Abstract
A Gram-negative, strictly aerobic, non-motile, slightly curved rod-shaped bacterial strain, designated as HL-RS19T, was isolated from a sea surface microlayer (SML) sample of the brackish Lake Shihwa. Here, we characterized the new strain HL-RS19T using a polyphasic approach to determine its taxonomic position. A phylogenetic analysis of its 16S rRNA gene sequence revealed that strain HL-RS19T belonged to the genus Lacinutrix and was closely related to L. mariniflava AKS432T (97.9%), L. algicola AKS293T (97.8%), and other Lacinutrix species (<97.3%). The complete genome sequence of strain HL-RS19T comprised a circular chromosome of 3.9 Mbp with a DNA G+C content of 35.2%. Genomic comparisons based on the average nucleotide identity and digital DNA-DNA hybridization showed that strain HL-RS19T was consistently discriminated from its closely related taxa in the genus Lacinutrix. Strain HL-RS19T showed optimal growth at 20–25 °C, pH 6.5–7.0, and 3.0–3.5% (w/v) sea salts. The major fatty acids (>5%) of strain HL-RS19T were identified as iso-C15:1 G (16.5%), iso-C16:0 3-OH (12.9%), anteiso-C15:1 A (9.9%), anteiso-C15:0 (9.7%), iso-C15:0 (9.0%), and iso-C15:0 3-OH (8.3%). The polar lipids consisted of phosphatidylethanolamine, three unidentified aminolipids, an unidentified phospholipid, and two unidentified lipids. The major respiratory quinone was MK-6. Based on phylogenetic, genomic, phenotypic, and chemotaxonomic data, strain HL-RS19T represents a novel species belonging to the genus Lacinutrix, for which the name Lacinutrix neustonica sp. nov. is proposed. The type strain is HL-RS19T (=KCCM 90497T = JCM 35710T). The genome sequence analysis of strain HL-RS19T suggests that it may be well adapted to a harsh SML environment and is likely involved in arsenic cycling, potentially contributing to the bioremediation of anthropogenic arsenic pollution.
1. Introduction
The sea surface microlayer (SML) represents the uppermost boundary layer of the ocean, where the exchange of gases and particles takes place between the ocean and the atmosphere [1,2]. With a total thickness ranging from 1 to 1000 μm, the SML exhibits distinct physicochemical and biological characteristics compared to the underlying water (UW) [3,4]. The unique characteristics of the SML can be attributed to several factors, including strong ultraviolet (UV) irradiation, the high activity of photoreactions, and the intense accumulation of autochthonous/allochthonous organic or inorganic matter [5,6]. Previous studies have reported the significant enrichment of anthropogenic pollutants, such as hydrocarbon compounds and heavy metals, in the coastal SML near urban, agricultural, and industrial areas [1,5,7,8,9]. Therefore, the bacterial inhabitants of the SML, known as bacterioneuston, present an intriguing natural model for studying the restoration of damages caused by UV radiation or radical oxidants, as well as for obtaining valuable biological resources for the bioremediation of toxic substances. In fact, a new bacterial species in diverse lineages has been successfully isolated from SML samples collected from pristine to industrial environments [10,11,12,13,14,15].
The genus Lacinutrix was first described alongside Lacinutrix copepodicola [16], and it currently belongs to the family Flavobacteriaceae in the phylum Bacteroidota. At the time of writing, twelve species of Lacinutrix have been reported, with validly published names [17]. Members of the genus Lacinutrix are known to be neutrophilic, Gram-negative, non-motile or motile by gliding, strictly aerobic, straight, slightly rod-shaped, or coccoid cells that are catalase- and oxidase-positive and that produce golden-yellow, orange, or orange-yellow pigments [18,19,20]. The type strains of the genus Lacinutrix have been isolated from diverse marine habitats, such as seawater, sediment, macroalgae, copepod, clam, crab, and flounder, from temperate to polar environments [16,19,20,21,22,23,24,25,26,27,28]. During a year-round investigation of the SML at a temperate coast near an industrial complex, we isolated a neustonic bacterium identified as belonging to the genus Lacinutrix and designated as HL-RS19T. In this study, we conducted a comprehensive characterization of strain HL-RS19T, including its phylogenetic, genomic, phenotypic, and chemotaxonomic properties to accurately position it within the genus Lacinutrix in terms of taxonomic classification.
2. Materials and Methods
2.1. Habitat, Isolation and Cultivation Conditions
The SML sample was collected using a glass plate sampler [4] in the brackish Lake Shihwa (37°18′3.6″ N, 126°43′58.8″ E), South Korea, in January 2020. This research area receives the riverine discharge from a nearby industrial complex. During the sampling, the water temperature and salinity were 3 °C and 36.3 ‰, respectively. For cultivation, an aliquot (100 µL) of the SML sample was spread onto a saline Reasoner’s 2A agar (R2A; BD Difco, Franklin Lakes, NJ, USA) medium supplemented with 3% (w/v) sea salts (Sigma-Aldrich, St. Louis, MO, USA). The saline R2A medium was incubated aerobically at 25 °C for 4 days. Strain HL-RS19T was isolated and purified via subculturing more than four times. In a preliminary test, strain HL-RS19T grew better on the marine agar (MA; BD Difco, Franklin Lakes, NJ, USA) than on the saline R2A agar. After, strain HL-RS19T was routinely cultivated on an MA at 25 °C. The strain was preserved in a marine broth (MB; BD Difco, Franklin Lakes, NJ, USA) supplemented with 20 % (v/v) glycerol and stored at −70 °C. Over the course of four seasonal surveys conducted throughout a year in the study area, 212 bacterial strains were isolated from SML samples, and identified by the sequencing the 16S rRNA genes, as detailed in Section 2.2. Among them, strain HL-RS19T was discovered as a singular occurrence in the genus Lacinutrix, appearing only once during the entire sampling period.
2.2. 16S rRNA Gene Sequencing and Phylogenetic Analysis
Genomic DNA was extracted from a single colony using the boiling method as previously described [29]. The 16S rRNA gene of the strain HL-RS19T was amplified through polymerase chain reaction (PCR) utilizing the universal primers 27F and 1492R [30]. Subsequently, the PCR product was subjected to purification using ExoSAP-IT (ThermoFisher Scientific, Waltham, MA, USA). Direct sequencing of the purified PCR products was performed using four sequencing primers—27F, 337F, 907R, and 1492R [30,31]—via an Applied Biosystems sequencer (ABI 3730XL) at Cosmo Genetech (Seoul, Republic of Korea). Almost the full length of the 16S rRNA gene sequence of strain HL-RS19T (1419 bp) was assembled with CodonCode Aligner version 10.0.2 (CodonCode Co., Centerville, OH, USA) and analyzed using GenBank BLAST searches and EzBioCloud databases [32]. The validly published phylogenetic neighbors and the complete 16S rRNA gene sequences of strain HL-RS19T were aligned using the EzEditor2 [33] considering the secondary structure of the bacterial 16S rRNA. Subsequently, the phylogenetic analyses were performed using MEGA version 11 [34]. A neighbor-joining (NJ) tree [35] was reconstructed using the Jukes-Cantor model [36] with uniform rates and pairwise deletion options. A maximum-likelihood (ML) tree [37] was reconstructed using the Kimura two-parameter model [38] with gamma distributed with invariant sites (G+I), using all sites option for gaps/missing data. A maximum-parsimony (MP) tree [39] was constructed using the Subtree-Pruning-Regrafting (SPR) search method [40] with the number of initial trees (random addition) as 10, using all sites options. The robustness of the phylogenetic trees was evaluated by performing a bootstrap analysis based on 1000 replicates [41].
2.3. Genome Sequencing, Assembly, Annotation, and Phylogenomic Analysis
For genome comparison, the genomic DNA of strain HL-RS19T was extracted following the manufacturer’s protocol using a DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany). The whole genome sequencing was performed using a sequencing library for the MinION Mk1C sequencer (Oxford Nanopore Technologies; ONT, Oxford, UK) with a native barcoding expansion kit 13-24 (EXP-NBD114; ONT, Oxford, UK) and the ligation sequencing kit (SQK-LSK109; ONT, Oxford, UK). Raw reads were base-called, demultiplexed, and adapter-trimmed using MinKNOW version 22.05.6 and Guppy version 6.3.8. De novo genome assembly was performed using Flye version 2.9.1 [42] and polished using Medaka version 1.7.2 https://github.com/nanoporetech/medaka (accessed on 21 November 2022). The genome size, N50, and DNA G+C content were calculated using QUAST version 5.2.0 [43]. Genome coverage was measured using SAMtools version 1.11 [44]. The overall genome relatedness index (OGRI) values, including average nucleotide identity (ANI) [45] and digital DNA-DNA hybridization (dDDH) by the genome-to-genome distance calculator (GGDC) [46], were obtained for all pairwise comparisons. Additionally, a complete 16S rRNA gene sequence was retrieved from the genome sequence of strain HL-RS19T using the ContEst16S program [47].
A phylogenomic analysis of strain HL-RS19T and the type strains of related species was performed based on the Genome Taxonomy Database (GTDB) taxonomy using GTDB-Tk [48]. The genome of strain HL-RS19T was compared with the type strains of related species, including six validly named Lacinutrix species and other taxonomically differentiated Flavobacteriaceae family members, which were available in the NCBI Genome database (Table S1). The amino acid sequences of 120 concatenated marker genes from the genomes were detected and aligned using the GTDB-Tk tool. Phylogenomic trees were reconstructed using the NJ, MP, and ML methods, with 1000 replications of the bootstrap analysis using MEGA version 11 [34].
A genome annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) [49]. The Metabolic Pathway/Genome Database (PGDB) was generated computationally using the PathoLogic component of Pathway Tools software version 26.0 and MetaCyc version 23.0 [50,51]. A search for the biosynthetic gene clusters of secondary metabolites was performed using antiSMASH version 6.1.1 [52] with the option of strict detection. For a comparative genomic investigation, genome sequences of Lacinutrix available in the NCBI Genome database were retrieved and analyzed to search for protein-coding genes using tBLASTn version 2.9.0 with an e-value threshold of 1e−5. The genomic structure and variations within the Lacinutrix genus were evaluated by utilizing the Integrated Prokaryotes Genome and Pan-genome Analysis (IPGA) web server https://nmdc.cn/ipga (accessed on 25 May 2023) [53] with default settings.
2.4. Physiological, Morphological and Biochemical Characteristics
Based on its 16S rRNA gene sequence, Lacinutrix mariniflava AKS432T (=KCCM 42306T) [25], which was most similar to strain HL-RS19T by 97.9%, was purchased from the Korean Culture Center of Microorganisms (KCCM) and used as a reference strain. Unless otherwise specified, L. mariniflava KCCM 42306T and strain HL-RS19T were incubated on an MA for 3–4 days at 20 °C and 25 °C, respectively, for phenotypic tests under the exponential growth phase. According to the minimal standards for describing a new taxon of the family Flavobacteriaceae [54], all experiments for the physiological characteristics of strain HL-RS19T were carried out in duplicates, along with the type strain of L. mariniflava KCCM 42306T.
The temperature range for growth was tested on an MA at 5–40 °C (5 °C intervals) for 2 weeks. A salt-tolerance test was carried out using synthetic ZoBell broth (Bacto peptone 5 g, yeast extract 1 g, and ferric citrate 0.1 g per liter of distilled water) [55] supplemented with 0–4% (0.5% intervals), 5–10% (1% intervals), 12%, and 15% (w/v) of sea salts (Sigma-Aldrich, St. Louis, MO, USA). The growth under different pH values (pH 5.0–10.0) was investigated via inoculation in MB adjusted using pH buffer systems (MES, pH 5.0–6.5; MOPS, pH 7.0–7.5; AMPD, pH 8.0–9.5; CAPS, pH 10.0). Growth was monitored by measuring the optical density at 600 nm (SPECTRostar Nano spectrophotometer, BMG Labtech, Ortenberg, Germany) at 1–3 days intervals for 2 weeks. Anaerobic growth was assessed on both the MA and MA supplemented with potassium nitrate (0.1%, w/v) as an electron acceptor [56] and incubated in an anaerobic jar with AnaeroPack (Mitsubishi Gas Chemical Co., Tokyo, Japan) for 2 weeks. Gram-staining was performed using a Gram-Staining kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions. Motility was tested using a semi-solid MA (0.4% agar, w/v) method [57] and the hanging drop method [54,58]. Cell morphology and size were examined via transmission electron microscopy (LIBRA 120; Carl Zeiss, Oberkochen, Germany) using the strain grown on an MA at 25 °C for 3 days. The presence of carotenoid for the cells incubated in both light and dark conditions for 3 days was examined using the spectrophotometric test, as previously described [59]. Flexirubin-type pigments were determined using two different methods, as previously described [59,60].
In addition, the biochemical test and enzyme activities of strain HL-RS19T and L. mariniflava KCCM 42306T were determined by using API 20E, API 20NE, and API ZYM kits (bioMérieux) according to the manufacturer’s recommendations, except that the cells were resuspended in distilled water containing 3% (w/v) sea salts and were incubated at the optimal growth temperature for each bacterial strain. Catalase and oxidase tests were performed as previously described [61]. The hydrolysis of starch; Tweens 40, 60, 80; casein; gelatin; and urea was tested on an MA supplemented with the corresponding substrates [62]. The hydrolysis of xanthine, hypoxanthine, and L-tyrosine was examined using the method as described [63]. The hydrolysis of aesculin and the nitrate reduction test was determined by the method as described [64]. The ability to utilize a sole carbon source was tested by inoculating bacterial suspension into the basal medium consisting of NaCl 2.36 g, KCl 0.06 g, MgCl2·6H2O 4.5 g, MgSO4·7H2O 5.9 g, CaCl2·2H2O 1.3 g, NaNO3 0.2 g, NH4Cl 0.2 g, and yeast extract 0.05 g per liter of distilled water, supplemented with a final concentration of 0.4% (w/w) tested carbon sources [11]. Carbon utilization was determined as being negative when the growth was equal to or less than that in the negative control without a carbon source. The growth was determined by monitoring changes in the OD600 for 3 weeks.
The chemotaxonomic characteristics of strain HL-RS19T and L. mariniflava KCCM 42306T were determined using the cells grown on the MA for 3 days at 20 °C. The cellular fatty acid composition was analyzed via gas chromatography based on the Microbial Identification System (MIDI, Microbial ID) and RTSBA 6 version 6.21 at the KCCM. The polar lipids of strain HL-RS19T and L. mariniflava KCCM 42306T were identified via thin-layer chromatography (TLC) followed by spraying with appropriate detection reagents [65,66] at the KCCM. The isoprenoid quinone composition of strain HL-RS19T was determined as previously described [66,67] and analyzed via HPLC at the KCCM.
3. Results and Discussion
3.1. 16S rRNA Gene Sequencing and Phylogenetic Analysis
The 16S rRNA gene sequence of strain HL-RS19T (1419 bp), determined via direct sequencing, was almost identical (99.8–99.9%) to the three copies of the complete 16S rRNA gene sequences (1518 bp) retrieved from the genome sequence of strain HL-RS19T. At the 16S rRNA gene sequence level, strain HL-RS19T was most closely related to L. mariniflava AKS432T with a similarity of 97.9% and followed by Lacinutrix algicola AKS293T with a similarity of 97.8%. The 16S rRNA gene similarity values between strain HL-RS19T and other type strains of Lacinutrix species were below 97.3%. The phylogenetic analyses of the 16S rRNA gene sequences revealed that strain HL-RS19T formed a robust clade with L. mariniflava AKS432T and L. algicola AKS293T, which was consistently recovered in all phylogenetic trees (NJ, ML, and MP) (Figure 1 and Figure S1). Therefore, the phylogenetic position of strain HL-RS19T showed that the strain could be assigned to a novel species in the genus Lacinutrix.
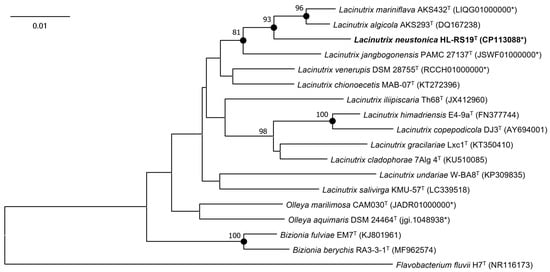
Figure 1.
Neighbor-joining tree based on the 16S rRNA gene sequences of HL-RS19T and related taxa in the family Flavobacteriaceae. Flavobacterium fluvii H7T was used as an outgroup. Bootstrap values at nodes indicate a percentage higher than 70% (based on 1000 replicates). Filled circles indicate that the corresponding nodes were recovered in the maximum-likelihood and the maximum-parsimony trees. Bar, 0.01 substitutions per nucleotide position. The asterisk mark in the parentheses indicates 16S rRNA gene sequence retrieved from the genome sequence of the type strain.
3.2. Genome Analysis and Genomic Features
The complete genome size of strain HL-RS19T was 3.9 Mbp, with a DNA G+C content of 35.2 mol% (Table S1). The ANI values between strain HL-RS19T and the closely related Lacinutrix species (i.e., L. mariniflava AKS432T and L. algicola AKS293T) were 75.2–75.3% (Table 1). The genomic relatedness analysis based on genome-to-genome distance showed that HL-RS19T was related to L. mariniflava AKS432T and L. algicola AKS293T via dDDH values of 19.7% and 20.2%, respectively (Table 1). This level is obviously below the proposed ANI and dDDH cut-off values (95–96% and 70%, respectively) for delineating bacterial species [45,68]. These results indicate that strain HL-RS19T is a new member of a distinct species of the genus Lacinutrix.

Table 1.
Results of genomic relatedness analyses based on the average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values. 1, Lacinutrix neustonica HL-RS19T (GCA_026625145.1); 2, Lacinutrix mariniflava KCCM 42306T (GCA_001418015.1); 3, Lacinutrix algicola AKS293T (GCA_001418085.1); 4, Lacinutrix jangbogonensis PAMC 27137T (GCA_000797445.1); 5, Lacinutrix venerupis DSM 28755T (GCA_003663945.1); 6, Lacinutrix himadriensis E4-9aT (GCA_001418105.1). The ANI values are indicated in the region above the diagonal grey area (values of 100%). The dDDH values are depicted below the diagonal grey area (values of 100%).
In the phylogenomic tree, strain HL-RS19T formed a discriminated clade with L. algicola AKS293T, L. mariniflava AKS432T, L. jangbogonensis PAMC 27137T, and L. venerupis DSM 28755T (Figure 2 and Figure S2), which were differed in the phylogenetic trees of the 16S rRNA gene sequences (Figure 1), resulting in somewhat different tree topologies. An incongruity between 16S rRNA gene- and genome-based trees in the genus Lacinutrix was found in this study.
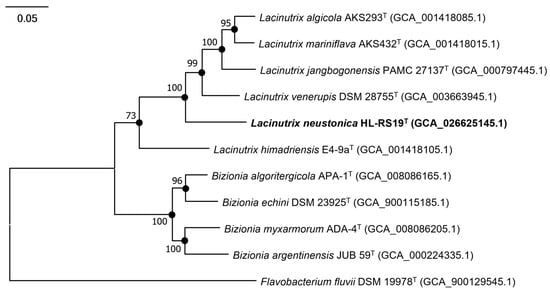
Figure 2.
Maximum-likelihood tree based on the amino acid sequences for 120 concatenated marker genes of strain HL-RS19T and related taxa in the family Flavobacteriaceae. Flavobacterium fluvii H7T was used as an outgroup. Bootstrap values at nodes indicate a percentage higher than 70% (based on 1000 replicates). Filled circles indicate that the corresponding nodes were recovered in the neighbor-joining and the maximum-parsimony trees. Bar, 0.05 substitutions per amino acid position.
The genomic analyses revealed that strain HL-RS19T possesses genes that might increase its fitness to a harsh SML environment at a coastal industrial complex, such as strong UV irradiation and the relative enrichment of the heavy metals from anthropogenic and/or natural sources [4,5]. A carotenoid biosynthetic gene cluster (BGC) was detected in the genome sequence of strain HL-RS19T (Figure 3), and the production of a carotenoid pigment was experimentally confirmed in the present study (Figure S3). Carotenoid pigments are known to be effective in UV absorption and screening in heterotrophic bacteria [69,70].
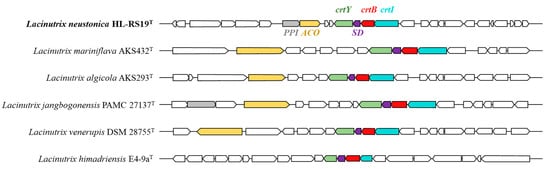
Figure 3.
Schematic biosynthesis gene cluster of carotenoids predicted from the genome sequence of Lacinutrix type strains identified using antiSMASH [52]. Each color represents the corresponding gene. PPI, peptidylprolyl isomerase (grey); ACO, aconitate hydratase (yellow); crtY, lycopene cyclase (green); SD, sterol desaturase (purple); crtB, phytoene/squalene synthase (red); crtI, phytoene desaturase (light blue).
In addition, the carotenoid biosynthetic gene cluster was identified in all Lacinutrix species through the analysis of genomic sequences using tBLASTn (Figure 3). The genus Lacinutrix was speculated to possess the capability to biosynthesize β-carotene through the utilization of key genetic elements, namely phytoene synthase (crtB), phytoene desaturase (crtI), and lycopene cyclase (crtY). The initial step in carotenoid production involves the enzymatic conversion of the immediate precursor, geranylgeranyl diphosphate (GGPP) [71], into phytoene via the activity of crtB [72]. Subsequently, the crtI gene facilitates the synthesis of lycopene from phytoene [72], and ultimately, the crtY gene governs the conversion of lycopene into β-carotene [72]. Although certain Lacinutrix species may lack specific components of the carotenoid gene cluster, the majority of them possess the essential crtB, crtI, and crtY genes. The presence of these carotenoid biosynthetic genes in their genomes indicates the presumed production of β-carotene as the ultimate product of the biosynthetic pathway.
Multiple DNA repair systems to restore UV-induced DNA damage were found in strain HL-RS19T, including light-dependent photoreactivation (DNA photolyase), nucleotide excision repair (UvrABC excinuclease complex), and homologous recombination repair (Holliday junction helicase complex) [73,74]. To cope with arsenic-rich conditions in an SML environment, strain HL-RS19T has an arsenic detoxification system, which comprises essential genes encoding ArsR transcriptional regulator, arsenate reductase (ArsC), and arsenite efflux transport protein ArsB [75], suggestive of its participation in the biogeochemical cycling of arsenic in such an environment. In practical applications, arsenic-resistant bacteria have been utilized for bioremediation as effective agents for reducing harmful metal concentrations [76,77,78,79]. The existence of the ars operon (arsRCB) in strain HL-RS19T potentially signifies its capability as a promising bioremediation tool [76], whether deployed as a wild-type strain or as genetically engineered microbes, to address environmental arsenic contamination across a broad spectrum of salinity levels.
The orthologous genes present in six genomes of Lacinutrix species were systematically classified into core, accessory, and unique gene clusters using IPGA (Figure 4). Among these, 8.4% were classified as core gene clusters, indicating their presence across all genomes, while 70.8% were classified as unique gene clusters specific to particular genes in different genomes (Figure 4). Notably, strain HL-RS19T exhibited the highest number of unique gene clusters (3214) among all currently known species within the genus Lacinutrix. Most of the unique genes were classified as unknown, with only 263 genes (8%) being annotated. Specifically, genes associated with antimicrobial resistance (tetM and catB), restriction and modification system (hsdS, yhdJ, and mcrC), and DNA repair and recombination proteins (uvrA, uvrC, recN, and gyrB) are present in strain HL-RS19T.
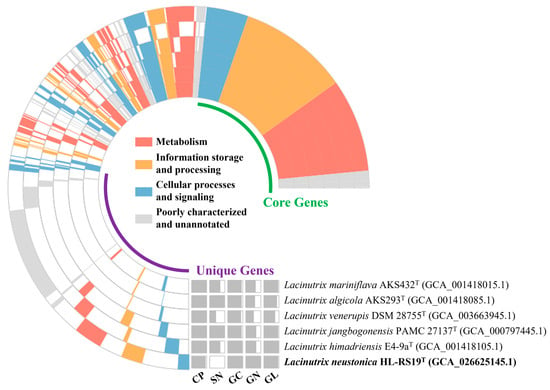
Figure 4.
Pan-genome profile of Lacinutrix species. Orthologous genes of individual genomes were clustered into three groups: core genes, accessory genes and unique genes. CP, genome completeness; SN, scaffold number; GC, GC content; GN, gene number; GL, genome length.
3.3. Physiological, Morphological and Biochemical Characteristics
Strain HL-RS19T grew at 10–30 °C, with its optimum at 20–25 °C (Table 2). In addition, strain HL-RS19T was able to grow at pH 6.0–8.5, with its optimum at pH 6.5–7.0. Strain HL-RS19T required salt to grow and was tolerant up to 7.0% (w/v) (Table 2). Strain HL-RS19T was strictly aerobic, Gram-negative, non-motile, and slightly curved rod-shaped cells (Figure 5) that displayed activities of catalase and oxidase, and were capable of carotenoid formation, which are common properties of the genus Lacinutrix [18,19,20]. Its other physiological and biochemical characteristics are summarized in Table 2, along with the species description.

Table 2.
Physiological characteristics that distinguish strain HL-RS19T from other closely related species of the genus Lacinutrix. 1: Lacinutrix neustonica HL-RS19T (this study), 2: Lacinutrix mariniflava KCCM 42306T (this study), and 3: Lacinutrix algicola AKS293T [25]. +, Positive; −, negative.
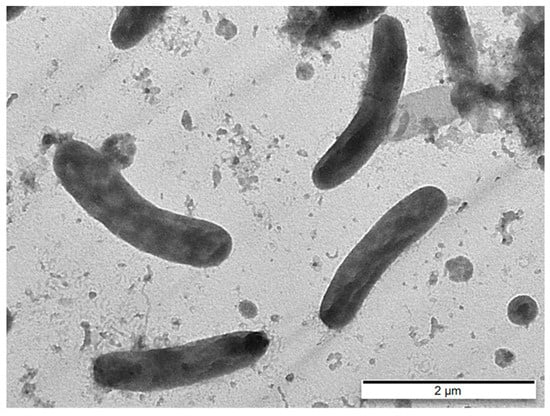
Figure 5.
Transmission electron micrograph (TEM) of negatively stained cells of strain HL-RS19T grown on marine agar at 25 °C for 3 days. Bar, 2.0 µm.
The major fatty acids (>5%) of strain HL-RS19T were iso-C15:1 G (16.5%), iso-C16:0 3-OH (12.9%), anteiso-C15:1 A (9.9%), anteiso-C15:0 (9.7%), iso-C15:0 (9.0%), and iso-C15:0 3-OH (8.3%); a detailed fatty acid composition is given in Table S2. The fatty acid profile of strain HL-RS19T was very similar to that of L. mariniflava KCCM 42306T (Table S2), except some minor fatty acids (cyclopropane fatty acids and some unsaturated fatty acids) were not detected in strain HL-RS19T. The polar lipids of strain HL-RS19T were phosphatidylethanolamine (PE), three unidentified aminolipids (AL1–3), an unidentified phospholipid (PL), and two unidentified lipids (L1–2), which were nearly identical to those of L. mariniflava KCCM 42306T, except for the absence of an unidentified lipid (L3) (Figure S4). MK-6 was identified as the only menaquinone present in strain HL-RS19T, which is the same as that found in other Lacinutrix spp. [16,19,20,21,22,24,25,26,27,28].
Strain HL-RS19T could be phenotypically differentiated from its most closely related phylogenetic neighbor L. mariniflava KCCM 42306T as follows: The temperature range for the growth of strain HL-RS19T (10–30 °C) was higher than that of L. mariniflava KCCM 42306T (5–20 °C; Table 2). The salt tolerance range of strain HL-RS19T (1.0–7.0%) was broader than that of L. mariniflava KCCM 42306T (1.5–6.0%; Table 2). In addition, strain HL-RS19T could not hydrolyze casein, gelatin, and Tween 40, which were different characteristics from those of L. mariniflava KCCM 42306T (Table 2). Strain HL-RS19T could also be distinguished from its other phylogenetical relative L. algicola AKS293T, for example, the inability to grow at 0–5 °C, the ability to grow in the presence of 3–7% (w/v) sea salts, the ability to hydrolyze Tween 80, the inability to hydrolyze urea, and the absence of β-galactosidase activity (Table 2).
4. Conclusions
Based on the phylogenetic, genomic, phenotypic, and chemotaxonomic characteristics described above, strain HL-RS19T should be placed in the genus Lacinutrix as representing a novel species, for which the name Lacinutrix neustonica sp. nov. is proposed. The presence of a suite of essential genes encoding arsenic detoxification processes in the genome of strain HL-RS19T displays its potential for bioremediation in arsenic-contaminated saline environments.
Description of Lacinutrix neustonica sp. nov.:
Lacinutrix neustonica (neus.to’ni.ca. N.L. fem. adj. neustonica pertaining to and living in the neuston).
Cells are strictly aerobic, Gram-negative, non-motile, and slightly curved rod-shaped (0.3–0.8 μm wide and 1.4–4.2 μm long. Colonies are circular, shiny, golden-yellow, and convex with entire margins after 7 days of incubation on MA plates. They are positive for oxidase and catalase activities. Growth occurs at 10–30 °C (optimum at 20–25 °C), at pH 6.0–8.5 (optimum at 6.5–7.0), and in the presence of sea salts with a concentration of 1.0–7.0% (w/v) (optimum 3.0–3.5%). Starch, aesculin, Tweens 60 and 80 are hydrolyzed, but casein, gelatin, hypoxanthine, xanthine, L-tyrosine, Tween 40, and urea are not. Nitrate is not reduced. Carotenoid pigments are produced. Flexirubin-type pigments are not produced. In the API ZYM system, they are positive for acid phosphatase, alkaline phosphatase, α-chymotrypsin, cystine arylamidase, esterase (C4), esterase lipase (C8), leucine arylamidase, naphthol-AS-BI-phosphohydrolase, trypsin, and valine arylamidase, but negative for N-acetyl-β-glucosaminidase, α-fucosidase, α- and β-galactosidases, α- and β- glucosidases, β-glucuronidase, lipase (C14), and α-mannosidase. In the API 20E system, they are positive for the Voges-Proskauer test, but negative for arginine dihydrolase, β-galactosidase, gelatinase, lysine decarboxylase, ornithine decarboxylase, citrate utilization, urease, tryptophan deaminase, and the production of hydrogen sulfide and indole. In the API 20NE system, they are positive for esculin hydrolysis and paranitrophenyl-β-D-galactopyranosidase (weakly), but negative for glucose fermentation, hydrolysis of L-arginine, gelatin, urea, indole production, and nitrate reduction. D-Maltose, D-mannose, L-proline, and trisodium citrate are utilized as sole carbon sources, but acetate, N-acetyl-D-glucosamine, L-arabinose, D-glucose, inositol, L-lysine, malic acid, mannitol, potassium gluconate, pyruvate, and raffinose are not utilized. The major fatty acids are iso-C15:1 G, iso-C16:0 3-OH, anteiso-C15:1 A, anteiso-C15:0, iso-C15:0, and iso-C15:0 3-OH. The polar lipids are phosphatidylethanolamine, three unidentified aminolipids, an unidentified phospholipid, and two unidentified lipids. The menaquinone present is MK-6.
The type strain HL-RS19T (=KCCM 90497T = JCM 35710T) was isolated from the surface microlayer sample of brackish Lake Shihwa. The GenBank/EMBL/DBBJ accession numbers for the 16S rRNA gene sequence and the genome sequence of strain HL-RS19T are MZ820004 and CP113088, respectively. The DNA G+C content is 35.2%, determined via genome analysis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15091004/s1, Figure S1: (a) Maximum-likelihood (ML) and (b) maximum-parsimony (MP) phylogenetic tree based on 16S rRNA gene sequences of strain HL-RS19T and related taxa in the family Flavobacteriaceae; Figure S2: (a) Neighbor-joining (NJ) and (b) maximum-parsimony (MP) phylogenomic tree based on the amino acid sequences for 120 concatenated marker genes of strain HL-RS19T and related taxa in the family Flavobacteriaceae; Figure S3: UV-VIS spectra of diverse carotenoids detected in strain HL-RS19T incubated under light and dark conditions; Figure S4: Two-dimensional thin-layer chromatography (TLC) of the polar lipids of (a) Lacinutrix neustonica HL-RS19T and (b) Lacinutrix mariniflava KCCM 42306T; Table S1: List of whole genome sequences of Lacinutrix spp.; Table S2: Cellular fatty acid composition (%) of strain HL-RS19T and L. mariniflava KCCM 42306T.
Author Contributions
Isolation, morphological, and biochemical characterization of strains, J.Y.C., S.Y.K. and Y.W.H.; chemotaxonomic characterization, S.Y.K.; Sanger sequencing, J.Y.C. and J.K.K.; genome sequencing, phylogenetic, and phylogenomic analyses, J.Y.C., B.J.K. and D.Y.S.; writing—original draft preparation, J.Y.C. and C.Y.H.; manuscript editing, B.C.C. and C.Y.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1F1A1059526) and by the “Development of Advanced Science and Technology for Marine Environmental Impact Assessment” of the Korea Institute of Marine Science and Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (202104272), Korea.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequence and the complete genome sequence of strain HL-RS19T are MZ820004 and CP113088, respectively. Strain HL-RS19T was deposited in the Korean Culture Center of Microorganisms (KCCM) and Japan Collection of Microorganisms (JCM) under the number KCCM 90497T and JCM 35710T, respectively.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hunter, K.A. Processes affecting particulate trace metals in the sea surface microlayer. Mar. Chem. 1980, 9, 49–70. [Google Scholar] [CrossRef]
- Zancker, B.; Cunliffe, M.; Engel, A. Bacterial community composition in the sea surface microlayer off the peruvian coast. Front. Microbiol. 2018, 9, 2699. [Google Scholar] [CrossRef] [PubMed]
- Carlucci, A.F.; Wolgast, D.M.; Craven, D.B. Microbial populations in surface films: Amino acid dynamics in nearshore and offshore waters off Southern California. J. Geophys. Res. Oceans 1992, 92, 5271–5280. [Google Scholar] [CrossRef]
- Cunliffe, M.; Engel, A.; Frka, S.; Gasparovic, B.; Guitart, C.; Murrell, J.C.; Salter, M.; Stolle, C.; Upstill-Goddard, R.; Wurl, O. Sea surface microlayers: A unified physicochemical and biological perspective of the air-ocean interface. Prog. Oceanogr. 2013, 109, 104–116. [Google Scholar] [CrossRef]
- Wurl, O.; Obbard, J.P. A review of pollutants in the sea-surface microlayer (SML): A unique habitat for marine organisms. Mar. Pollut. Bull. 2004, 48, 1016–1030. [Google Scholar] [CrossRef] [PubMed]
- Cincinelli, A.; Stortini, A.M.; Checchini, L.; Martellini, T.; Del Bubba, M.; Lepri, L. Enrichment of organic pollutants in the sea surface microlayer (SML) at Terra Nova Bay, Antarctica: Influence of SML on superfacial snow composition. J. Environ. Monitor. 2005, 7, 1305–1312. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.T.; Crecelius, E.A.; Antrim, L.D.; Kiesser, S.L.; Broadhurst, V.L.; Boehm, P.D.; Steinhauer, W.G.; Coogan, T.H. Aquatic surface microlayer contamination in Chesapeake Bay. Mar. Chem. 1990, 28, 333–351. [Google Scholar] [CrossRef]
- Williams, P.M.; Carlucci, A.F.; Henrichs, S.M.; Vanvleet, E.S.; Horrigan, S.G.; Reid, F.M.H.; Robertson, K.J. Chemical and microbiological studies of sea-surface films in the Southern Gulf of California and off the West-Coast of Baja-California. Mar. Chem. 1986, 19, 17–98. [Google Scholar] [CrossRef]
- Hardy, J.T.; Apts, C.W.; Crecelius, E.A.; Bloom, N.S. Sea-surface microlayer metals enrichments in an urban and rural bay. Estuar. Coast. Shelf Sci. 1985, 20, 299–312. [Google Scholar] [CrossRef]
- Lee, I.; Jang, G.I.; Cho, Y.; Yoon, S.J.; Pham, H.M.; Nguyen, A.V.; Lee, Y.M.; Park, H.; Rhee, T.S.; Kim, S.H.; et al. Sandaracinobacter neustonicus sp. nov., isolated from the sea surface microlayer in the Southwestern Pacific Ocean, and emended description of the genus Sandaracinobacter. Int. J. Syst. Evol. Microbiol. 2020, 70, 4698–4703. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.I.; Lee, I.; Ha, T.T.; Yoon, S.J.; Hwang, Y.J.; Yi, H.; Yun, S.; Lee, W.S.; Hwang, C.Y. Pseudomonas neustonica sp. nov., isolated from the sea surface microlayer of the Ross Sea (Antarctica). Int. J. Syst. Evol. Microbiol. 2020, 70, 3832–3838. [Google Scholar] [CrossRef]
- Hwang, C.Y.; Lee, I.; Hwang, Y.J.; Yoon, S.J.; Lee, W.S.; Cho, B.C. Pseudoalteromonas neustonica sp. nov., isolated from the sea surface microlayer of the Ross Sea (Antarctica), and emended description of the genus Pseudoalteromonas. Int. J. Syst. Evol. Microbiol. 2016, 66, 3377–3382. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.K.; Yoshizawa, S.; Nakajima, Y.; Cuadra, M.J.; Nogi, Y.; Nakamura, K.; Takami, H.; Ogura, Y.; Hayashi, T.; Chiura, H.X.; et al. Amylibacter kogurei sp. nov., a novel marine alphaproteobacterium isolated from the coastal sea surface microlayer of a marine inlet. Int. J. Syst. Evol. Microbiol. 2018, 68, 2872–2877. [Google Scholar] [CrossRef]
- Urios, L.; Agogue, H.; Intertaglia, L.; Lesongeur, F.; Lebaron, P. Melitea salexigens gen. nov., sp. nov., a gammaproteobacterium from the Mediterranean Sea. Int. J. Syst. Evol. Microbiol. 2008, 58, 2479–2483. [Google Scholar] [CrossRef]
- Urios, L.; Intertaglia, L.; Lesongeur, F.; Lebaron, P. Haliea salexigens gen. nov., sp. nov., a member of the Gammaproteobacteria from the Mediterranean Sea. Int. J. Syst. Evol. Microbiol. 2008, 58, 1233–1237. [Google Scholar] [CrossRef]
- Bowman, J.P.; Nichols, D.S. Novel members of the family Flavobacteriaceae from Antarctic maritime habitats including Subsaximicrobium wynnwilliamsii gen. nov., sp. nov., Subsaximicrobium saxinquilinus sp. nov., Subsaxibacter broadyi gen. nov., sp. nov., Lacinutrix copepodicola gen. nov., sp. nov., and novel species of the genera Bizionia, Gelidibacter and Gillisia. Int. J. Syst. Evol. Microbiol. 2005, 55, 1471–1486. [Google Scholar] [CrossRef]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Goker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Bowman, J.P. Lacinutrix. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–3. [Google Scholar]
- Nedashkovskaya, O.I.; Kim, S.G.; Zhukova, N.V.; Lee, J.S.; Mikhailov, V.V. Lacinutrix cladophorae sp. nov., a flavobacterium isolated from the green alga Cladophora stimpsonii, transfer of Flavirhabdus iliipiscaria Shakeela et al. 2015 to the genus Lacinutrix as Lacinutrix iliipiscaria comb. nov. and emended description of the genus Lacinutrix. Int. J. Syst. Evol. Microbiol. 2016, 66, 4339–4346. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, T.N.; Prasad, S.; Manasa, P.; Sailaja, B.; Begum, Z.; Shivaji, S. Lacinutrix himadriensis sp. nov., a psychrophilic bacterium isolated from a marine sediment, and emended description of the genus Lacinutrix. Int. J. Syst. Evol. Microbiol. 2013, 63, 729–734. [Google Scholar] [CrossRef]
- Huang, Z.; Li, G.; Lai, Q.; Gu, L.; Shao, Z. Lacinutrix gracilariae sp. nov., isolated from the surface of a marine red alga Gracilaria sp. Int. J. Syst. Evol. Microbiol. 2016, 66, 587–591. [Google Scholar] [CrossRef]
- Kim, H.; Yoon, S.C.; Choi, K.H.; Kim, S.T.; Lee, J.B.; Kim, D.S.; Han, H.L.; Bae, K.S.; Park, D.S. Lacinutrix chionocetis sp. nov., isolated from gut of a red snow crab. Arch. Microbiol. 2017, 199, 597–603. [Google Scholar] [CrossRef]
- Lasa, A.; Dieguez, A.L.; Romalde, J.L. Description of Lacinutrix venerupis sp. nov.: A novel bacterium associated with reared clams. Syst. Appl. Microbiol. 2015, 38, 115–119. [Google Scholar] [CrossRef]
- Lee, Y.M.; Hwang, C.Y.; Lee, I.; Jung, Y.J.; Cho, Y.; Baek, K.; Hong, S.G.; Kim, J.; Chun, J.; Lee, H.K. Lacinutrix jangbogonensis sp. nov., a psychrophilic bacterium isolated from Antarctic marine sediment and emended description of the genus Lacinutrix. Anton. Leeuw. 2014, 106, 527–533. [Google Scholar] [CrossRef]
- Nedashkovskaya, O.I.; Kwon, K.K.; Yang, S.H.; Lee, H.S.; Chung, K.H.; Kim, S.J. Lacinutrix algicola sp. nov. and Lacinutrix mariniflava sp. nov., two novel marine alga-associated bacteria and emended description of the genus Lacinutrix. Int. J. Syst. Evol. Microbiol. 2008, 58, 2694–2698. [Google Scholar] [CrossRef]
- Park, S.; Park, J.M.; Jung, Y.T.; Kang, C.H.; Yoon, J.H. Lacinutrix undariae sp. nov., isolated from a brown algae reservoir. Int. J. Syst. Evol. Microbiol. 2015, 65, 2696–2701. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yoon, J.; Lee, J.S.; Lee, K.C. Description of Lacinutrix salivirga sp. nov., a marine member of the family Flavobacteriaceae isolated from seawater. Arch. Microbiol. 2018, 200, 1159–1165. [Google Scholar] [CrossRef] [PubMed]
- Shakeela, Q.; Shehzad, A.; Zhang, Y.; Tang, K.; Zhang, X.H. Flavirhabdus iliipiscaria gen. nov., sp. nov., isolated from intestine of flounder (Paralichthys olivaceus) and emended descriptions of the genera Flavivirga, Algibacter, Bizionia and Formosa. Int. J. Syst. Evol. Microbiol. 2015, 65, 1347–1353. [Google Scholar] [CrossRef]
- Englen, M.D.; Kelley, L.C. A rapid DNA isolation procedure for the identification of Campylobacter jejuni by the polymerase chain reaction. Lett. Appl. Microbiol. 2000, 31, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematics, 2nd ed.; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: Chichester, UK, 1991; pp. 115–175. [Google Scholar]
- Anzai, Y.; Kudo, Y.; Oyaizu, H. The phylogeny of the genera Chryseomonas, Flavimonas, and Pseudomonas supports synonymy of these three genera. Int. J. Syst. Bacteriol. 1997, 47, 249–251. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.S.; Lee, K.; Park, S.C.; Kim, B.S.; Cho, Y.J.; Ha, S.M.; Chun, J. EzEditor: A versatile sequence alignment editor for both rRNA- and protein-coding genes. Int. J. Syst. Evol. Microbiol. 2014, 64, 689–691. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Jukes, T.H.; Cantor, C.R. Evolution of Protein Molecules. In Mammalian Protein Metabolism; Academic Press: New York, NY, USA, 1969; Volume 3, pp. 21–132. [Google Scholar]
- Felsenstein, J. Evolutionary trees from DNA-Sequences—A maximum-likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.A. Simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Fitch, W.M. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst. Biol. 1971, 20, 406–416. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, 142–150. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Anton. Leeuw. 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2021, 50, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Chalita, M.; Ha, S.M.; Na, S.I.; Yoon, S.H.; Chun, J. ContEst16S: An algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2017, 67, 2053–2057. [Google Scholar] [CrossRef]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc database of metabolic pathways and enzymes—A 2019 update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef] [PubMed]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinform. 2019, 20, 1085–1093. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, Y.; Fan, G.; Sun, D.; Zhang, X.; Yu, Z.; Wang, J.; Wu, L.; Shi, W.; Ma, J. IPGA: A handy integrated prokaryotes genome and pan-genome analysis web service. iMeta 2022, 1, e55. [Google Scholar] [CrossRef]
- Bernardet, J.F.; Nakagawa, Y.; Holmes, B. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int. J. Syst. Evol. Microbiol. 2002, 52, 1049–1070. [Google Scholar] [CrossRef]
- Lee, Y.S.; Lee, D.H.; Kahng, H.Y.; Sohn, S.H.; Jung, J.S. Polaribacter gangjinensis sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 2011, 61, 1425–1429. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Kang, S.J.; Jung, Y.T.; Lee, M.H.; Oh, T.K. Alkalibacillus flavidus sp. nov., isolated from a marine solar saltern. Int. J. Syst. Evol. Microbiol. 2010, 60, 434–438. [Google Scholar] [CrossRef]
- Jordan, E.O.; Caldwell, M.E.; Reiter, D. Bacterial motility. J. Bacteriol. 1934, 27, 165–174. [Google Scholar] [CrossRef]
- Skerman, V.B.D. A Guide to the Identification of the Genera of Bacteria, with Methods and Digests of Generic Characteristics, 2nd ed.; Williams & Wilkins Co.: Baltimore, MD, USA, 1967. [Google Scholar]
- Gosink, J.J.; Woese, C.R.; Staley, J.T. Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov., and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga-Flavobacterium-Bacteroides group and reclassification of ‘Flectobacillus glomeratus’ as Polaribacter glomeratus comb. nov. Int. J. Syst. Evol. Microbiol. 1998, 48, 223–235. [Google Scholar] [CrossRef]
- Fautz, E.; Reichenbach, H. A simple test for flexirubin-type pigments. Fems Microbiol. Lett. 1980, 8, 87–91. [Google Scholar] [CrossRef]
- Tindall, B.J.; Sikorski, J.; Smibert, R.A.; Krieg, N.R. Phenotypic Characterization and the Principles of Comparative Systematics. In Methods for General and Molecular Microbiology; Reddy, C.A., Beveridge, T.J., Breznak, J.A., Marzluf, G., Schmidt, T.M., Snyder, L.R., Eds.; American Society for Microbiology: Washington, DC, USA, 2007; pp. 330–393. [Google Scholar] [CrossRef]
- Smibert, R.M.; Krieg, N.R. Methods for General and Molecular Bacteriology; American Society for Microbiology: Washington, DC, USA, 1994; pp. 607–654. [Google Scholar]
- Cowan, S.T.; Steel, K.J. Manual for the Identification of Medical Bacteria; Cambridge University Press: London, UK, 1965. [Google Scholar]
- Lányi, B. Classical and Rapid Identification Methods for Medically Important Bacteria. In Current Methods for Classification and Idenfication of Mircroorganisms; Colwell, R.R., Grigorova, R., Eds.; Academic Press: London, UK, 1988; Volume 19, pp. 1–67. [Google Scholar]
- Komagata, K.; Suzuki, K. Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol. 1987, 19, 161–207. [Google Scholar] [CrossRef]
- Minnikin, D.E.; Odonnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Meth. 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Collins, M.D. Analysis of isoprenoid quinones. Methods Microbiol. 1985, 18, 329–366. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Dieser, M.; Greenwood, M.; Foreman, C.M. Carotenoid pigmentation in Antarctic heterotrophic bacteria as a strategy to withstand environmental stresses. Arct. Antarct. Alp. Res. 2010, 42, 396–405. [Google Scholar] [CrossRef]
- Stafsnes, M.H.; Josefsen, K.D.; Kildahl-Andersen, G.; Valla, S.; Ellingsen, T.E.; Bruheim, P. Isolation and characterization of marine pigmented bacteria from Norwegian coastal waters and screening for carotenoids with UVA-blue light absorbing properties. J. Microbiol. 2010, 48, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, G.A. Genetics of eubacterial carotenoid biosynthesis: A colorful tale. Annu. Rev. Microbiol. 1997, 51, 629–659. [Google Scholar] [CrossRef] [PubMed]
- Misawa, N.; Satomi, Y.; Kondo, K.; Yokoyama, A.; Kajiwara, S.; Saito, T.; Ohtani, T.; Miki, W. Structure and functional-analysis of a marine bacterial carotenoid biosynthesis gene-cluster and astaxanthin biosynthetic pathway proposed at the gene level. J. Bacteriol. 1995, 177, 6575–6584. [Google Scholar] [CrossRef]
- Crowley, D.J.; Boubriak, I.; Berquist, B.R.; Clark, M.; Richard, E.; Sullivan, L.; DasSarma, S.; McCready, S. The uvrA, uvrB and uvrC genes are required for repair of ultraviolet light induced DNA photoproducts in Halobacterium sp. NRC-1. Saline Syst. 2006, 2, 11. [Google Scholar] [CrossRef][Green Version]
- Donaldson, J.R.; Courcelle, C.T.; Courcelle, J. RuvABC is required to resolve Holliday junctions that accumulate following replication on damaged templates in Escherichia coli. J. Biol. Chem. 2006, 281, 28811–28821. [Google Scholar] [CrossRef]
- Rosen, B.P. Families of arsenic transporters. Trends Microbiol. 1999, 7, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Mateos, L.M.; Villadangos, A.F.; de la Rubia, A.G.; Mourenza, A.; Marcos-Pascual, L.; Letek, M.; Pedre, B.; Messens, J.; Gil, J.A. The arsenic detoxification system in Corynebacteria: Basis and application for bioremediation and redox control. Adv. Appl. Microbiol. 2017, 99, 103–137. [Google Scholar] [CrossRef]
- Villadangos, A.F.; Ordóñez, E.; Pedre, B.; Messens, J.; Gil, J.A.; Mateos, L.M. Engineered coryneform bacteria as a bio-tool for arsenic remediation. Appl. Microbiol. Biotechnol. 2014, 98, 10143–10152. [Google Scholar] [CrossRef]
- Feo, J.C.; Ordonez, E.; Letek, M.; Castro, M.A.; Munoz, M.I.; Gil, J.A.; Mateos, L.M.; Aller, A.J. Retention of inorganic arsenic by coryneform mutant strains. Water Resour. Res. 2007, 41, 531–542. [Google Scholar] [CrossRef]
- Sousa, T.; Branco, R.; Piedade, A.P.; Morais, P.V. Hyper accumulation of arsenic in mutants of Ochrobactrum tritici silenced for arsenite efflux pumps. PLoS ONE 2015, 10, e0131317. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).