2(1); Springer: New York, NY, USA, 2016; pp. 1–189. [Google Scholar]
- Eberl, R.; Cohen, S.; Cipriano, F.; Carpenter, E.J. Genetic diversity of the pelagic harpacticoid copepod Macrosetella gracilis on colonies of the cyanobacterium Trichodesmium spp. Aquatic Biology 2007, 1, 33–43. [Google Scholar] [CrossRef]
- Hirai, J.; Tsuda, A.; Goetze, E. Extensive genetic diversity and endemism across the global range of the oceanic copepod Pleuromamma abdominalis. Prog. Oceanogr. 2015, 138, 77–90. [Google Scholar] [CrossRef]
- Roe, H.S.J.; Angel, M.V.; Badcock, J.; Domanski, P.; James, P.T.; Pugh, P.R.; Thurston, M.H. The diel migrations and distributions within a mesopelagic community in the North East Atlantic. 1. Introduction and sampling procedures. Prog. Oceanogr 1984, 13, 3–4. [Google Scholar] [CrossRef]
- Omori, M. The biology of pelagic shrimps in the ocean. Adv. Mar. Biol. 1975, 12, 233–324. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Hughes, A.L. Evidence for abundant slightly deleterious polymorphisms in bacterial populations. Genetics 2005, 169, 533–538. [Google Scholar] [CrossRef]
- Ely, B.; Viñas, J.; Bremer, J.R.A.; Black, D.; Lucas, L.; Covello, K.; Labrie, A.V.; Thelen, E. Consequences of the historical demography on the global population structure of two highly migratory cosmopolitan marine fishes: The yellowfin tuna (Thunnus albacares) and the skipjack tuna (Katsuwonus pelamis). BMC Evol. Biol. 2005, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Hoelzel, A.R.; Shivji, M.S.; Magnussen, J.; Francis, M.P. Low worldwide genetic diversity in the basking shark (Cetorhinus maximus). Biol. Lett. 2006, 2, 639–642. [Google Scholar] [CrossRef] [PubMed]
- Haro-Bilbao, I.; Riginos, C.; Baldwin, J.D.; Zischke, M.; Tibbetts, I.R.; Thia, J.A. Global connections with some genomic differentiation occur between Indo-Pacific and Atlantic Ocean wahoo, a large circumtropical pelagic fish. J. Biogeogr. 2021, 48, 2053–2067. [Google Scholar] [CrossRef]
- Papetti, C.; Zane, L.; Bortolotto, E.; Bucklin, A.; Patarnello, T. Genetic differentiation and local temporal stability of population structure in the euphausiid Meganyctiphanes norvegica. Mar. Ecol. Prog. Ser. 2005, 289, 225–235. [Google Scholar] [CrossRef]
- Peijnenburg, K.T.C.A.; Fauvelot, C.; Breeuwer, J.A.J.; Menken, S.B.J. Spatial and temporal genetic structure of the planktonic Sagitta setosa (Chaetognatha) in European seas as revealed by mitochondrial and nuclear DNA markers. Mol. Ecol. 2006, 15, 3319–3338. [Google Scholar] [CrossRef]
- Bucklin, A.; Wiebe, P.H. Low mitochondrial diversity and small effective population sizes of the copepods Calanus finmarchicus and Nannocalanus minor: Possible impact of climatic variation during recent glaciation. J. Hered. 1998, 89, 383–392. [Google Scholar] [CrossRef]
- Peijnenburg, K.T.C.A.; van Haastrecht, E.K.; Fauvelot, C. Present-day genetic composition suggests contrasting demographic histories of two dominant chaetognaths of the North-East Atlantic, Sagitta elegans and S. setosa. Mar. Biol. 2005, 147, 1279–1289. [Google Scholar] [CrossRef]
- Villar, E.; Farrant, G.K.; Follows, M.; Garczarek, L.; Speich, S.; Audic, S. Lucie Bittner. Environmental characteristics of Agulhas rings affect interocean plankton transport. Science. 2015, 348, 1261447. [Google Scholar] [CrossRef]
- Crosnier, A. Oplophoridae (Crustacea Decapoda) récoltés de 1971 à 1982 par les navires français dans Lócean Indien occidental sud. Bull. Mus. Natl. Hist. Nat. 1987, 9, 695–726. [Google Scholar]
- Burridge, A.K.; Van Der Hulst, R.; Goetze, E.; Peijnenburg, K.T.C.A. Assessing species boundaries in the open sea: An integrative taxonomic approach to the pteropod genus Diacavolinia. Zool. J. Linn. Soc. 2019, 187, 16–40. [Google Scholar] [CrossRef]
- Aznar-Cormano, L.; Brisset, J.; Chan, T.Y.; Corbari, L.; Puillandre, N.; Utge, J.; Zbinden, M.; Zuccon, D.; Samadi, S. An improved taxonomic sampling is a necessary but not sufficient condition for resolving inter-families relationships in Caridean decapods. Genetica 2015, 143, 195–205. [Google Scholar] [CrossRef]
- Sha, Z.; Wang, Y. A new deep-sea species of the genus Systellaspis (Decapoda, Caridea, Oplophoridae) in the Western Pacific. Crustaceana 2015, 88, 1181–1192. [Google Scholar] [CrossRef]
- Iacchei, M.; Gaither, M.R.; Bowen, B.W.; Toonen, R.J. Testing dispersal limits in the sea: Range-wide phylogeography of the pronghorn spiny lobster Panulirus penicillatus. J. Biogeogr. 2016, 43, 1032–1044. [Google Scholar] [CrossRef]
- Ketmaier, V.; Argano, R.; Caccone, A. Phylogeography and molecular rates of subterranean aquatic stenasellid isopods with a peri-tyrrhenian distribution. Mol. Ecol. 2003, 12, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Knowlton, N.; Weigt, L.A. New dates and new rates for divergence across the Isthmus of Panama. Proc. R. Soc. B Biol. Sci. 1998, 265, 2257–2263. [Google Scholar] [CrossRef]
- Morrison, C.L.; Ríos, R.; Duffy, J.E. Phylogenetic evidence for an ancient rapid radiation of Caribbean sponge-dwelling snapping shrimps (Synalpheus). Mol. Phylogenet. Evol. 2004, 30, 381–563. [Google Scholar] [CrossRef]
- Quan, J.; Zhuang, Z.; Deng, J.; Dai, J.; Zhang, Y.P. Phylogenetic relationships of 12 Penaeoidea shrimp species deduced from mitochondrial DNA sequences. Biochem. Genet. 2004, 42, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.J.; Creer, S.; Dos Santos, A.; Costa, A.C.; Cunha, M.R.; Costa, F.O.; Carvalho, G.R. Systematic and evolutionary insights derived from mtDNA COI barcode diversity in the Decapoda (Crustacea: Malacostraca). PLoS ONE 2011, 6, e19449. [Google Scholar] [CrossRef]
- Dudoit, A.A.; Iacchei, M.; Coleman, R.R.; Gaither, M.R.; Browne, W.E.; Bowen, B.W.; Toonen, R.J. The little shrimp that could: Phylogeography of the circumtropical Stenopus hispidus (Crustacea: Decapoda), reveals divergent Atlantic and Pacific lineages. PeerJ 2018, 6, e4409. [Google Scholar] [CrossRef]
- Vereshchaka, A.; Kulagin, D.; Lunina, A. Discovery of a new species provides a deeper insight into taxonomic grouping of the deep-sea genus Acanthephyra (Crustacea: Decapoda). Diversity 2022, 14, 907. [Google Scholar] [CrossRef]
- DeHart, H.M.; Blanco-Bercial, L.; Passacantando, M.; Questel, J.M.; Bucklin, A. Pathways of pelagic connectivity: Eukrohnia hamata (Chaetognatha) in the Arctic Ocean. Front. Mar. Sci. 2020, 7, 396. [Google Scholar] [CrossRef]


| # | Character Description | Abbreviation | Unit of Measure | North Atlantic | South Atlantic | Indian | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CARAPACE | min | max | Average ± SD | min | max | Average ± SD | min | max | Average ± SD | |||
| 1 | Carapace height | CH | mm | 2.5 | 9 | 6.2 ± 1.94 | 7 | 9 | 7.7 ± 0.84 | 2 | 9.0 | 5.8 ± 2.05 |
| 2 | Carapace length | CL | mm | 4 | 14 | 10.2 ± 2.56 | 11 | 14 | 12.1 ± 1.34 | 3.5 | 14.0 | 9.8 ± 2.97 |
| 3 | Dorsal teeth | DT | n | 10 | 19 | 14.4 ± 1.52 | 12 | 16 | 14.4 ± 1.52 | 13 | 24.0 | 14.6 ± 2.28 |
| 4 | Postorbital dorsal teeth | PDT | n | 0 | 4 | 2.6 ± 0.73 | 2 | 3 | 2.6 ± 0.55 | 2 | 6.0 | 2.8 ± 1.01 |
| 5 | Ventral teeth | VT | n | 0 | 13 | 8.6 ± 1.95 | 7 | 9 | 8.4 ± 0.89 | 7 | 11.0 | 8.5 ± 1.16 |
| PLEON | ||||||||||||
| 6 | Third pleonic somite. Dorsal carina | Carina | +/− | + | + | + | ||||||
| 7 | Fourth pleon. Serrations on lateral margin-right side | 4_som_ser_r | n | 1 | 8 | 5.2 ± 1.79 | 5 | 8 | 6.8 ± 1.3 | 1 | 9.0 | 5.2 ± 2.34 |
| 8 | Fourth pleonic serrations on lateral margin-left side | 4_som_ser_l | n | 1 | 9 | 5 ± 1.84 | 5 | 8 | 6.6 ± 1.14 | 1 | 9.0 | 5.2 ± 2.35 |
| 9 | Fifth pleonic serrations on lateral margin-right side | 5_som_ser_r | n | 0 | 6 | 3.6 ± 1.17 | 4 | 5 | 4.6 ± 0.55 | 1 | 5.0 | 3.5 ± 1.32 |
| 10 | Fifth pleonic. serrations on lateral margin-left side | 5_som_ser_l | n | 1 | 5 | 3.4 ± 1.06 | 3 | 6 | 4.6 ± 1.14 | 1 | 5.0 | 3.3 ± 1.27 |
| 11 | Fifth pleonic somite. Sharp tooth on posterior margin of pleuron-left side | 5_pleur_tooth_l | −/+ | + | + | + | ||||||
| 12 | Fifth pleonic somite. Sharp tooth on posterior margin of pleuron-right side | 5_pleur_tooth_r | −/+ | + | + | + | ||||||
| TELSON | ||||||||||||
| 13 | Telson. Pairs of dorsolateral spines | t_dv_spines | n | 3 | 11 | 5.4 ± 1.51 | 5 | 5 | 5 ± 0 | 4 | 6.0 | 5 ± 0.42 |
| 14 | Telson. Numerous lateral spines arranged in two or more rows | t_lat_spines | −/+ | - | - | - | ||||||
| ANTENNA | ||||||||||||
| 15 | Scaphocerite. Medial dorsal groove | scaph | −/+ | + | + | + | ||||||
| PEREOPODS | ||||||||||||
| 16 | Third pereopod. Ischium. Anterior row of spines. movable spines | 3_pereopod_ischium_ant_spines | n | 0 | 0 | 0 ± 0 | 0 | 0 | 0 ± 0 | 0 | 0.0 | 0 ± 0 |
| 17 | Third pereopod. Ischium. Posterior row of movable spines, number of spines | 3_pereopod_ischium_post_spines | n | 2 | 6 | 3.3 ± 0.66 | 3 | 4 | 3.8 ± 0.45 | 3 | 4.0 | 3.3 ± 0.44 |
| 18 | Third pereopod. Merus. Anterior row of movable spines, number of spines | 3_pereopod_merus_ant_spines | n | 0 | 5 | 1.2 ± 0.71 | 1 | 2 | 1.8 ± 0.45 | 1 | 3.0 | 1.3 ± 0.53 |
| 19 | Third pereopod. Merus. Posterior row of movable spines, number of spines | 3_pereopod_merus_post_spines | n | 10 | 16 | 11.8 ± 1.64 | 12 | 14 | 12.8 ± 0.84 | 5 | 16.0 | 12.4 ± 2.55 |
| 20 | Third pereopod. Carpus. Anterior row of movable spines, number of spines | 3_pereopod_carpus _ant_spines | n | 0 | 1 | 0 ± 0.15 | 0 | 0 | 0 ± 0 | 0 | 0.0 | 0 ± 0 |
| 21 | Third pereopod. Carpus. Posterior row of movable spines, number of spines | 3_pereopod_carpus _post_spines | n | 0 | 1 | 1 ± 0.15 | 1 | 1 | 1 ± 0 | 1 | 1.0 | 1.0 ± 0 |
| 22 | Forth pereopod. Ischium. Anterior row of movable spines, number of spines | 4_pereopod_ischium_ant_spines | n | 0 | 3 | 1.1 ± 0.72 | 0 | 2 | 1 ± 0.71 | 0 | 5.0 | 1.4 ± 1.02 |
| 23 | Forth pereopod. Ischium. Posterior row of movable spines, number of spines | 4_pereopod_ischium_post_spines | n | 0 | 5 | 3.5 ± 1 | 3 | 6 | 4.4 ± 1.14 | 3 | 5.0 | 3.6 ± 0.72 |
| 24 | Forth pereopod. Merus. Anterior row of movable spines, number of spines | 4_pereopod_merus_ant_spines | n | 0 | 7 | 4.6 ± 1.44 | 5 | 6 | 5.2 ± 0.45 | 1 | 7.0 | 4.8 ± 1.85 |
| 25 | Forth pereopod. Merus. Posterior row of movable spines, number of spines | 4_pereopod_merus_post_spines | n | 0 | 13 | 9.8 ± 2 | 10 | 14 | 12.2 ± 1.48 | 4 | 16.0 | 10.4 ± 2.96 |
| 26 | Forth pereopod. Carpus. Anterior row of movable spines, number of spines | 4_pereopod_carpus _ant_spines | n | 0 | 1 | 0 ± 0.15 | 0 | 1 | 0.2 ± 0.45 | 0 | 1.0 | 0.0 ± 0.2 |
| 27 | Forth pereopod. Carpus. Posterior row of movable spines, number of spines | 4_pereopod_carpus _post_spines | n | 0 | 1 | 1 ± 0.21 | 0 | 1 | 0.8 ± 0.45 | 1 | 1.0 | 1.0 ± 0 |
| 28 | Fifth pereopod. Ischium. Anterior row of movable spines, number of spines | 5_pereopod_ischium_ant_spines | n | 0 | 1 | 0.4 ± 0.5 | 0 | 1 | 0.6 ± 0.55 | 0 | 1.0 | 0.5 ± 0.51 |
| 29 | Fifth pereopod. Ischium. Posterior row of movable spines, number of spines | 5_pereopod_ischium_post_spines | n | 0 | 3 | 1.2 ± 0.52 | 1 | 1 | 1 ± 0 | 1 | 2.0 | 1.1 ± 0.34 |
| 30 | Fifth pereopod. Merus. Anterior row of movable spines, number of spines | 5_pereopod_merus_ant_spines | n | 0 | 5 | 2 ± 0.89 | 3 | 3 | 3 ± 0 | 1 | 3.0 | 2.2 ± 0.83 |
| 31 | Fifth pereopod. Merus. Posterior row of movable spines, number of spines | 5_pereopod_merus_post_spines | n | 2 | 6 | 4.3 ± 0.83 | 2 | 5 | 4.2 ± 1.3 | 1 | 8.0 | 4.3 ± 1.39 |
| 32 | Fifth pereopod. Carpus. Anterior row of movable spines, number of spines | 5_pereopod_carpus _ant_spines | n | 0 | 4 | 1 ± 0.51 | 1 | 1 | 1 ± 0 | 1 | 1.0 | 1.0 ± 0 |
| Group | Number of Specimens | Number of Haplotypes | Haplotype Diversity (Hd ± Sd) | Nucleotide Diversity (π ± Sd) | Tajima’s D |
|---|---|---|---|---|---|
| Clade 1 | 102 | 21 | 0.547 ± 0.059 | 0.0016 ± 0.000 | −233,753 ** |
| North Atlantic | 69 | 15 | 0.611 ± 0.064 | 0.0020 ± 0.000 | −1.91338 * |
| South Atlantic | 6 | 6 | 1.000 ± 0.096 | 0.0056 ± 0.000 | −1.42284 |
| Indian Ocean | 27 | 10 | 0.726 ± 0.089 | 0.0027 ± 0.001 | −144,135 |
| Clade 2 | 5 | 5 | 1.000 ± 0.126 | 0.0122 ± 0.003 | −0.60926 |
| In total | 107 | 26 | 0.589 ± 0.056 | 0.0071 ± 0.002 | −2.05858 * |
| Axis Type | Constrained | Unconstrained | ||
|---|---|---|---|---|
| Axis | RDA1 | RDA1 | PC1 | PC2 |
| Eigenvalue | 5.0983 | 0.2853 | 7.4947 | 3.6436 |
| Proportion of Variance Explained | 0.1728 | 0.0097 | 0.254 | 0.1235 |
| Cumulative Proportion of Variance explained | 0.1825 | 0.3775 | ||
| 0.56 |
| S. braueri | S. cristata | S. curvispina | S. debilis Clade 1 | S. debilis Clade 2 | S. guillei | S. liui | S. paucispinosa | S. pellucida | |
|---|---|---|---|---|---|---|---|---|---|
| S. braueri | 12.76% | ||||||||
| S. cristata | 23.52% | 11.31% | |||||||
| S. curvispina | 24.50% | 12.76% | 0.34% | ||||||
| S. debilis clade 1 | 31.59% | 29.45% | 27.95% | 0.40% | |||||
| S. debilis clade 2 | 31.39% | 29.35% | 27.38% | 6.91% | 1.06% | ||||
| S. guillei | 24.12% | 22.42% | 22.21% | 31.80% | 29.91% | NA | |||
| S. liui | 31.56% | 29.30% | 27.51% | 6.52% | 1.59% | 29.84% | NA | ||
| S. paucispinosa | 8.14% | 22.24% | 23.40% | 29.98% | 30.26% | 23.92% | 30.55% | NA | |
| S. pellucida | 24.33% | 18.97% | 19.75% | 29.62% | 28.80% | 20.99% | 29.56% | 23.09% | 10.98% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shapkina, A.; Kulagin, D.; Khaitov, V.; Lunina, A.; Vereshchaka, A. Exploring Genetic and Morphological Integrity across Ocean Basins: A Case Study of the Mesopelagic Shrimp Systellaspis debilis (Decapoda: Oplophoridae). Diversity 2023, 15, 1008. https://doi.org/10.3390/d15091008
Shapkina A, Kulagin D, Khaitov V, Lunina A, Vereshchaka A. Exploring Genetic and Morphological Integrity across Ocean Basins: A Case Study of the Mesopelagic Shrimp Systellaspis debilis (Decapoda: Oplophoridae). Diversity. 2023; 15(9):1008. https://doi.org/10.3390/d15091008
Chicago/Turabian StyleShapkina, Anna, Dmitry Kulagin, Vadim Khaitov, Anastasiia Lunina, and Alexander Vereshchaka. 2023. "Exploring Genetic and Morphological Integrity across Ocean Basins: A Case Study of the Mesopelagic Shrimp Systellaspis debilis (Decapoda: Oplophoridae)" Diversity 15, no. 9: 1008. https://doi.org/10.3390/d15091008
APA StyleShapkina, A., Kulagin, D., Khaitov, V., Lunina, A., & Vereshchaka, A. (2023). Exploring Genetic and Morphological Integrity across Ocean Basins: A Case Study of the Mesopelagic Shrimp Systellaspis debilis (Decapoda: Oplophoridae). Diversity, 15(9), 1008. https://doi.org/10.3390/d15091008








