Sexual Selection and Proteinaceous Diversity in the Femoral Gland Secretions of Lacertid Lizards
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
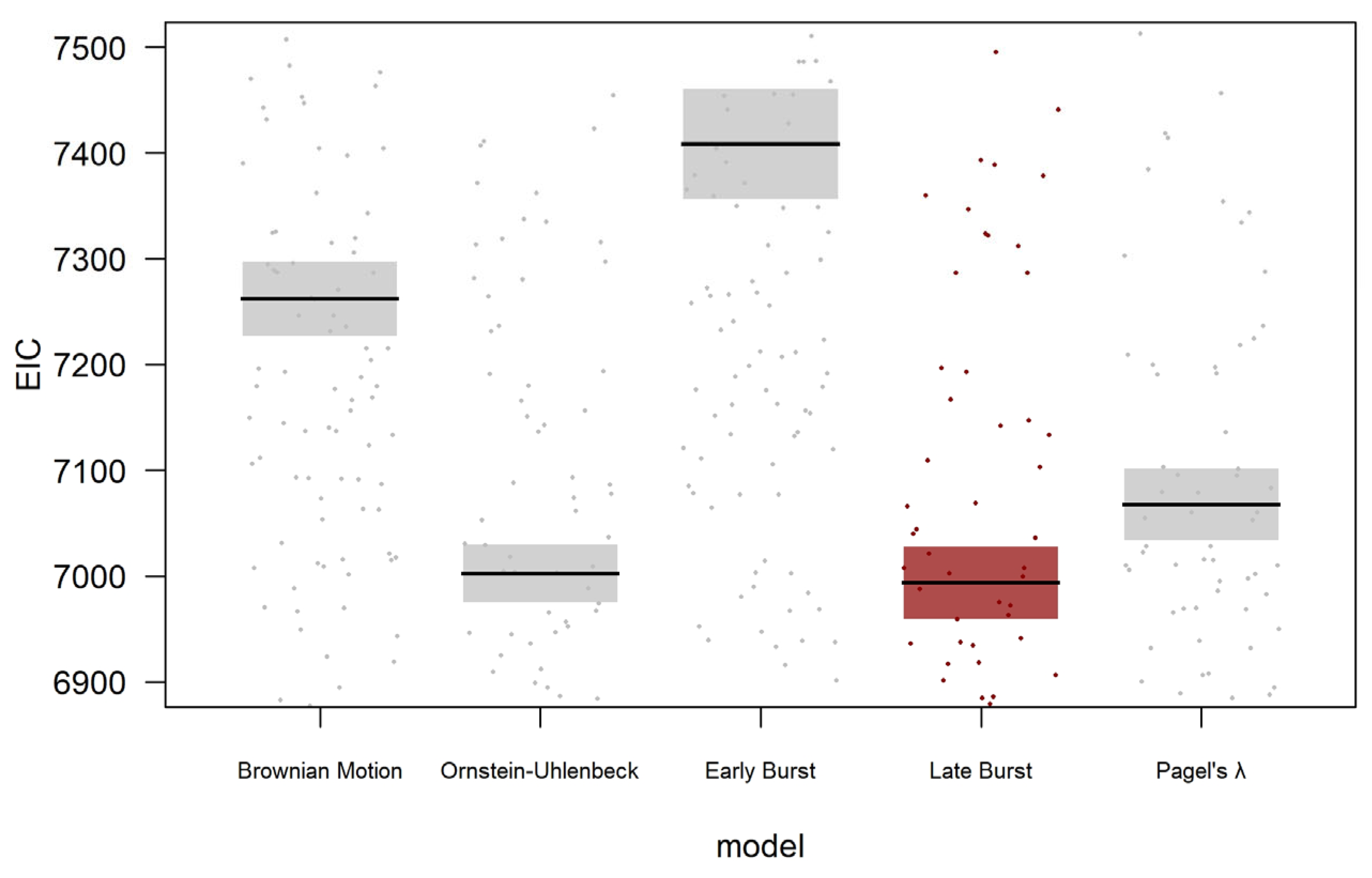
2.2. Statistical Analysis
2.3. SSD-Related Protein Identification
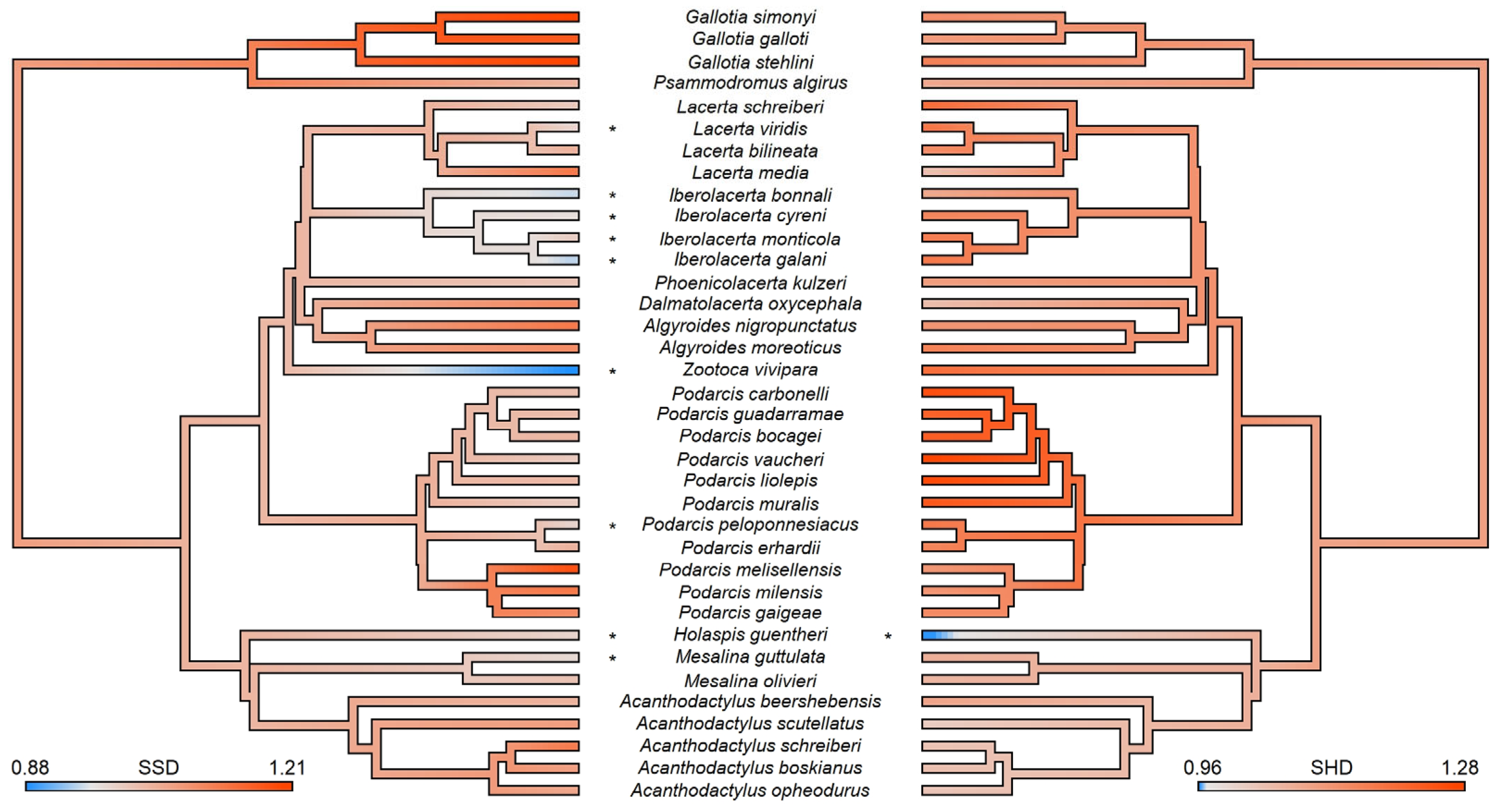
3. Results
SSD-Related Protein Identification
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Apps, P.J.; Weldon, P.J.; Kramer, M. Chemical Signals in Terrestrial Vertebrates: Search for Design Features. Nat. Prod. Rep. 2015, 32, 1131–1153. [Google Scholar] [CrossRef]
- Weldon, P.J.; Flachsbarth, B.; Schulz, S. Natural Products from the Integument of Nonavian Reptiles. Nat. Prod. Rep. 2008, 25, 738–756. [Google Scholar] [CrossRef]
- Bradbury, J.W.; Vehrencamp, S.L. Principles of Animal Communication; Sinauer Associates, Inc.: Sunderland, UK, 2011; ISBN 978-0-87893-045-6. [Google Scholar]
- Wyatt, T.D. Pheromones and Signature Mixtures: Defining Species-Wide Signals and Variable Cues for Identity in Both Invertebrates and Vertebrates. J. Comp. Physiol. A 2010, 196, 685–700. [Google Scholar] [CrossRef]
- Wiens, J.J.; Tuschhoff, E. Songs versus Colours versus Horns: What Explains the Diversity of Sexually Selected Traits? Biol. Rev. 2020, 95, 847–864. [Google Scholar] [CrossRef] [PubMed]
- Tibbetts, E.A.; Mullen, S.P.; Dale, J. Signal Function Drives Phenotypic and Genetic Diversity: The Effects of Signalling Individual Identity, Quality or Behavioural Strategy. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160347. [Google Scholar] [CrossRef]
- Symonds, M.R.E.; Elgar, M.A. The Evolution of Pheromone Diversity. Trends Ecol. Evol. 2008, 23, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Simmons, L.W. Sexual Selection and Mate Choice. Trends Ecol. Evol. 2006, 21, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.G.; Mitko, L.; Eltz, T.; Ramírez, S.R. Macroevolution of Perfume Signalling in Orchid Bees. Ecol. Lett. 2016, 19, 1314–1323. [Google Scholar] [CrossRef]
- Steiger, S.; Stökl, J. The Role of Sexual Selection in the Evolution of Chemical Signals in Insects. Insects 2014, 5, 423–438. [Google Scholar] [CrossRef]
- Hunt, J.; Snook, R.R.; Mitchell, C.; Crudgington, H.S.; Moore, A.J. Sexual Selection and Experimental Evolution of Chemical Signals in Drosophila Pseudoobscura. J. Evol. Biol. 2012, 25, 2232–2241. [Google Scholar] [CrossRef]
- Schwenk, K. Of Tongues and Noses: Chemoreception in Lizards and Snakes. Trends Ecol. Evol. 1995, 10, 7–12. [Google Scholar] [CrossRef]
- Baeckens, S.; Herrel, A.; Broeckhoven, C.; Vasilopoulou-Kampitsi, M.; Huyghe, K.; Goyens, J.; Van Damme, R. Evolutionary Morphology of the Lizard Chemosensory System. Sci. Rep. 2017, 7, 10141. [Google Scholar] [CrossRef]
- Martín, J.; López, P. Condition-Dependent Chemosignals in Reproductive Behavior of Lizards. Horm. Behav. 2015, 68, 14–24. [Google Scholar] [CrossRef]
- Baeckens, S. Evolution of Animal Chemical Communication: Insights from Non-Model Species and Phylogenetic Comparative Methods. Belg. J. Zool. 2019, 149, 63–93. [Google Scholar] [CrossRef]
- Baeckens, S.; Whiting, M.J. Investment in Chemical Signalling Glands Facilitates the Evolution of Sociality in Lizards. Proc. R. Soc. B Biol. Sci. 2021, 288, 20202438. [Google Scholar] [CrossRef] [PubMed]
- Kopena, R.; López, P.; Martín, J. Immune Challenged Male Iberian Green Lizards May Increase the Expression of Some Sexual Signals If They Have Supplementary Vitamin, E. Behav. Ecol. Sociobiol. 2017, 71, 173. [Google Scholar] [CrossRef]
- López, P.; Martín, J. Female Iberian Wall Lizards Prefer Male Scents That Signal a Better Cell-Mediated Immune Response. Biol. Lett. 2005, 1, 404–406. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.; Civantos, E.; Amo, L.; López, P. Chemical Ornaments of Male Lizards Psammodromus algirus May Reveal Their Parasite Load and Health State to Females. Behav. Ecol. Sociobiol. 2007, 62, 173–179. [Google Scholar] [CrossRef]
- Labra, A. Chemoreception and the Assessment of Fighting Abilities in the Lizard Liolaemus monticola. Ethology 2006, 112, 993–999. [Google Scholar] [CrossRef]
- Martín, J.; López, P. Scent May Signal Fighting Ability in Male Iberian Rock Lizards. Biol. Lett. 2007, 3, 125–127. [Google Scholar] [CrossRef]
- López, P.; Martín, J. Male Iberian Rock Lizards May Reduce the Costs of Fighting by Scent Matching of the Resource Holders. Behav. Ecol. Sociobiol. 2011, 65, 1891–1898. [Google Scholar] [CrossRef]
- Aragón, P.; López, P.; Martín, J. Size-Dependent Chemosensory Responses to Familiar and Unfamiliar Conspecific Faecal Pellets by the Iberian Rock-Lizard, Lacerta monticola. Ethology 2000, 106, 1115–1128. [Google Scholar] [CrossRef]
- Aragón, P.L.; López, P.; Martín, J. Chemosensory Discrimination of Familiar and Unfamiliar Conspecifics by Lizards: Implications of Field Spatial Relationships between Males. Behav. Ecol. Sociobiol. 2001, 50, 128–133. [Google Scholar] [CrossRef]
- Carazo, P.; Font, E.; Desfilis, E. Beyond “nasty Neighbours” and “Dear Enemies”? Individual Recognition by Scent Marks in a Lizard (Podarcis hispanica). Anim. Behav. 2008, 76, 1953–1963. [Google Scholar] [CrossRef]
- Mangiacotti, M.; Gaggiani, S.; Coladonato, A.J.; Scali, S.; Zuffi, M.A.L.; Sacchi, R. First Experimental Evidence That Proteins from Femoral Glands Convey Identity-Related Information in a Lizard. Acta Ethologica 2019, 22, 57–65. [Google Scholar] [CrossRef]
- López, P.; Gabirot, M.; Martín, J. Immune Activation Affects Chemical Sexual Ornaments of Male Iberian Wall Lizards. Naturwissenschaften 2009, 96, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Cole, C.J. Femoral Glands in Lizards: A Review. Herpetologica 1966, 22, 199–206. [Google Scholar]
- Mayerl, C.; Baeckens, S.; Van Damme, R. Evolution and Role of the Follicular Epidermal Gland System in Non-Ophidian Squamates. Amphib.-Reptil. 2015, 36, 185–206. [Google Scholar] [CrossRef]
- Mangiacotti, M.; Pezzi, S.; Fumagalli, M.; Coladonato, A.J.; d’Ettorre, P.; Leroy, C.; Bonnet, X.; Zuffi, M.A.L.; Scali, S.; Sacchi, R. Seasonal Variations in Femoral Gland Secretions Reveals Some Unexpected Correlations Between Protein and Lipid Components in a Lacertid Lizard. J. Chem. Ecol. 2019, 45, 673–683. [Google Scholar] [CrossRef]
- Alberts, A.C.; Sharp, T.R.; Werner, D.I.; Weldon, P.J. Seasonal Variation of Lipids in Femoral Gland Secretions of Male Green Iguanas (Iguana iguana). J. Chem. Ecol. 1992, 18, 703–712. [Google Scholar] [CrossRef]
- Martins, E.P.; Ord, T.J.; Slaven, J.; Wright, J.L.; Housworth, E.A. Individual, Sexual, Seasonal, and Temporal Variation in the Amount of Sagebrush Lizard Scent Marks. J. Chem. Ecol. 2006, 32, 881–893. [Google Scholar] [CrossRef]
- Baeckens, S.; Martín, J.; García-Roa, R.; Van Damme, R. Sexual Selection and the Chemical Signal Design of Lacertid Lizards. Zool. J. Linn. Soc. 2018, 183, 445–457. [Google Scholar] [CrossRef]
- Sindaco, R.; Jeremcenko, V.K. The Reptiles of the Western Palearctic: Annotated Checklist and DistriButional Atlas of the Turtles, Crocodiles, Amphisbaenians and Lizards of Europe, North Africa, Middle East and Central Asia; Edizioni Belvedere: Latina, Italy, 2008; Volume 1, ISBN 978-88-89504-14-7. [Google Scholar]
- Roll, U.; Feldman, A.; Novosolov, M.; Allison, A.; Bauer, A.M.; Bernard, R.; Böhm, M.; Castro-Herrera, F.; Chirio, L.; Collen, B.; et al. The Global Distribution of Tetrapods Reveals a Need for Targeted Reptile Conservation. Nat. Ecol. Evol. 2017, 1, 1677–1682. [Google Scholar] [CrossRef]
- Garcia-Porta, J.; Irisarri, I.; Kirchner, M.; Rodríguez, A.; Kirchhof, S.; Brown, J.L.; MacLeod, A.; Turner, A.P.; Ahmadzadeh, F.; Albaladejo, G.; et al. Environmental Temperatures Shape Thermal Physiology as Well as Diversification and Genome-Wide Substitution Rates in Lizards. Nat. Commun. 2019, 10, 4077. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.; López, P. Pheromones and Chemical Communication in Lizards. In Reproductive Biology and Phylogeny of Lizards and Tuatara; Rheubert, J.L., Siegel, D.S., Trauth, S.E., Eds.; Taylor and Francis Group USA: New York, NY, USA, 2014; pp. 54–88. [Google Scholar]
- Mangiacotti, M.; Fumagalli, M.; Scali, S.; Zuffi, M.A.L.; Cagnone, M.; Salvini, R.; Sacchi, R. Inter- and Intra-Population Variability of the Protein Content of Femoral Gland Secretions from a Lacertid Lizard. Curr. Zool. 2017, 63, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Alberts, A.C.; Werner, D.I. Chemical Recognition of Unfamiliar Conspecifics by Green Iguanas: Functional Significance of Different Signal Components. Anim. Behav. 1993, 46, 197–199. [Google Scholar] [CrossRef]
- Mangiacotti, M.; Martín, J.; López, P.; Reyes-Olivares, C.V.; Rodríguez-Ruiz, G.; Coladonato, A.J.; Scali, S.; Zuffi, M.A.L.; Sacchi, R. Proteins from Femoral Gland Secretions of Male Rock Lizards Iberolacerta cyreni Allow Self—But Not Individual—Recognition of Unfamiliar Males. Behav. Ecol. Sociobiol. 2020, 74, 68. [Google Scholar] [CrossRef]
- Martín, J.; López, P. Pheromones and Reproduction in Reptiles. In Hormones and Reproduction of Vertebrates; Lopez, K.H., Norris, D.O., Eds.; Academic Press: London, UK, 2011; pp. 141–167. ISBN 978-0-12-374930-7. [Google Scholar]
- Alberts, A.C.; Phillips, J.A.; Werner, D.I. Sources of Intraspecific Variability in the Protein Composition of Lizard Femoral Gland Secretions. Copeia 1993, 1993, 775–781. [Google Scholar] [CrossRef]
- Ibáñez, A.; Skupien-rabian, B.; Jankowska, U.; Kędracka-krok, S.; Zając, B. Functional Protein Composition in Femoral Glands of Sand Lizards (Lacerta agilis). Molecules 2022, 27, 2371. [Google Scholar] [CrossRef]
- Wyatt, T.D. Proteins and Peptides as Pheromone Signals and Chemical Signatures. Anim. Behav. 2014, 97, 273–280. [Google Scholar] [CrossRef]
- Tellkamp, F.; Lang, F.; Ibáñez, A.; Abraham, L.; Quezada, G.; Günther, S.; Looso, M.; Tann, F.J.; Müller, D.; Cemic, F.; et al. Proteomics of Galápagos Marine Iguanas Links Function of Femoral Gland Proteins to the Immune System. Mol. Cell. Proteom. 2020, 19, 1523–1532. [Google Scholar] [CrossRef]
- Mangiacotti, M.; Baeckens, S.; Scali, S.; Martín, J.; Van Damme, R.; Sacchi, R. Evolutionary and Biogeographical Support for Species-Specific Proteins in Lizard Chemical Signals. Biol. J. Linn. Soc. 2021, 134, 912–928. [Google Scholar] [CrossRef]
- García-Roa, R.; Jara, M.; Baeckens, S.; López, P.; Van Damme, R.; Martín, J.; Pincheira-Donoso, D. Macroevolutionary Diversification of Glands for Chemical Communication in Squamate Reptiles. Sci. Rep. 2017, 7, 9288. [Google Scholar] [CrossRef]
- Martín, J.; Castilla, A.M.; López, P.; Al-Jaidah, M.; Al-Mohannadi, S.F.; Al-Hemaidi, A.A.M. Chemical Signals in Desert Lizards: Are Femoral Gland Secretions of Male and Female Spiny-Tailed Lizards, Uromastyx aegyptia microlepis Adapted to Arid Conditions? J. Arid. Environ. 2016, 127, 192–198. [Google Scholar] [CrossRef]
- Khannoon, E.R.; Flachsbarth, B.; El-Gendy, A.; Mazik, K.; Hardege, J.D.; Schulz, S. New Compounds, Sexual Differences, and Age-Related Variations in the Femoral Gland Secretions of the Lacertid Lizard Acanthodactylus boskianus. Biochem. Syst. Ecol. 2011, 39, 95–101. [Google Scholar] [CrossRef]
- Garfin, D.E. One-Dimensional Gel Electrophoresis. In Methods in Enzymology; Burgess, R.R., Deutscher, M.P.B.T., Eds.; Academic Press: Cambridge, MA, USA, 2009; Volume 463, pp. 497–513. ISBN 978-0-12-374536-1. [Google Scholar]
- Alberts, A.C. Phylogenetic and Adaptive Variation in Lizard Femoral Gland Secretions. Copeia 1991, 1991, 69–79. [Google Scholar] [CrossRef]
- Eng, J.K.; Searle, B.C.; Clauser, K.R.; Tabb, D.L. A Face in the Crowd: Recognizing Peptides Through Database Search. Mol. Cell. Proteom. 2011, 10, R111.009522. [Google Scholar] [CrossRef]
- Lovich, J.E.; Gibbons, J.W. Review of Techniques for Quantifying Sexual Size Dimorphism. Growth Dev. Aging 1992, 56, 269–281. [Google Scholar]
- Smith, R.J. Statistics of Sexual Size Dimorphism. J. Hum. Evol. 1999, 36, 423–458. [Google Scholar] [CrossRef]
- Braña, F. Sexual Dimorphism in Lacertid Lizards: Male Head Increase vs Female Abdomen Increase? Oikos 1996, 75, 511. [Google Scholar] [CrossRef]
- Cox, R.M.; Skelly, S.L.; John-Alder, H.B. A Comparative Test of Adaptive Hypotheses for Sexual Size Dimorphism in Lizards. Evolution 2003, 57, 1653–1669. [Google Scholar] [CrossRef] [PubMed]
- Sacchi, R.; Mangiacotti, M.; Scali, S.; Sannolo, M.; Zuffi, M.A.L.; Pellitteri-Rosa, D.; Bellati, A.; Galeotti, P.; Fasola, M. Context-Dependent Expression of Sexual Dimorphism in Island Populations of the Common Wall Lizard (Podarcis muralis). Biol. J. Linn. Soc. 2015, 114, 552–565. [Google Scholar] [CrossRef]
- Sacchi, R.; Pupin, F.; Gentilli, A.; Rubolini, D.; Scali, S.; Fasola, M.; Galeotti, P. Male-Male Combats in a Polymorphic Lizard: Residency and Size, but Not Color, Affect Fighting Rules and Contest Outcome. Aggress. Behav. 2009, 35, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Titone, V.; Marsiglia, F.; Mangiacotti, M.; Sacchi, R.; Scali, S.; Zuffi, M.A.L. Better to Be Resident, Larger or Coloured? Experimental Analysis on Intraspecific Aggression in the Ruin Lizard. J. Zool. 2018, 304, 260–267. [Google Scholar] [CrossRef]
- Gvoždík, L.; Van Damme, R. Evolutionary Maintenance of Sexual Dimorphism in Head Size in the Lizard Zootoca vivipara: A Test of Two Hypotheses. J. Zool. 2003, 259, 7–13. [Google Scholar] [CrossRef]
- Huyghe, K.; Vanhooydonck, B.; Scheers, H.; Molina-Borja, M.; Van Damme, R. Morphology, Performance and Fighting Capacity in Male Lizards, Gallotia galloti. Funct. Ecol. 2005, 19, 800–807. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. Ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Clavel, J.; Aristide, L.; Morlon, H. A Penalized Likelihood Framework for High-Dimensional Phylogenetic Comparative Methods and an Application to New-World Monkeys Brain Evolution. Syst. Biol. 2019, 68, 93–116. [Google Scholar] [CrossRef]
- Aitchison, J. The Statistical Analysis of Compositional Data. J. R. Stat. Society. Ser. B (Methodol.) 1982, 44, 139–177. [Google Scholar] [CrossRef]
- van den Boogaart, K.G.; Tolosana-Delgado, R. Analyzing Compositional Data with R; Springer: Berlin/Heidelberg, Germany, 2013; Volume 122, ISBN 978-3-642-36809-7. [Google Scholar]
- Clavel, J.; Morlon, H. Reliable Phylogenetic Regressions for Multivariate Comparative Data: Illustration with the MANOVA and Application to the Effect of Diet on Mandible Morphology in Phyllostomid Bats. Syst. Biol. 2020, 69, 927–943. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Clavel, J.; Escarguel, G.; Merceron, G. MvMORPH: An R Package for Fitting Multivariate Evolutionary Models to Morphometric Data. Methods Ecol. Evol. 2015, 6, 1311–1319. [Google Scholar] [CrossRef]
- Mangiacotti, M.; Fumagalli, M.; Cagnone, M.; Viglio, S.; Bardoni, A.M.; Scali, S.; Sacchi, R. Morph-Specific Protein Patterns in the Femoral Gland Secretions of a Colour Polymorphic Lizard. Sci. Rep. 2019, 9, 8412. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I. Proteogenomics: Concepts, Applications and Computational Strategies. Nat. Methods 2014, 11, 1114–1125. [Google Scholar] [CrossRef]
- Kim, S.; Gupta, N.; Pevzner, P.A. Spectral Probabilities and Generating Functions of Tandem Mass Spectra: A Strike against Decoy Databases. J. Proteome Res. 2008, 7, 3354–3363. [Google Scholar] [CrossRef]
- Kim, S.; Pevzner, P.A. MS-GF+ Makes Progress towards a Universal Database Search Tool for Proteomics. Nat. Commun. 2014, 5, 5277. [Google Scholar] [CrossRef]
- Creasy, D.M.; Cottrell, J.S. Unimod: Protein Modifications for Mass Spectrometry. Proteomics 2004, 4, 1534–1536. [Google Scholar] [CrossRef]
- Schittmayer, M.; Fritz, K.; Liesinger, L.; Griss, J.; Birner-Gruenberger, R. Cleaning out the Litterbox of Proteomic Scientists Favorite Pet: Optimized Data Analysis Avoiding Trypsin Artifacts. J. Proteome Res. 2016, 15, 1222–1229. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Pedersen, T. MzID: An MzIdentML Parser for R, v. 1.28.0. 2021. [CrossRef]
- Pagès, H.; Aboyoun, P.; Gentleman, R.; DebRoy, S. Biostrings: Efficient Manipulation of Biological Strings, R package version 2.46.0. 2021. [CrossRef]
- Carpentler, S.C.; Panis, B.; Vertommen, A.; Swennen, R.; Sergeant, K.; Renaut, J.; Laukens, K.; Witters, E.; Samyn, B.; Devreese, B. Proteome Analysis of Non-Model Plants: A Challenging but Powerful Approach. Mass. Spectrom. Rev. 2008, 27, 354–377. [Google Scholar] [CrossRef] [PubMed]
- Everett, L.J.; Bierl, C.; Master, S.R. Unbiased Statistical Analysis for Multi-Stage Proteomic Search Strategies. J. Proteome Res. 2010, 9, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Schäffer, A.A.; Aravind, L.; Madden, T.L.; Shavirin, S.; Spouge, J.L.; Wolf, Y.I.; Koonin, E.V.; Altschul, S.F. Improving the Accuracy of PSI-BLAST Protein Database Searches with Composition-Based Statistics and Other Refinements. Nucleic Acids Res. 2001, 29, 2994–3005. [Google Scholar] [CrossRef] [PubMed]
- Frost, S.C. Physiological Functions of the Alpha Class of Carbonic Anhydrases. In Carbonic Anhydrase: Mechanism, Regulation, Links to Disease, and Industrial Applications; Frost, S.C., McKenna, R., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 9–30. ISBN 978-94-007-7359-2. [Google Scholar]
- Kopena, R.; Martín, J.; López, P.; Herczeg, G. Vitamin E Supplementation Increases the Attractiveness of Males’ Scent for Female European Green Lizards. PLoS ONE 2011, 6, e19410. [Google Scholar] [CrossRef]
- Martín, J.; López, P. Chemoreception, Symmetry and Mate Choice in Lizards. Proc. R. Soc. B Biol. Sci. 2000, 267, 1265–1269. [Google Scholar] [CrossRef]
- Martín, J.; López, P. Links between Male Quality, Male Chemical Signals, and Female Mate Choice in Iberian Rock Lizards. Funct. Ecol. 2006, 20, 1087–1096. [Google Scholar] [CrossRef]
- López, P.; Martín, J. Chemosensory Exploration of Male Scent by Female Rock Lizards Result from Multiple Chemical Signals of Males. Chem. Senses 2012, 37, 47–54. [Google Scholar] [CrossRef] [PubMed]
- López, P.; Martín, J.; Cuadrado, M. Pheromone-Mediated Intrasexual Aggression in Male Lizards, Podarcis hispanicus. Aggress. Behav. 2002, 28, 154–163. [Google Scholar] [CrossRef]
- Font, E.; Barbosa, D.; Sampedro, C.; Carazo, P. Social Behavior, Chemical Communication, and Adult Neurogenesis: Studies of Scent Mark Function in Podarcis Wall Lizards. Gen. Comp. Endocrinol. 2012, 177, 9–17. [Google Scholar] [CrossRef]
- Carazo, P.; Font, E.; Desfilis, E. Chemosensory Assessment of Rival Competitive Ability and Scent-Mark Function in a Lizard, Podarcis hispanica. Anim. Behav. 2007, 74, 895–902. [Google Scholar] [CrossRef]
- López, P.; Martín, J. Chemical Rival Recognition Decreases Aggression Levels in Male Iberian Wall Lizards, Podarcis hispanica. Behav. Ecol. Sociobiol. 2002, 51, 461–465. [Google Scholar] [CrossRef]
- Martín, J.; Moreira, P.L.; López, P. Status-Signalling Chemical Badges in Male Iberian Rock Lizards. Funct. Ecol. 2007, 21, 568–576. [Google Scholar] [CrossRef]
- Whiting, M.J.; While, G.M. Sociality in Lizards. In Comparative Social Evolution; Rubenstein, D.R., Abbot, P., Eds.; Cambridge University Press: Cambridge, MA, USA, 2017; pp. 354–389. ISBN 978-1-107-33831-9. [Google Scholar]
- Kikuyama, S.; Toyoda, F.; Ohmiya, Y.; Matsuda, K.; Tanaka, S.; Hayashi, H. Sodefrin: A Female-Attracting Peptide Pheromone in Newt Cloacal Glands. Science 1995, 267, 1643–1645. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, F.; Yamamoto, K.; Iwata, T.; Hasunuma, I.; Cardinali, M.; Mosconi, G.; Polzonetti-Magni, A.M.; Kikuyama, S. Peptide Pheromones in Newts. Peptides 2004, 25, 1531–1536. [Google Scholar] [CrossRef]
- Mucignat-Caretta, C.; Caretta, A. Message in a Bottle: Major Urinary Proteins and Their Multiple Roles in Mouse Intraspecific Chemical Communication. Anim. Behav. 2014, 97, 255–263. [Google Scholar] [CrossRef]
- López, P.; Martin, J. Sexual Selection and Chemoreception in Lacertid Lizards. In The Biology of Lacertid Lizards. Evolutionary and Ecological Perspectives; Pérez-Mellado, V., Riera, V., Perera, A., Eds.; Recerca; Institut Menorquí d’Estudis: Balearic Islands, Spain, 2004; pp. 119–137. [Google Scholar]
- Tibbetts, E.A.; Dale, J. Individual Recognition: It Is Good to Be Different. Trends Ecol. Evol. 2007, 22, 529–537. [Google Scholar] [CrossRef]
- McKenna, R.; Frost, S.C. Overview of the Carbonic Anhydrase Family. In Carbonic Anhydrase: Mechanism, Regulation, Links to Disease, and Industrial Applications; Frost, S.C., McKenna, R., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 3–5. ISBN 978-94-007-7359-2. [Google Scholar]
- Supuran, C.T. Structure and Function of Carbonic Anhydrases. Biochem. J. 2016, 473, 2023–2032. [Google Scholar] [CrossRef]
- Kitade, K.; Nishita, T.; Yamato, M.; Sakamoto, K.; Hagino, A.; Katoh, K.; Obara, Y. Expression and Localization of Carbonic Anhydrase in Bovine Mammary Gland and Secretion in Milk. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2003, 134, 349–354. [Google Scholar] [CrossRef]
- Yrjänäinen, A.; Patrikainen, M.S.; Azizi, L.; Tolvanen, M.E.E.; Laitaoja, M.; Jänis, J.; Hytönen, V.P.; Nocentini, A.; Supuran, C.T.; Parkkila, S. Biochemical and Biophysical Characterization of Carbonic Anhydrase VI from Human Milk and Saliva. Protein J. 2022, 41, 489–503. [Google Scholar] [CrossRef]
- Lazar, J.; Rasmussen, L.E.L.; Greenwood, D.R.; Bang, I.S.; Prestwich, G.D. Elephant Albumin: A Multipurpose Pheromone Shuttle. Chem. Biol. 2004, 11, 1093–1100. [Google Scholar] [CrossRef]
- Hurst, J.L. Female Recognition and Assessment of Males through Scent. Behav. Brain Res. 2009, 200, 295–303. [Google Scholar] [CrossRef] [PubMed]
- García-Roa, R.; Jara, M.; López, P.; Martín, J.; Pincheira-Donoso, D. Heterogeneous Tempo and Mode of Evolutionary Diversification of Compounds in Lizard Chemical Signals. Ecol. Evol. 2017, 7, 1286–1296. [Google Scholar] [CrossRef] [PubMed]
- Mangiacotti, M.; Baeckens, S.; Fumagalli, M.; Martín, J.; Scali, S.; Sacchi, R. Protein–Lipid Association in Lizard Chemical Signals. Integr. Org. Biol. 2023, 5, obad016. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.; López, P. Vitamin D Supplementation Increases the Attractiveness of Males’ Scent for Female Iberian Rock Lizards. Proc. R. Soc. B Biol. Sci. 2006, 273, 2619–2624. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.M.; Butler, M.A.; John-Alder, H.B. The Evolution of Sexual Size Dimorphism in Reptiles. In Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism; Fairbairn, D.J., Blanckenhorn, W.U., Székely, T., Eds.; Oxford University Press: Oxford, UK, 2007; ISBN 978-0-19-920878-4. [Google Scholar]
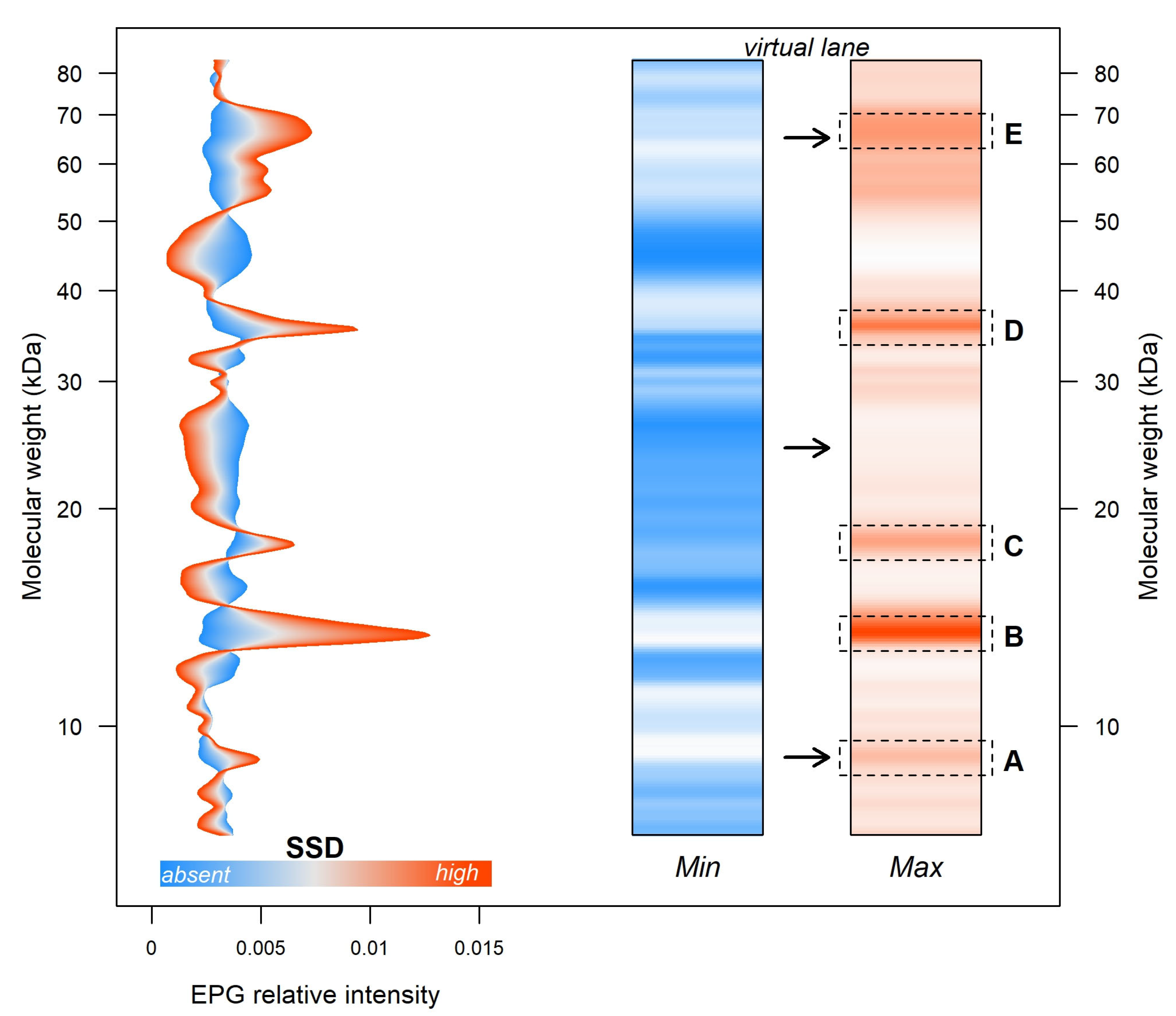
| Band | ID | Peptide | Score | Error | UniProtID | Description | Coverage (%) | MW |
|---|---|---|---|---|---|---|---|---|
| B | 3544 | SCIDTELCDVGYGSASITSSMYIQSK | 9.374 | 0.036 | A0A670IP55 | UPAR/Ly6 domain-containing protein | 17.81 | 15.8 |
| 2948 | AHDGIR | 8.026 | −0.009 | A0A670JC88 | Zinc finger protein 436-like | 1.16 | 58.8 | |
| 2948 | AHDGLR | 8.026 | −0.009 | A0A670JYQ1 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 | 0.61 | 109.1 | |
| C | 3168 | QMIKINFK | 8.341 | −0.005 | A0A670KI84 | COP9 signalosome complex subunit 2 | 1.78 | 52.4 |
| D | 3157 | YSMELHIVHTK | 14.856 | 0.005 | A0A670JE51 | Carbonic anhydrase | 4.25 | 28.4 |
| Band | ID | Peptide | Score | Error | UniProtID | Identity (%) | Description | Organism | Coverage (%) | MW |
|---|---|---|---|---|---|---|---|---|---|---|
| B | 3544 | SCIDTELCDVGYGSASITSSMYIQSK | 9.434 | 0.036 | A0A670IP55 | 100.0 | UPAR/Ly6 domain- containing protein | Podarcis muralis | 17.81 | 15.8 |
| 2908 | REERPR | 5.558 | −0.008 | A0A8C5QXT2 | 37.4 | Leptobrachium leishanense | 2.76 | 23.1 | ||
| D | 3157 | YSMELHIVHTK | 14.836 | 0.005 | A0A670JE51 | 100.0 | Carbonic anhydrase | Podarcis muralis | 4.25 | 28.4 |
| 3184 | EPITHYIPACRQVNR | 6.191 | 0.970 | A0A8C1L806 | 41.4 | Cyprinus carpio | 4.44 | 36.9 | ||
| 2893 | MELHVVNK | 6.191 | 1.020 | A0A6P6QLI1 | 38.2 | Carassius auratus | 2.48 | 35.8 | ||
| 3816 | ANDSSALAVLGFFIEGTDEADK | 5.667 | 0.982 | Q08C20 | 38.6 | Danio rerio | 6.79 | 35.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mangiacotti, M.; Baeckens, S.; Fumagalli, M.; Martín, J.; Scali, S.; Sacchi, R. Sexual Selection and Proteinaceous Diversity in the Femoral Gland Secretions of Lacertid Lizards. Diversity 2023, 15, 777. https://doi.org/10.3390/d15060777
Mangiacotti M, Baeckens S, Fumagalli M, Martín J, Scali S, Sacchi R. Sexual Selection and Proteinaceous Diversity in the Femoral Gland Secretions of Lacertid Lizards. Diversity. 2023; 15(6):777. https://doi.org/10.3390/d15060777
Chicago/Turabian StyleMangiacotti, Marco, Simon Baeckens, Marco Fumagalli, José Martín, Stefano Scali, and Roberto Sacchi. 2023. "Sexual Selection and Proteinaceous Diversity in the Femoral Gland Secretions of Lacertid Lizards" Diversity 15, no. 6: 777. https://doi.org/10.3390/d15060777
APA StyleMangiacotti, M., Baeckens, S., Fumagalli, M., Martín, J., Scali, S., & Sacchi, R. (2023). Sexual Selection and Proteinaceous Diversity in the Femoral Gland Secretions of Lacertid Lizards. Diversity, 15(6), 777. https://doi.org/10.3390/d15060777







