Abstract
Rain-fed mountain granite rock basins are temporary habitats conditioned by a fluctuating environment and the unpredictability of precipitation or flooding rates. These small highland freshwater habitats remain largely unexplored at the microbial level. The aim of this work is to report the presence in these habitats of genetic sequences of microbial eukaryotes that are pathogens and potential pathogens of humans, wildlife, cattle, crops as well as of other microorganisms. We sequenced the hypervariable region v4 of the 18S rDNA gene from environmental DNA of sediments taken from 21 rock basins in a National Park in Spain. More than a fifth (21%) of the eukaryotic Operational Taxonomic Units (OTUs) found are ascribed to pathogenic (within 11 Phyla) and potential pathogenic (within 1 phylum, the Chytridiomycota) microorganisms. Some OTUs retrieved are of agro-economic and public health importance (e.g., Pythium spp., Lagenidium spp., Candida spp. and Vermamoeba vermiformis). In 86% of the basins, the most abundant OTUs were affiliated to Chytridiomycota, a broad fungal group including saprozoic and parasitic taxa. Two OTUs affiliated to chytrids were significantly correlated with high concentrations of heavy metals. The high proportion of chytrid-like microbial sequences found emphasises the role of these freshwater habitats for adding knowledge regarding the ecological trade-offs of the still rather unknown Chytridiomycota. Our results show that rain-fed rock basins may be model habitats for the study and surveillance of microbial community dynamics and genetics of (mainly opportunistic) microbial pathogens.
1. Introduction
The pathogenic (including parasitic and parasitoid) lifestyle is one of the earliest and most widespread strategies in nature, occurring in almost all environments [1,2,3]. Pathogens are drivers of evolutionary processes. They can promote genetic diversity and speciation in host populations, coevolving with hosts [4,5,6,7,8]. Pathogens have a pivotal role in ecological interactions, as they are involved in many trophic links. They control host population dynamics, species succession, competition for resources and energy flows [1,9,10]. Moreover, pathogens can also be preyed upon by other organisms [11,12]. The identification of pathogenic organisms including parasites is therefore important for biodiversity conservation and to predict the future distribution of diseases. Host–pathogens dynamics are complex in many microbial groups. Changes in ecological conditions can induce a switch in the lifestyle in these groups from a pathogenic to a saprotrophic form and vice versa [1]. Moreover, some fungi traditionally regarded as saprophytic environmental fungi and nonpathogenic have virulence factors similar to those of their pathogenic relatives, and are described as emerging pathogens [13]. The drivers that underlie these dynamics remain challenging to characterise because most potential pathogens have high ecological versatility and still unresolved taxonomy [14,15,16,17]. At present, there is no reliable estimate of the number (species richness and abundance) of microbial pathogens occurring in natural ecosystems [18].
In recent years, new fungi and fungus-like protists that cause disease in fish, amphibians, reptiles, mammals and microorganisms were discovered [19,20,21]. Hidden diversity, including parasitic lifestyle, was revealed by 18S rDNA environmental surveys, especially within the picoplanktonic size-fraction [16,22,23]. Microbial eukaryotic parasites were found in pristine oligotrophic habitats [24], in neotropical forests [7], in mountain lakes [25] and in the oceans [17]. Well-known groups of microbial eukaryotes that are pathogens or may frequently include pathogens were shown to be particularly diverse in the environment, including the Apicomplexa [7,26], Peronosporomycetes (traditionally Oomycota), [24,27], Perkinsea [28] and Chytridiomycota [15]. Their succession and dynamics of co-occurrence with hosts were also studied through environmental DNA surveys [26,29]. Chytridiomycota include a large number of known parasites and are considered probably the dominant group of microbial pathogens in freshwater ecosystems worldwide [20,30]. They can act both as a food source for zooplankton and control phytoplankton dynamics and blooms, and thus the primary production of aquatic systems (see [20] for a review). Environmental DNA surveys can provide a new insight into their diversity and some hints on their relative abundance. In that respect, a wider variety of microbial habitats need to be explored worldwide to obtain a comprehensive understanding of the diversity and role of potential pathogens in nature.
Freshwater mountain ecosystems are prone to climate vulnerability and hydrological extremes [31]: they are subject to temperatures that cause both freezing and desiccation. Mountain granite rock basins are temporary freshwater habitats usually on top of granite outcrops and conditioned by the precipitation regime. The largely fluctuating environment and the unpredictability of the flooding rates subject the biological communities growing in these habitats to high variability and are adapted to survive the dry periods by the production of resistant stages or by active or passive migration [32,33]. These small highland freshwater habitats remain largely unexplored at the microbial level, especially in comparison to large waterbodies [25,34,35].
We have recently shown the high eukaryotic microbial heterogeneity and diversity at the community level in the sediments of ombrotrophic mountain rock basins, i.e., which receive all of their water and nearly all nutrients from precipitation, rather than from streams or springs. Eukaryotic microbial communities in the majority of pools were composed of opportunistic species with a great tendency to enter dormancy to withstand adverse conditions [36].
In this study, we focus on the pathogenic treats of these communities in the rock basin sediments. The aim of the work is to display the diversity and distribution of the microbial OTUs with both pathogenic and potential pathogenic activity on humans, wildlife, cattle, crops as well as on other microorganisms. Microbes with effective pathogenic potential must have successful mechanisms of dispersal and resistance in order to find hosts in adverse habitats. The ability to form spores as vehicles of passive dispersal mediated by outside forces, i.e., the movement of water, wind, soils, animals or even other microbes [37], as well as the capacity for active dispersal in response to a rapid change in resource (host) availability, for example, are an advantage in habitats subjected to highly fluctuating conditions. It was proposed that some pathogenicity-associated traits may be a result of fungal adaptation to growing under highly variable, and sometimes extreme, biotic and abiotic stress [38]; and significant associations were found between the ability to survive in multiple different types of extreme conditions and opportunistic pathogenicity [39].
We therefore hypothesise that pathogenic eukaryotic microorganisms with the ability to form spores should be numerous and diverse in the sediments deposited in the mountain outcrop basins studied, as these are habitats often subjected to drastic seasonal and diurnal environmental stress.
2. Material and Methods
2.1. Habitats Studied and Sample Collection
The ombrotrophic basins studied are located in La Pedriza del Manzanares (UTM: 30N 425279 4511417, DATUM: ETRS89), within the Sierra de Guadarrama National Park (Madrid, Spain). Twenty-one basins (20 active, temporarily flooded basins and one nonactive basin as control) were randomly selected in an area of 0.2 km2 with a variation between basins of 170 m in altitude (Figure 1). A detailed description of the area and the morphometric characterisation of the basins are presented in [36]. Samples of the sediments were taken after summer (September–October 2015). Sediment of each basin was homogenised with a spatula, and one sample per basin was taken and stored in sterile polypropylene containers. In the laboratory, they were spread on sterile Petri dishes protected from light and left to dry further at room temperature (21 °C ± 0.2) until DNA analysis.
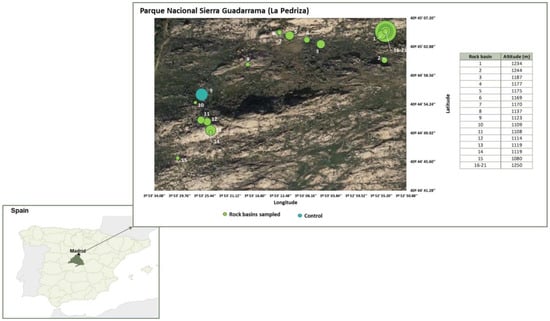
Figure 1.
Location of the rock basins (1–21) sampled in the Sierra de Guadarrama National Park (La Pedriza). Basins are represented by their relative areas versus the basin with the largest area (basin 17). Altitude is shown for each basin.
2.2. Chemical Analyses
At each basin, ~4 g of additional sediment was collected with a sterile flask to determine the following variables (analytical methods used are indicated in parentheses): total organic carbon (combustion analysis method and nondispersive infrared detection), Kjeldahl nitrogen (Kjeldahl method), anions—phosphates, nitrites, nitrates, sulphates, chlorides (ionic chromatography)—and metals (inductively coupled plasma-optic emission spectrometry—ICP-OES).
2.3. DNA Extraction and Sequencing
The molecular approach is described in detail in [36]. Briefly, environmental DNA was extracted with the PowerSoil® DNA Isolation Kit (MoBIO, Carlsbad, CA, USA) according to the manufacturer’s instructions. The V4 variable region of the 18S rRNA gene was amplified using the universal primers V4F (TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCAGCASCYGCGGTAATTCC) and V4R (GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGACTTTCGTTCTGATYRATGA) [40]. PCR amplifications and purification were according to the manufacturer’s instructions. The reads were obtained using a MiSeq (Illumina) platform (Fasteris, Geneva, Switzerland).
2.4. Bioinformatic Analyses
We followed the bioinformatics pipeline developed by [17] to: (1) correct sequencing errors, (2) find reads without rDNA sequence and (3) remove chimeras and other erroneous sequences. In order to reduce noise, singletons were also eliminated [41]. The remaining reads were clustered into OTUs using the algorithm SWARM v2 [42]. The dominant reads of each OTU were taxonomically assigned using the GGSearch algorithm from the FASTA package [43] using both the curated ribosomal eukaryotic database PR2 [44] and the SILVA bacterial and archaeal database. Microbial OTUs with a sequence similarity threshold (OTUs vs. PR2) of ≥90% were selected after eliminating the Prokaryotic, Metazoa and Embryophyta OTUs [36].
2.5. Taxonomic Classification of Pathogen and Potentially Pathogenic Microorganisms
The original database of microbial eukaryotic OTUs was analysed, one by one, to select the OTUs affiliated to pathogenic and potentially pathogenic microorganisms. Due to the drastic effect of pathogenic chytrids on the global biodiversity of many species [45], and with the aim of not missing any possible pathogenic sequence, a conservative criterion was used for the phylum Chytridiomycota: all the chytrid sequences were selected and included as “potential microbial pathogens sensu lato” for our analysis, as it was shown that in this phylum there may be a continuum between saprotrophic and pathogenic/parasite style of life depending on environmental conditions [1].
With the selected OTUs, we created a new database where the OTUs were classified in function of: (i) their status as either fungi or protists; (ii) their affiliation to the taxonomic level of phylum; (iii) their potential host (“Vertebrate”, “Invertebrate”, “Plant”, “Microbial Eukaryote”, “Unknown”). An additional category was created for those per se nonparasitic OTUs but that are known to act as a “Host” of pathogenic microorganisms (i.e., some Amoebozoa OTUs).
The complete database of pathogenic OTUS and potential pathogenic OTUs (Chytridiomycota) is included in the Supplementary Materials file (Table S1). All V4-rDNA sequences are deposited at the European Nucleotide Archive (Reference number PRJEB24091).
2.6. Statistical Analyses
Alpha diversity (Gini–Simpson, Shannon–Weaver and Evenness indexes) and beta diversity estimates were computed based on raw reads using the R packages Vegan 2.5-2 and Beta part 1.5.0. For beta diversity, both incidence-based and abundance-based dissimilarities were calculated according to [46,47], respectively. Distance-decay similarity was obtained for both components of beta diversity following [48]. The frequency of desiccation of the basins was measured indirectly using the Area/Volume (A/V) ratio (the higher the A/V, the higher the evaporation rate and the probability of desiccation, [49,50]). Student’s t-test was applied to compare the alpha diversity indexes and richness in two groups of basins according to their A/V ratio (basin with A/V ≥ 1 and basins with A/V < 1). To visualise comparatively the intra- and interbasin proportions of OTU richness and abundance, mosaic charts were produced with R mekko package (vs 0.1.0). A Marimekko/Mosaic chart is a variable-width (axis x) plot in which all the bars are of equal height (axis y). Both axes represent categorical data and are shown relative to 100%. Canonical correspondence analysis (CCA) was performed (package Vegan 2.5-2) to relate the abundance of the OTUs present in all the basins to the matrix of chemical (explanatory) variables. These last were split into three groups in function of their correlative content in the basins: micronutrients (Zn, Na, Al, Mn, Fe, Ti, Si), heavy metals (Cr, Ni, Cu, Pb) and macronutrients (C, N, P, S, Mg). Significance was obtained by permutation tests (number of permutations: 2000) for both the whole model and the CCA axes. The significance of the CCA axes was examined further only if the overall CCA model was found to be significant after permutation. Analyses were conducted using the R software (R Development Core Team, 2013) (vs 3.5.0).
3. Results
3.1. Chemical Characterisation of Rock Basins
A heatmap representing the relative abundance of the chemical elements analysed is illustrated in Figure 2. Element values are detailed in Table S2. Generally, basins had high values of nitrogen and phosphorous, which are characteristic for eutrophic environments. No distinctive pattern was observed in the chemical composition for any of the basins (or groups of basins), apart from the clear separation of basin 9 (the control basin, as its depth equals zero and therefore cannot hold water) from the rest of the basins by its very different chemical profile (Figure 2). Most of the metal concentrations were within the certified values for granite constituents (http://www.mintek.co.za, accessed on 19 January March 2021), except for Ti, Cu, Ni, Pb and Cr (Figure 2). Cu, Ni, Cr and Pb were present in the majority of the sediments, and their concentration in some of the basins exceeded the reference values permitted in soils [51]. High concentrations of these metals may have a potential toxic effect on microorganisms and for biological communities.
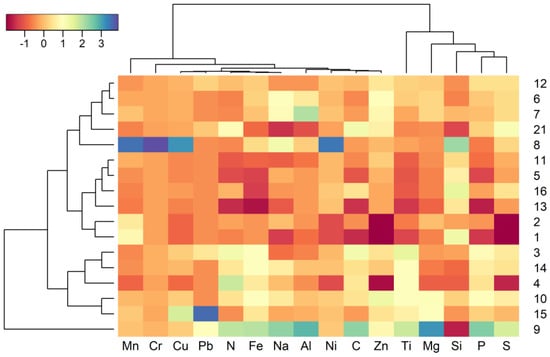
Figure 2.
Heatmap depicting the relative values of the elemental concentrations in each rock basin. Only the basins with recorded values for all the parameters were included. Elemental concentrations were standardised between basins (mean = 0, sd = 1), so colour scale represents standardised values of each element at each basin. Dendrograms represent the clustering of rock basins (left dendrogram) or chemical elements (top dendrogram) Euclidean distances (hierarchical clustering with complete agglomerative method in both cases).
3.2. Community Structure of Pathogenic and Potentially Pathogenic OTUs
Potential pathogenic microorganisms represented 158,127 reads. These reads accounted for 15% of all microbial V4 reads found in the basins (1,059,658 reads; see [36]). The reads were clustered into 577 OTUs, representing 21% of the total microbial OTUs (2761 OTUs; see [36]), of which 54% belonged to pathogens and 46% belonged to the only group classified as potential pathogens, Chytridiomycota. Rarefaction curves showed that the OTU richness was not wholly retrieved, as the curves did not reach saturation in any of the basins (Figure S1). The distribution of the richness and abundance of OTUs (Figure 3) was nonuniform in the rock basins (Chi-squared = 357.5; d.f = 20; p-value < 10−4 for differences in OTU richness; K-W = 467.7; d.f = 20; p-value < 10−4 for differences in reads), with contrasting values ranging from 270 reads in basin 18 to 21,295 reads in basin 3. OTU richness was also highly variable, from 61 OTUs in basin 13 to 219 OTUs in basin 3.
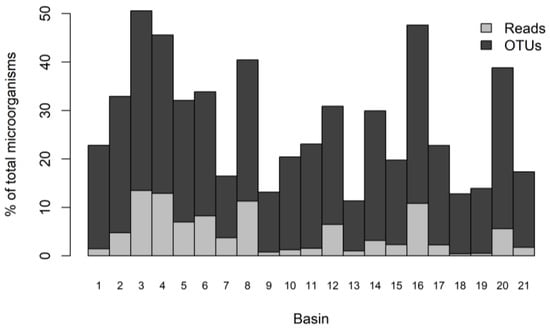
Figure 3.
Relative proportions (%) of reads and number of OTUs of pathogenic and potentially pathogenic microbial eukaryotes in each of the 21 rock basins.
OTUs were classified according to their relative abundance in the basins: Rare (R): OTUs with abundance values ≤ 0.01%; Abundant (A): OTUs with abundance values ≥ 1%; Nonrare nonabundant (NRA): OTUs with abundance values > 0.01% and <1% (based on [52]). Within each category, the average OTU abundance (K-W, p-value < 10−4) and richness (χ2-Square, p-value < 0.05) were not distributed homogeneously in the basins (Figure 4).
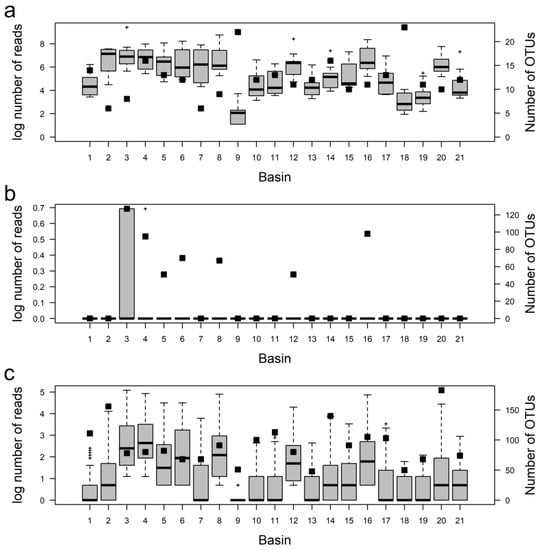
Figure 4.
Variation of the reads (box-plots: median and standard error) and total OTU number (black squares) in the 21 basins for each OTU abundance category. (a) Abundant; (b) Rare; (c) Nonrare nonabundant. See Results for a full description of the categories.
These abundance categories (R, A and NRA) were analysed in function of their frequencies of occurrence in the basins. No potential pathogenic OTU was found to be exclusively abundant (A) when appearing in the basins, while 52.2% of the OTUs (302) were exclusively rare (R). However, more than half of these (57.2%) only appeared in 3 out of the 20 basins studied, and there was not a Rare OTU that appeared in all basins studied. A total of 3.4% of the OTUs (20 OTUs) were found to be exclusively NRA and appeared only in three or fewer basins. A proportion of 44.4% of the total OTUs (257) were found with variable abundances depending on the basin (A, R and NRA, i.e., ARNRA); only 12 of these last OTUs were present in all the basins (Figure 5).
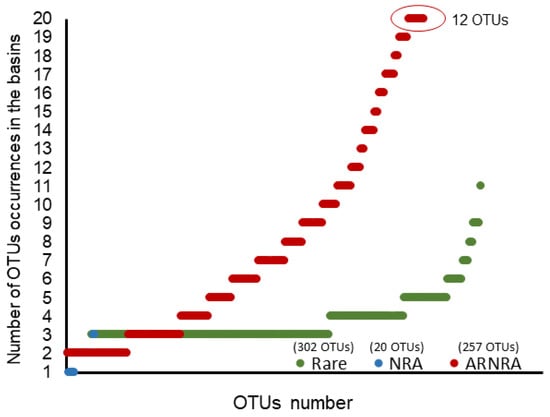
Figure 5.
Number of occurrences of pathogenic and potentially pathogenic microbial eukaryotic OTUs in the basins grouped by abundance. X axis shows the number of OTUs within each abundance category; Y axis shows the number of times (occurrences) the OTUs of each category appeared in the 20 basins (all but the control). Rare: OTUs always within the Rare category. NRA: OTUs always within the Nonrare nonabundant category. ARNRA: OTUs with values in the basins ranging within all three categories (Abundant, Rare and Nonrare nonabundant). See Results section for a detailed description of the categories.
3.3. Taxonomic Affiliation of OTUs and Abundance and Richness of Phyla
Pathogens found in the basins belonged to 11 eukaryotic Phyla, 3 of them within Fungi and 8 within Protists (Figure 6 and Table 1). Fungi were represented by a higher number of OTUs than Protists in all the basins, and their abundances were also higher in 18 out of 21 basins. The potential pathogens were represented for just one phylum, the Chytridiomycota, which was the most abundant group in all but three of the basins. In five of the basins, Ascomycota were the second most abundant group.
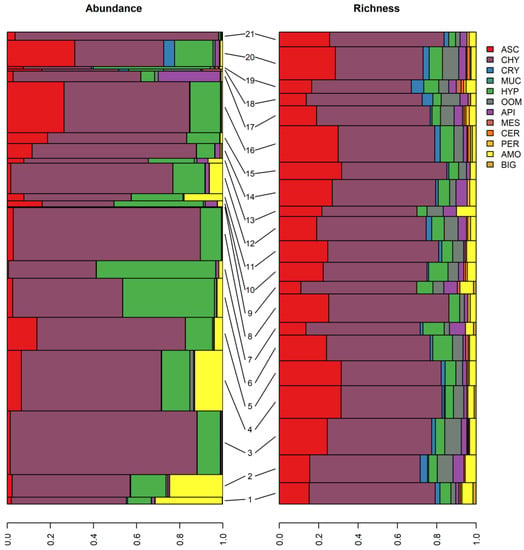
Figure 6.
Marimekko charts showing the relative frequencies in the inter- and intrabasin abundance and richness of pathogenic and potentially pathogenic OTUs (for the 21 basins). Both axes represent categorical data and are shown relative to 100%. ASC: Ascomycota; CHY: Chytridiomycota; CRY: Cryptomycota; MUC: Mucoromycota; HYP: Hyphochytriomycota; OOM: Oomycota; API: Apicomplexa; MES: Mesomycetozoa; CER: Cercozoa; PER: Perkinsozoa; AMO: Amoebozoa; BIG: Bigyra.

Table 1.
Abundance per basin (%), richness and putative host type [Host] of pathogenic and potential microbial pathogenic OTUs. ASC: Ascomycota; CHY: Chytridiomycota; CRY: Cryptomycota; MUC: Mucoromycota; HYP: Hyphochytriomycota; OOM: Oomycota; API: Apicomplexa; MES: Mesomycetozoa; CER: Cercozoa; PER: Perkinsozoa; AMO: Amoebozoa; BIG: Bigyra. Host type: 1: Vertebrate; 2: Invertebrate; 3: Plant; 4: Microbial Eukaryote; 5: Unknown; 0: Human pathogen host.
In terms of OTU richness, Chytridiomycota was also the most represented group, but the second richest were always Ascomycota. Some phyla (Mucoromycota and Bigyra) appeared in very few basins and with very low OTU abundances and richness (Table 1). In two basins, Amoebozoa (here represented by potential “host” species closely related to Vermamoeba vermiformis) were the second most abundant group.
Some of the OTUs had a wide span of potential hosts, as those belonging to the phyla Ascomycota and Oomycota, while others were more host-group specific (Apicomplexa, Mesomycetozoa). For some groups, such as Chytridiomycota, the potential host type could not be named, owing to the lack of taxonomic resolution below family rank.
3.4. Relationship of OTUs with Basin Morphometry and Chemical Parameters
Alpha diversity descriptors were compared between the two groups of basins grouped in function of their Area/Volume (A/V) ratios (A/V ≥ 1; n = 12 basins and A/V < 1; n = 8 basins), used as a proxy of the likelihood of basin desiccation. No statistically significant differences (p-value > 0.05; Student’s t-test) were found for the diversity indexes, which showed very similar values in both groups of basins (Figure 7). However, the basin A/V ratio seemed to affect the OTU richness, as the number of OTUs was correlated with higher frequency of desiccation (p-value < 0.05; Student’s t-test) (Figure 7), as predicted. Any other morphometric indicators of the basins (length, width, depth, area or volume separately) did not show correlation with the alpha diversity metrics of the microbial pathogens (Rho Spearman correlations’ p-value > 0.05; results not shown).
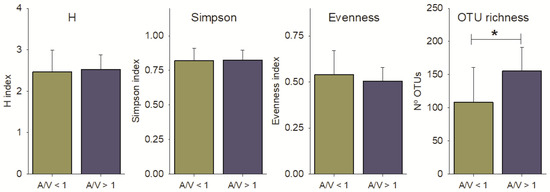
Figure 7.
Values for the diversity indexes of Shannon (H), Simpson, Pielou Evenness and OTUs richness in basins with Area/Volume ratio (A/V) ≥ 1 or basins with A/V < 1. Significance was tested by Student’s t-test; p < 0.05. * means significant differences.
Moreover, both the OTU composition and the chemical variables were ordinated by Nonmetric Multidimensional Scaling (NMDS) in function of the two A/V basin groups (see Figure S3 in Supplementary Materials). Results showed no relationship of any of the two A/V clusters of basins with either the composition in OTUs or the chemical variables, as indicated by the overlapped Ordihull convex figures of both groups of basins (Figure S3).
To evaluate the influence of the chemical environment on the variation of OTU composition, we then performed a canonical correspondence analysis (CCA), first on all nonpathogenic OTUs present in the basins (47 OTUs Table S3, Supplementary Materials), to set up a framework for comparison, and then for the common pathogenic and potential pathogenic OTUs (12 OTUs, highlighted in red in Table S3, Supplementary Materials). The CCA model did not show any significant association between the abundance of nonpathogenic microbial OTUs and the macronutrients (C, N, P, S and Mg) or the heavy metals (Cr, Ni, Cu and Pb) (p-value = 0.34 and p-value = 0.07, respectively, for the CCA models). Nevertheless, the abundance of these microbial OTUs had a significant relation to micronutrients (Na, Al, Mn, Fe, Ti and Si) (p-value = 0.003). Three groups of OTUs could be observed (Figure 8a). Basin 21 is associated with OTU 38 (affiliated to a nonidentified oxytrichid ciliate). Another group clustered with OTUs affiliated to Chlorophyta Stephanosphera sp. (OTU 2), ciliate Halteria grandinella (OTU 8) and Cercozoan Limnofila anglica (OTU 68) is associated with basin numbers 1, 7 and 13. Finally, a third group contained the OTUs related to the streptophyte Staurastrum punctulatum, the ciliate Colpoda magna, the chlorophyte Monomastix opisthostigma and a dinoflagellate (Order Suessiales) (OTUs 49, 50, 67, 78) with the basin number 16. These two last groups of OTUs had their highest abundance for medium-high values of Si, Mn and Al, and medium-low values for Na, Ti and Fe (Figure 8a).
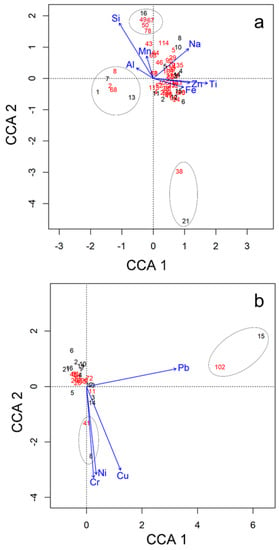
Figure 8.
Statistically significant canonical correspondence analysis (CCA) between the chemicals and the 47 nonpathogenic microbial eukaryotic OTUs present in all the basins (a) and the 12 potentially pathogenic microbial eukaryotic OTUs present in all the basins (b). Colour code: Basins (black), OTUs (red), chemicals (blue).
The CCA model of pathogenic and potentially pathogenic OTUs showed statistically significant association (p-value = 0.002) with some heavy metals (Pb, Cr, Cu and Ni). The OTU 102, related to the chytrid genus Powellomyces, was associated with high Pb concentrations, and the OTU 41, linked to another chytrid genus (Rhizophlyctis), with high levels of Ni, Cr and Cu (Figure 8b).
4. Discussion
This study reveals that about a fifth (21%) of the eukaryotic OTUs present in the basin sediments are affiliated with pathogenic and potential pathogenic microbes, and some of them obligate parasites. Their sequences represent 15% of all microbial reported in these granite basins [36]. These proportions are within the range of values reported in previous studies for other continental microbial habitats. Lepère et al. [53] indicated that up to 30% of the sequences found in a mesotrophic lake came from chytrids and Perkinsozoa. Their results are in line with our study, even if the sequencing methods differ (Sanger vs. Illumina). Another study [54] showed that putative parasites accounted for 6.1% of the reads and 12.4% of the OTUs found in Lake Baikal. Proportions of pathogenic eukaryotic micro-organisms may be even higher, as found by [7] in a study of the molecular diversity of protists in neotropical rainforest soils where the parasitic Apicomplexa were the dominant communities.
The resilience and adaptation of fungi living in extreme habitats, supported by their high genetic plasticity, may cause some of the fungi in harsh environments to develop the capacity to infect other organisms, even in the absence of virulence factors [38,55]. Many fungi display versatile ecological strategies to adapt and survive in environments with extreme temperatures, periods of low nutrient availability, prolonged desiccation and solar irradiation: all these are characteristics of the granite rock basin habitats studied. Therefore, unknown fungi present in unexplored habitats may have potential and emergent pathogenic virulence factors that threaten plants, animals and humans [55]. In fact, most fungi are considered to be facultative parasites (with no need to passage through a host as an obligate step in their life cycle), and whose opportunistic pathogenicity derives mostly from strategies developed to counteract environmental stresses [13]. In this context, most of the pathogens we encountered in these unpredictable habitats could be considered to be opportunists. Ascomycota, Oomycota, Hyphochytridiomycota and Chytridiomycota are composed of opportunistic pathogens that can potentially also live as decomposers. Only the Mesomycetozoa and the Apicomplexa have at least one parasitic life stage (i.e., one life stage obligatorily includes life inside a host); these organisms, however, were generally in low abundance (0.08% and 1.39%, respectively, of all pathogenic reads).
The most abundant reads of pathogenic and potential pathogenic microbes (up to 53%) corresponded to OTUs affiliated with Chytridiomycota. This group is largely composed of opportunistic pathogens of plants, animals and algae, as well as saprozoic species. This corroborates our hypothesis that granitic basins should host a majority of opportunistic pathogens. The approach of including all the Chytridiomycota as ‘potentially pathogens sensu lato’ means that our estimate of the number of pathogenic species within this phylum might possibly be affected by an inclusion bias, that is, classifying a chytrid fungus that cannot be pathogenic as a pathogen. As we are only beginning to understand the many roles of microbes in soils [2,56], the ecological role of many species is not known, and there are many chytrid species which cannot be described as having either pathogenic or nonpathogenic treats, but instead they may be living in a continuum between free living and parasite life strategies [1]. Therefore, we have here applied a conservative approach: if Chytridiomycota includes members that are pathogens, then all other clade members are likely to be at least potentially pathogenic under environmental stress, as is the case of the ecosystems studied. This approach was followed with the aim of not discounting any obligate or facultative species of pathogens within the Chytridiomycota.
The dominance of Chytridiomycota in freshwater habitats was already reported in several studies [15,16,20,30,53,57]. Chytridiomycota are putatively prime decomposers, often specialised in degrading refractory materials such as chitin, keratin and cellulose [58], which is reflected in the opportunistic pathogenic lifestyle that most species seem to have. These microbial fungi have motile, waterborne zoospores that can be resilient through cyclical desiccation periods. There is a growing interest in the study of chytrids due to their role as parasites of vertebrates and phytoplankton. A wealth of recent research has been devoted to phytoplankton chytrid parasites [10,59,60]. Although we did not find sequences of chytrid-amphibian parasites (i.e., Batrachochytridium spp.) in the granitic basins studied, their pathogenesis and negative influence on amphibian population survival were extensively investigated in other habitats of the same National Park studied here [61,62]. Chytrids were reported to be common in high altitude soils decomposing large amounts of pollen and also feeding on dead algae [63]. This may also be the case in the basin sediments studied here, where a high amount of Chlorophyta-like sequences were found [36]. Yet, it was recently suggested that chytrids may turn from decomposers to parasites and vice versa when environmental conditions change [1], which means that many decomposers are “facultative” (potential) pathogens. Undoubtedly, more studies are still needed to understand the ecological role of chytrids as pathogens of other microbial groups and the causes that trigger their pathogenicity [1].
Several other OTUs retrieved from the basins were also of agro-economic and public health importance. We found reads related to Pythium, a genus that include pathogens with a high incidence in crop plants and forest species [64,65], and on zooplankton such as cladocerans [66]. Other OTUs related to Oomycota belonged to Lagedinium. Although most are invertebrate-parasites, sequences such as L. giganteum, which may infect mammals including humans [67], were also detected. The reads affiliated with Apicomplexa corresponded to invertebrate (Gregarines, and Adelina and Stenophora-like sequences) and Eimeria-like OTUs, probably parasitising vertebrates. Regarding Ascomycota, OTUs with 100% homology with species of the genus Candida (C. haemulonis, C. orthopsilosis, C. parapsilosis) were identified; these are opportunistic pathogens that can affect humans [68]. Additionally, in Ascomycota, we retrieved reads related to Chrysosporium indicum, which causes secondary infections in pododermatitis-associated processes in cattle (http://vetbook.org, accessed on 4 December 2021), and phytopathogenic Podosphaera-like sequences. Within the Amoebozoa, OTUs associated with Vermamoeba vermiformis (Hartmanella vermiformis), a naked amoeba described as a host of human pathogenic bacteria and associated with cases of meningoencephalitis and bronchopneumonia, were identified [69,70]. Few and low-abundant OTUs with similarities to the parasitic Mesomycetozoa and Perkinsida were also retrieved from the basins.
Abundance of reads and richness of OTUs were differently distributed among the basins. Beta diversity dissimilarity was mostly due to spatial turnover, that is, to the replacement of OTUs from one basin to another. This dissimilarity was not correlated to the spatial distance between basins (see Supplementary Materials, Figure S2 and Table S4). All these features emulate those shown by our previous study on total microbial OTUs [36] and reflect a similar distribution pattern for free-living and pathogenic microbial communities in mountain rock basin habitats.
We found that richness of pathogens and putative pathogens was higher when the probability of basin desiccation was higher. Intuitively, a high basin desiccation frequency (high A/V) and the prevalence of inundations that do not last long enough to allow microbial reproduction should pose environmental stresses that may trigger mortality [50]. A possible explanation for our results is that the increase in desiccation provoked spore formation and dormancy rather than mortality. Indeed, (facultative) parasites are selected for producing large amounts of resistant propagules that can withstand long periods of inactivity while waiting for a host [71]. Therefore, it is anticipated that a higher richness of pathogenic OTUs will be recovered from the sediments in basins with higher frequency of desiccation. Such conditions may favour opportunist organisms that have considerable dispersion abilities and that can easily withstand adverse conditions. Alternatively, the stress of desiccation may not have been a strong community structure limiting factor, as the existence of competition-based traits among the hosts, and the consequent loss of hosts occurring when hydro periods extend, may therefore render more richness of pathogenic OTUs when the desiccation frequency increases. These hypotheses need to be further explored by direct measurements of basin hydrological regimes in long time series studies [72].
The most likely mechanism for pathogen arrival in basins is through the spores. In chytrids, flagellated zoospores are the key dispersal phase of the cell cycle, but they also are the free-living motile and infectious life stage and are believed to actively search for new hosts by chemotaxis [1]. The dispersal of chytrids can actually be a rather complex process and may involve passive and active dispersal. Passive dispersal involves mediation by outside forces, such as the movement of water, wind, soils, animal vectors or other microbes, and active dispersal is driven by organismal intrinsic traits, i.e., motility and response to chemical signalling or chemotaxis [39]. Therefore, the arrival of chytrids, and other pathogens, in rock basins and the surrounding environment, may occur following a random (stochastic) pattern, mediated by abiotic, mechanical mechanisms such as wind [73] and phoresis [74], but may also be the consequence of a deterministic process when collective traits, such as spore formation, active movement and dormancy can select groups of microbes, making them better suited for dispersal [39].
On the other hand, the numbers of chytrids or other pathogens isolated from the rock basins may not always reach the threshold of abundance or propagule frequency and size needed to become pathogenic. Therefore, it is plausible to think that some of the pathogenic OTUs found belong to organisms that did not actually develop their whole life cycle in the basins, but that just remain and accumulate inside these habitats.
The study of sediment chemistry revealed that the effect of chemical variables on the distribution of the OTUs inhabiting all the basins is different for nonpathogenic and for pathogenic (and potential pathogenic) microorganisms. Micronutrients were the most influential factor on the community composition of nonpathogenic OTUs, while heavy toxic metals were the most significant variable for the potential pathogens. Interestingly, two OTUs related to chytrid fungi, so far never described formally as microbial pathogens, appeared clearly related to high values of heavy and toxic metals: Powellomyces sp. and Rhizophlyctis rosea. Both organisms are known for their resilience, with spores able to withstand drought and extreme temperatures [75]. Some fungi have the ability of biosorption of metals accumulating in their cell wall, and several mechanisms of metal resistance were previously described (see [76] for a comprehensive review). Their probable resistance towards heavy metals is also a feature that gives them the opportunity to colonise new environments.
In conclusion, our results show that rain-fed rock basins might be genetic repositories for (mainly opportunistic) microbial pathogens that are favoured by the unstable conditions of these systems. Some may also have a bioindicator potential. Our study emphasizes the isolation and biotic/abiotic heterogeneity of these transitory aquatic environments and highlights them as habitats in which to explore the ecological relevance of potential pathogens and their impact on host population dynamics and the planktonic food web structure. The high proportion of chytrid-like sequences found advocates the study of these habitats as models for adding knowledge on the ecological and physiological trade-offs of the highly abundant but functionally still rather unknown Chytridiomycota.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15050594/s1, Table S1: Database sheet of pathogenic and potential pathogenic OTUs and their taxonomic resolution; Table S2: Numerical values for the chemical parameters analysed in the rock basins; Table S3: Taxonomic affiliation of the overall microbial eukaryotic OTUs with 100% of occurrence in the basins. Pathogen and potential pathogens are highlighted in red. Table S4: Values of Beta diversity components for the potential pathogenic microbial eukaryotes in the rock basins; Figure S1: Size-based rarefaction (solid line) and extrapolation (dashed line) for each rock basin sampled; Figure S2: Relationship (exponential decay model [48]) between Beta diversity components and the spatial distance among basins; Figure S3: Nonmetric Multidimensional Scaling (NMDS) ordination plot for the OTUs (top) and for the chemical variables (bottom) in function of the two A/V basin groups (A/V < 1, black; A/V > 1, red). A polygon for the convex hulls encircling each cluster was drawn. NMDS was conducted with two components, using Bray–Curtis distance in the case of OTUs and Euclidean distance on standardised values in the case of chemical variables.
Author Contributions
Conceptualisation, M.M.-C., A.S.-J. and I.V.-G.; Methodology, I.V.-G., E.L., D.S., A.S.-J., A.M. and M.M.-C.; Formal Analysis, I.V.-G., M.M.-C. and A.S.-J.; Investigation: M.M.-C., I.V.-G. and A.S.-J.; Resources: M.M.-C., M.G.-R., R.W. and A.d.C.-G.; Data Curation: I.V.-G., A.d.C.-G. and M.M.-C.; Writing—Original Draft Preparation: I.V.-G. and M.M.-C.; Writing—Review and Editing: E.L., R.W., B.P.-U., D.S. A.S.-J. and M.M.-C.; Supervision: M.M.-C. and A.S.-J.; Project Administration: M.M.-C. and B.P.-U.; Funding Acquisition: M.M.-C., E.L. and D.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ministerio de Economía y Competitividad (MINECO- Spain), Project MICROEPICS Ref: CGL2013-40851-P/BOS 2014–2018 and Project Santander-UCM Ref: PR44/21-29928 to PI M.M.-C. E.L. was funded by the project “Atracción de talento investigador” by the Consejería de Educación, Juventud y Deporte, Comunidad de Madrid (Spain) 2017-T1/AMB-5210 and by a grant from the Swiss National Foundation for Research (SNF 31003A_143960). D.S. was funded by the Swiss NSF (P2NEP3_178543). A.S., A.M., A.C.-G., B.P.-U. and M.M.-C. are funded by the research group “Modelling, Data Analysis and Computational Tools for Biology (UCM)”; A.M. is also funded by research groups “Neurocomputing and Neurorobotics (UCM)” and “Brain Plasticity Group” (Health Research Institute of the Hospital Clínico San Carlos, IdISSC).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
All V4-rDNA reads were deposited at the European nucleotide archive under the Project number PRJEB24091. Other data are available from the authors upon reasonable request.
Acknowledgments
Permits to collect samples and facilities provided by The Parque Nacional Sierra de Guadarrama are gratefully acknowledged. The authors are grateful to CAI-Técnicas Geológicas UCM for their contribution to chemical analyses.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Frenken, T.; Alacid, E.; Berger, S.A.; Bourne, E.C.; Gerphagnon, M.; Grossart, H.P.; Gsell, A.S.; Ibelings, B.W.; Kagami, M.; Küpper, F.C.; et al. Integrating chytrid fungal parasites into plankton ecology: Research gaps and needs. Environ. Microbiol. 2017, 19, 3802–3822. [Google Scholar] [CrossRef]
- Geisen, S.; Mitchell, E.A.D.; Adl, S.; Bonkowski, M.; Dunthorn, M.; Ekelund, F.; Fernández, L.D.; Jousset, A.; Krashevska, V.; Singer, D.; et al. Soil protists: A fertile frontier in soil biology research. FEMS Microbiol. Rev. 2018, 42, 293–323. [Google Scholar] [CrossRef] [PubMed]
- Selbach, C.; Soldánová, M.; Feld, C.K.; Kostadinova, A.; Sures, B. Hidden parasite diversity in a European freshwater system. Sci. Rep. 2020, 10, 2694. [Google Scholar] [CrossRef]
- Evison, S.E.; Fazio, G.; Chappell, P.; Foley, K.; Jensen, A.B.; Hughes, W.O. Host–parasite genotypic interactions in the honey bee: The dynamics of diversity. Ecol. Evol. 2013, 3, 2214–2222. [Google Scholar] [CrossRef]
- Hamilton, W.D. Pathogens as causes of genetic diversity in their host populations. In Population Biology of Infectious Diseases; Anderson, R.M., May, R.M., Eds.; Springer: Berlin/Heidelberg, Germany, 1982; pp. 269–296. [Google Scholar]
- Ibelings, B.W.; De Bruin, A.; Kagami, M.; Rijkeboer, M.; Brehm, M.; Donk, E.V. Host parasite interactions between freshwater phytoplankton and chytrid fungi (Chytridiomycota). J. Phycol. 2004, 40, 437–453. [Google Scholar] [CrossRef]
- Mahé, F.; de Vargas, C.; Bass, D.; Czech, L.; Stamatakis, A.; Lara, E.; Singer, D.; Mayor, J.; Bunge, J.; Sernaker, S.; et al. Parasites dominate hyperdiverse soil protist communities in Neotropical rainforests. Nat. Ecol. Evol. 2017, 1, 0091. [Google Scholar] [CrossRef]
- Weinbauer, M.G.; Rassoulzadegan, F. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 2004, 6, 1–11. [Google Scholar] [CrossRef]
- Amundsen, P.-A.; Lafferty, K.D.; Knudsen, R.; Primicerio, R.; Klemetsen, A.; Kuris, A.M. Food web topology and parasites in the pelagic zone of a subarctic lake. J. Anim. Ecol. 2009, 78, 563–572. [Google Scholar] [CrossRef]
- Sime-Ngando, T. Phytoplankton chytridiomycosis: Fungal parasites of phytoplankton and their imprints on the food web dynamics. Front. Microbiol. 2012, 3, 361. [Google Scholar] [CrossRef]
- Agha, R.; Saebelfeld, M.; Manthey, C.; Rohrlack, T.; Wolinska, J. Chytrid parasitism facilitates trophic transfer between bloom-forming cyanobacteria and zooplankton (Daphnia). Sci. Rep. 2016, 6, 35039. [Google Scholar] [CrossRef]
- Johnson, P.T.J.; Dobson, A.; Lafferty, K.D.; Marcogliese, D.J.; Memmott, J.; Orlofske, S.A.; Poulin, R.; Thieltges, D.W. When parasites become prey: Ecological and epidemiological significance of eating parasites. Trends Ecol. Evol. 2010, 25, 362–371. [Google Scholar] [CrossRef]
- De S Araújo, G.R.; de Souza, W.; Frases, S. The hidden pathogenic potential of environmental fungi. Fut. Microbiol. 2017, 12, 1533–1540. [Google Scholar] [CrossRef]
- Boisard, J.; Florent, I. Why the -omic future of Apicomplexa should include Gregarines. Biol. Cell 2020, 112, 173–185. [Google Scholar] [CrossRef]
- Comeau, A.M.; Vincent, W.F.; Bernier, L.; Lovejoy, C. Novel chytrid lineages dominate fungal sequences in diverse marine and freshwater habitats. Sci. Rep. 2016, 6, 30120. [Google Scholar] [CrossRef]
- Lefèvre, E.; Roussel, B.; Amblard, C.; Sime-Ngando, T. The molecular diversity of freshwater picoeukaryotes reveals high occurrence of putative parasitoids in the plankton. PLoS ONE 2008, 3, e2324. [Google Scholar] [CrossRef]
- de Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahé, F.; Logares, R.; Lara, E.; Berney, C.; Le Bescot, N.; Probert, I. Eukaryotic plankton diversity in the sunlit ocean. Science 2015, 348, 1261605. [Google Scholar] [CrossRef]
- Morand, S. (macro-) Evolutionary ecology of parasite diversity: From determinants of parasite species richness to host diversification. Int. J. Parasitol. Parasites Wildl. 2015, 4, 80–87. [Google Scholar] [CrossRef]
- Corsaro, D.; Walochnik, J.; Venditti, D.; Hauröder, B.; Michel, R. Solving an old enigma: Morellospora saccamoebae gen. nov., sp. nov. (Rozellomycota), a Sphaerita-like parasite of free-living amoebae. Parasitol. Res. 2020, 119, 925–934. [Google Scholar] [CrossRef]
- Gleason, F.H.; Kagami, M.; Lefevre, E.; Sime-Ngando, T. The ecology of chytrids in aquatic ecosystems: Roles in food web dynamics. Fungal Biol. Rev. 2008, 22, 17–25. [Google Scholar] [CrossRef]
- Reynolds, H.; Raudabaugh, D.; Lilje, O.; Allender, M.; Miller, A.; Gleason, F. Emerging mycoses and fungus-like diseases of vertebrate wildlife. In The Fungal Community: Its Organization and Role in the Ecosystem, 4th ed.; Dighton, J., White, J.F., Eds.; CRC Press: Boca Raton, FL, USA, 2017; pp. 385–404. [Google Scholar]
- Lefranc, M.; Thénot, A.; Lepère, C.; Debroas, D. Genetic diversity of small eukaryotes in lakes differing by their trophic status. Appl. Environ. Microbiol. 2005, 71, 5935–5942. [Google Scholar] [CrossRef] [PubMed]
- López-García, P.; Rodríguez-Valera, F.; Pedrós-Alió, C.; Moreira, D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 2001, 409, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Singer, D.; Lara, E.; Steciow, M.M.; Seppey, C.V.W.; Paredes, N.; Pillonel, A.; Oszako, T.; Belbahri, L. High-throughput sequencing reveals diverse oomycete communities in oligotrophic peat bog micro-habitat. Fungal Ecol. 2016, 23, 42–47. [Google Scholar] [CrossRef]
- Ortiz-Álvarez, R.; Triadó-Margarit, X.; Camarero, L.; Casamayor, E.O.; Catalan, J. High planktonic diversity in mountain lakes contains similar contributions of autotrophic, heterotrophic and parasitic eukaryotic life forms. Sci. Rep. 2018, 8, 4457. [Google Scholar] [CrossRef]
- Singer, D.; Duckert, C.; Heděnec, P.; Lara, E.; Hiltbrunner, E.; Mitchell, E.A.D. High-throughput sequencing of litter and moss eDNA reveals a positive correlation between the diversity of Apicomplexa and their invertebrate hosts across alpine habitats. Soil Biol. Biochem. 2020, 147, 107837. [Google Scholar] [CrossRef]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the classification, nomenclature, and diversity of Eukaryotes. J. Eukaryotic Microbiol. 2019, 66, 4–119. [Google Scholar] [CrossRef] [PubMed]
- Bråte, J.; Logares, R.; Berney, C.; Ree, D.K.; Klaveness, D.; Jakobsen, K.S.; Shalchian-Tabrizi, K. Freshwater Perkinsea and marine-freshwater colonizations revealed by pyrosequencing and phylogeny of environmental rDNA. ISME J. 2010, 4, 1144–1153. [Google Scholar] [CrossRef]
- Käse, L.; Metfies, K.; Neuhaus, S.; Boersma, M.; Wiltshire, K.H.; Kraberg, A.C. Host-parasitoid associations in marine planktonic time series: Can metabarcoding help reveal them? PLoS ONE 2021, 16, e0244817. [Google Scholar] [CrossRef]
- Kagami, M.; Miki, T.; Takimoto, G. Mycoloop: Chytrids in aquatic food webs. Front. Microbiol. 2014, 5, 166. [Google Scholar] [CrossRef] [PubMed]
- Schmeller, D.S.; Loyau, A.; Bao, K.; Brack, W.; Chatzinotas, A.; De Vleeschouwer, F.; Friesen, J.; Gandois, L.; Hansson, S.V.; Haver, M.; et al. People, pollution and pathogens—global change impacts in mountain freshwater ecosystems. Sci. Total Environ. 2018, 622, 756–763. [Google Scholar] [CrossRef]
- Jocque, M.; Vanschoenwinkel, B.; Brendonck, L. Freshwater rock pools: A review of habitat characteristics, faunal diversity and conservation value. Freshwater Biol. 2010, 55, 1587–1602. [Google Scholar] [CrossRef]
- Pérez Uz, M.B.; Velasco González, I.; Murciano, A.; Sánchez Jiménez, A.; García-Rodríguez, M.; Centeno Carrillo, J.d.D.; Montero González, E.; Muñoz Araújo, B.; Olmedo Salinas, C.; Quintela Alonso, P.; et al. Rain-fed granite rock pools in a national park: Extreme niches for protists. Limnetica 2021, 40, 1–18. [Google Scholar] [CrossRef]
- Birck, C.; Epaillard, I.; Leccia, F.; Crassous, C.; Morand, A.; Miaud, C.; Bertrand, C.; Cavalli, L.; Jacquet, S.; Moullec, P. Sentinel lakes: A network for the study and management of mountain lakes in the French Alps and in Corsica. Eco. Mont. 2013, 5, 63–69. [Google Scholar] [CrossRef]
- Oertli, B.; Biggs, J.; Céréghino, R.; Grillas, P.; Joly, P.; Lachavanne, J.-B. Conservation and monitoring of pond biodiversity: Introduction. Aquat. Conserv. Mar. Freshwater Ecosyst. 2005, 15, 535–540. [Google Scholar] [CrossRef]
- Velasco-González, I.; Sanchez-Jimenez, A.; Singer, D.; Murciano, A.; Díez-Hermano, S.; Lara, E.; Martín-Cereceda, M. Rain-fed granite rock basins accumulate a high diversity of dormant microbial eukaryotes. Microb. Ecol. 2020, 79, 882–897. [Google Scholar] [CrossRef] [PubMed]
- Custer, G.F.; Bresciani, L.; Dini-Andreote, F. Ecological and evolutionary implications of microbial dispersal. Front. Microbiol. 2022, 13, 855859. [Google Scholar] [CrossRef]
- Baumgardner, D.J. Soil-related bacterial and fungal infections. J. Am. Board Family Med. 2012, 25, 734–744. [Google Scholar] [CrossRef]
- Rokas, A. Evolution of the human pathogenic lifestyle in fungi. Nat. Microbiol. 2022, 7, 607–619. [Google Scholar] [CrossRef]
- Stoeck, T.; Bass, D.; Nebel, M.; Christen, R.; Jones, M.D.M.; Breiner, H.-W.; Richards, T.A. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 2010, 19, 21–31. [Google Scholar] [CrossRef]
- Schiaffino, M.R.; Lara, E.; Fernández, L.D.; Balagué, V.; Singer, D.; Seppey, C.C.W.; Massana, R.; Izaguirre, I. Microbial eukaryote communities exhibit robust biogeographical patterns along a gradient of Patagonian and Antarctic lakes. Environ. Microbiol. 2016, 18, 5249–5264. [Google Scholar] [CrossRef]
- Mahé, F.; Rognes, T.; Quince, C.; de Vargas, C.; Dunthorn, M. Swarm: Robust and fast clustering method for amplicon-based studies. PeerJ 2014, 2, e593. [Google Scholar] [CrossRef]
- Pearson, W.R. BLAST and FASTA similarity searching for multiple sequence alignment. In Multiple Sequence Alignment Methods; Russell, D.J., Ed.; Humana Press: Totowa, NJ, USA, 2014; pp. 75–101. [Google Scholar]
- Guillou, L.; Bachar, D.; Audic, S.; Bass, D.; Berney, C.; Bittner, L.; Boutte, C.; Burgaud, G.; de Vargas, C.; Decelle, J.; et al. The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2012, 41, D597–D604. [Google Scholar] [CrossRef]
- Scheele, B.C.; Pasmans, F.; Skerratt, L.F.; Berger, L.; Martel, A.; Beukema, W.; Acevedo, A.A.; Burrowes, P.A.; Carvalho, T.; Catenazzi, A.; et al. Amphibian fungal panzootic causes catastrophic and ongoing loss of biodiversity. Science 2019, 363, 1459–1463. [Google Scholar] [CrossRef] [PubMed]
- Baselga, A. Partitioning the turnover and nestedness components of beta diversity. Glob. Ecol. Biogeogr. 2010, 19, 134–143. [Google Scholar] [CrossRef]
- Baselga, A. Separating the two components of abundance-based dissimilarity: Balanced changes in abundance vs. abundance gradients. Methods Ecol. Evol. 2013, 4, 552–557. [Google Scholar] [CrossRef]
- Gómez-Rodríguez, C.; Baselga, A. Variation among European beetle taxa in patterns of distance decay of similarity suggests a major role of dispersal processes. Ecography 2018, 41, 1825–1834. [Google Scholar] [CrossRef]
- Bengtsson, J.; Ebert, D. Distributions and impacts of microparasites on Daphnia in a rockpool metapopulation. Oecologia 1998, 115, 213–221. [Google Scholar] [CrossRef]
- Therriault, T.W.; Kolasa, J. Desiccation frequency reduces species diversity and predictability of community structure in coastal rock pools. Israel J. Zool. 2001, 47, 477–489. [Google Scholar] [CrossRef]
- De Miguel, E. Determinación de Niveles de Fondo y Niveles de Referencia de Metales Pesados y Otros Elementos Traza en Suelos de la Comunidad de Madrid; Instituto Geológico y Minero de España: Madrid, Spain, 2002. [Google Scholar]
- Logares, R.; Mangot, J.-F.; Massana, R. Rarity in aquatic microbes: Placing protists on the map. Res. Microbiol. 2015, 166, 831–841. [Google Scholar] [CrossRef]
- Lepère, C.; Domaizon, I.; Debroas, D. Unexpected importance of potential parasites in the composition of the freshwater small-eukaryote community. Appl. Environ. Microbiol. 2008, 74, 2940–2949. [Google Scholar] [CrossRef]
- Yi, Z.; Berney, C.; Hartikainen, H.; Mahamdallie, S.; Gardner, M.; Boenigk, J.; Cavalier-Smith, T.; Bass, D. High-throughput sequencing of microbial eukaryotes in Lake Baikal reveals ecologically differentiated communities and novel evolutionary radiations. FEMS Microbiol. Ecol. 2017, 93, fix073. [Google Scholar] [CrossRef]
- de Sousa, J.R.; Goncalves, V.N.; de Holanda, R.A.; Santos, D.A.; Bueloni, C.F.; Costa, A.O.; Petry, M.V.; Rosa, C.A.; Rosa, L.H. Pathogenic potential of environmental resident fungi from ornithogenic soils of Antarctica. Fungal Biol. 2017, 121, 991–1000. [Google Scholar] [CrossRef]
- Geisen, S.; Mitchell, E.A.D.; Wilkinson, D.M.; Adl, S.; Bonkowski, M.; Brown, M.W.; Fiore-Donno, A.M.; Heger, T.J.; Jassey, V.E.J.; Krashevska, V.; et al. Soil protistology rebooted: 30 fundamental questions to start with. Soil Biol. Biochem. 2017, 111, 94–103. [Google Scholar] [CrossRef]
- Blaalid, R.; Khomich, M. Current knowledge of Chytridiomycota diversity in Northern Europe and future research needs. Fungal Biol. Rev. 2021, 36, 42–51. [Google Scholar] [CrossRef]
- Barr, D. Phylum Chytridiomycota. In Handbook of Protoctista; Margulis, L., Corliss, J., Melkonian, M., Chapman, D.J., Eds.; Jones & Barlett: Burlington, MA, USA, 1990; pp. 454–466. [Google Scholar]
- Gerphagnon, M.; Colombet, J.; Latour, D.; Sime-Ngando, T. Spatial and temporal changes of parasitic chytrids of cyanobacteria. Sci. Rep. 2017, 7, 6056. [Google Scholar] [CrossRef]
- Van den Wyngaert, S.; Rojas-Jimenez, K.; Seto, K.; Kagami, M.; Grossart, H.-P. Diversity and hidden host specificity of chytrids infecting colonial volvocacean algae. J. Eukaryotic Microbiol. 2018, 65, 870–881. [Google Scholar] [CrossRef]
- Bosch, J.; Martínez-Solano, I.; García-París, M. Evidence of a chytrid fungus infection involved in the decline of the common midwife toad (Alytes obstetricans) in protected areas of central Spain. Biol. Conserv. 2001, 97, 331–337. [Google Scholar] [CrossRef]
- Fernández-Beaskoetxea, S.; Bosch, J.; Bielby, J. Infection and transmission heterogeneity of a multi-host pathogen (Batrachochytrium dendrobatidis) within an amphibian community. Dise. Aquat. Organ. 2016, 118, 11–20. [Google Scholar] [CrossRef]
- Freeman, K.R.; Martin, A.P.; Karki, D.; Lynch, R.C.; Mitter, M.S.; Meyer, A.F.; Longcore, J.E.; Simmons, D.R.; Schmidt, S.K. Evidence that chytrids dominate fungal communities in high-elevation soils. Proc. Natl. Acad. Sci. USA 2009, 106, 18315–18320. [Google Scholar] [CrossRef]
- Hendrix, F.F.; Campbell, W.A. Pythium as plant pathogens. Ann. Rev. Phytopathol. 1973, 11, 77–98. [Google Scholar] [CrossRef]
- Levesque, C.A.; de Cock, A.W. Molecular phylogeny and taxonomy of the genus Pythium. Mycol. Res. 2004, 108, 1363–1383. [Google Scholar] [CrossRef]
- Wolinska, J.; Giessler, S.; Koerner, H. Molecular identification and hidden diversity of novel Daphnia parasites from European lakes. Appl. Environ. Microbiol. 2009, 75, 7051–7059. [Google Scholar] [CrossRef] [PubMed]
- Vilela, R.; Taylor, J.W.; Walker, E.D.; Mendoza, L. Lagenidium giganteum pathogenicity in mammals. Emerg. Infect. Dis. 2015, 21, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Martí Carrizosa, M. Candida parapsilosis, C. orthopsilosis y C. metapsilosis: Epidemiología de las Candidemias, Patrones de Sensibilidad y Mecanismos de Resistencia a las Equinocandinas. Ph.D. Thesis, Universitat Autònoma de Barcelona, Barcelona, Spain, 2015. [Google Scholar]
- Brieland, J.; McClain, M.; Heath, L.; Chrisp, C.; Huffnagle, G.; LeGendre, M.; Hurley, M.; Fantone, J.; Engleberg, C. Coinoculation with Hartmannella vermiformis enhances replicative Legionella pneumophila lung infection in a murine model of Legionnaires’ disease. Infect. Immun. 1996, 64, 2449–2456. [Google Scholar] [CrossRef] [PubMed]
- Centeno, M.; Rivera, F.; Cerva, L.; Tsutsumi, V.; Gallegos, E.; Calderón, A.; Ortiz, R.; Bonilla, P.; Ramírez, E.; Suárez, G. Hartmannella vermiformis isolated from the cerebrospinal fluid of a young male patient with meningoencephalitis and bronchopneumonia. Arch. Med. Res. 1996, 27, 579–586. [Google Scholar]
- Poulin, R. The evolution of life history strategies in parasitic animals. Adv. Parasitol. 1996, 37, 107–134. [Google Scholar]
- Altermatt, F.; Pajunen, V.I.; Ebert, D. Desiccation of rock pool habitats and its influence on population persistence in a Daphnia metacommunity. PLoS ONE 2009, 4, e4703. [Google Scholar] [CrossRef]
- Vanschoenwinkel, B.; Gielen, S.; Seaman, M.; Brendonck, L. Wind mediated dispersal of freshwater invertebrates in a rock pool metacommunity: Differences in dispersal capacities and modes. Hydrobiologia 2009, 635, 363–372. [Google Scholar] [CrossRef]
- Bogitsh, B.J.; Carter, C.E.; Oeltmann, T.N. Human Parasitology; Academic Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Gleason, F.H.; Letcher, P.M.; McGee, P.A. Some Chytridiomycota in soil recover from drying and high temperatures. Mycol. Res. 2004, 108, 583–589. [Google Scholar] [CrossRef]
- Gadd, G.M. Geomycology: Biogeochemical transformations of rocks, minerals, metals and radionuclides by fungi, bioweathering and bioremediation. Mycol. Res. 2007, 111, 3–49. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).