Morphology, Phylogeny, and Pathogenicity of Colletotrichum Species Causing Anthracnose in Camellia japonica in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Isolation
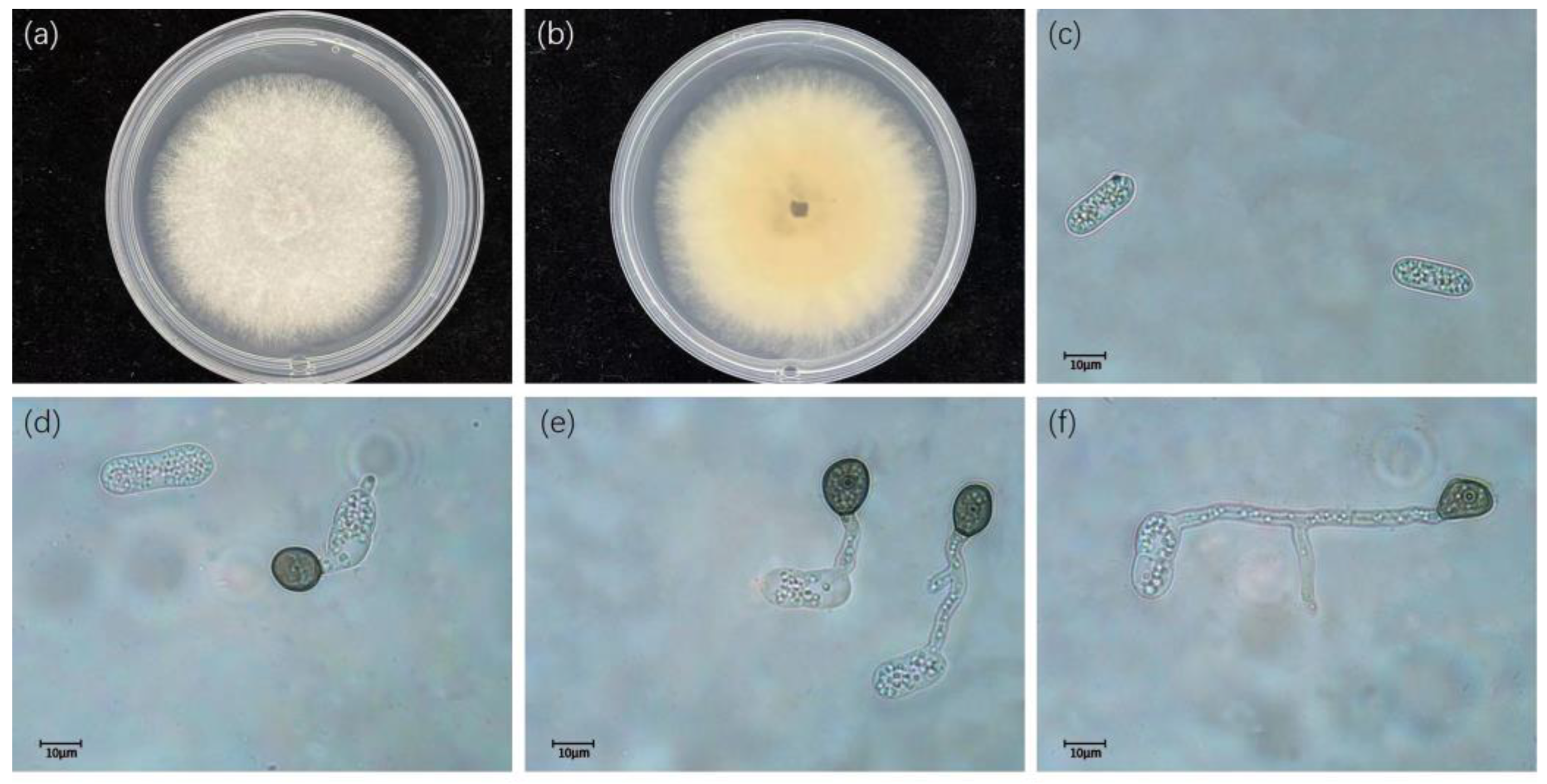
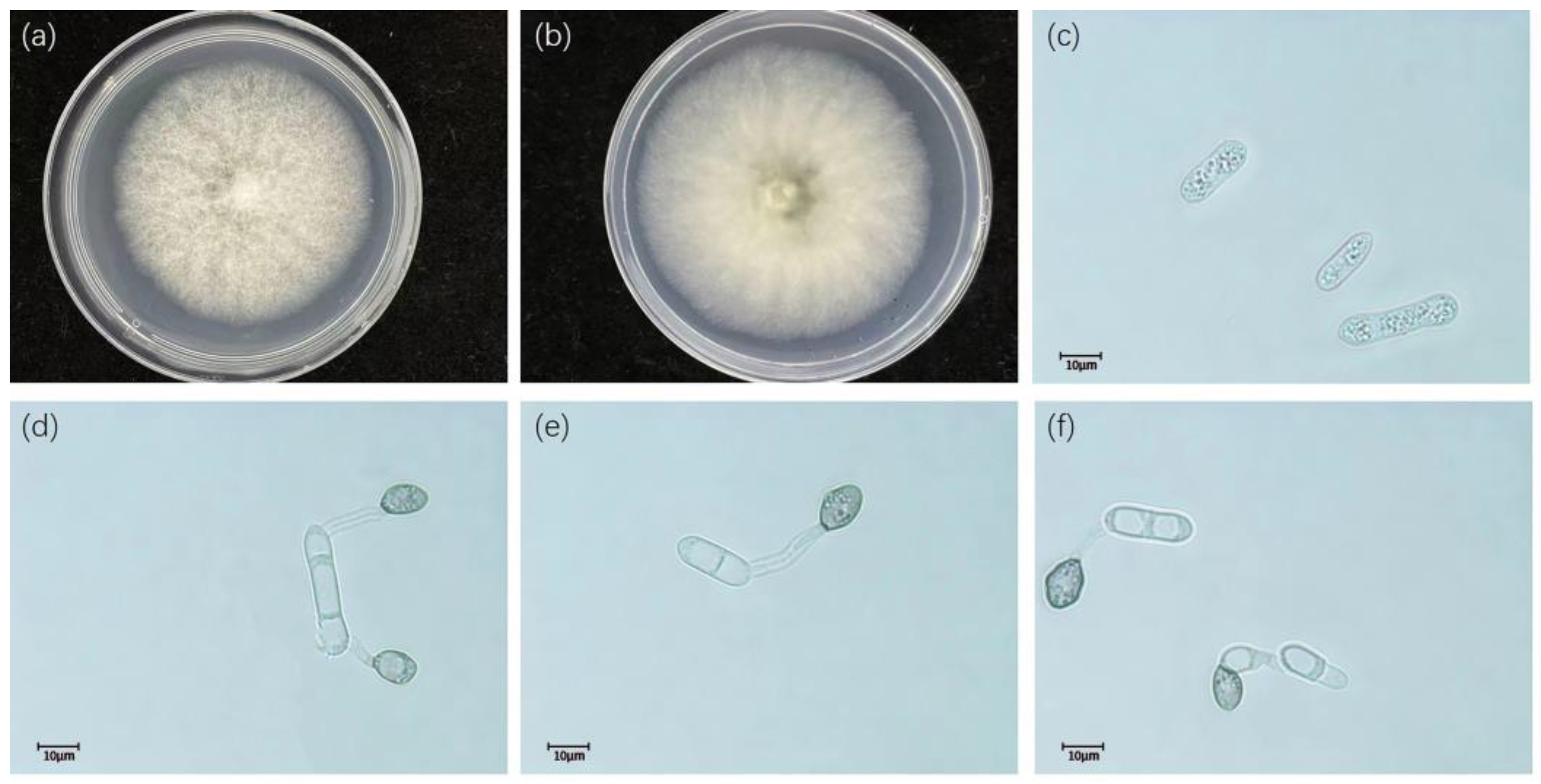
2.2. Morphological Characterization
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. Phylogenetic Analyses
2.5. Pathogenicity Testing
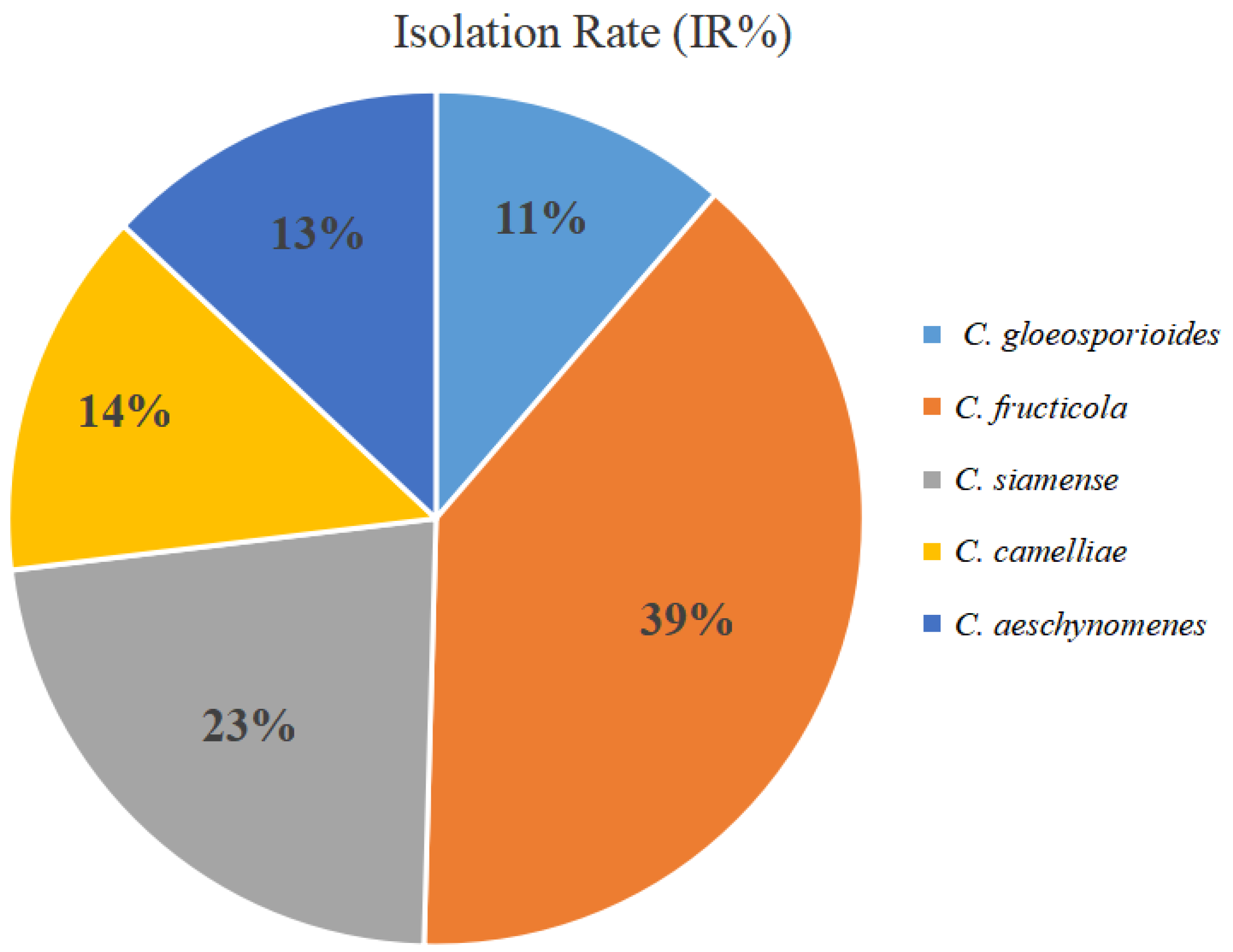
3. Results
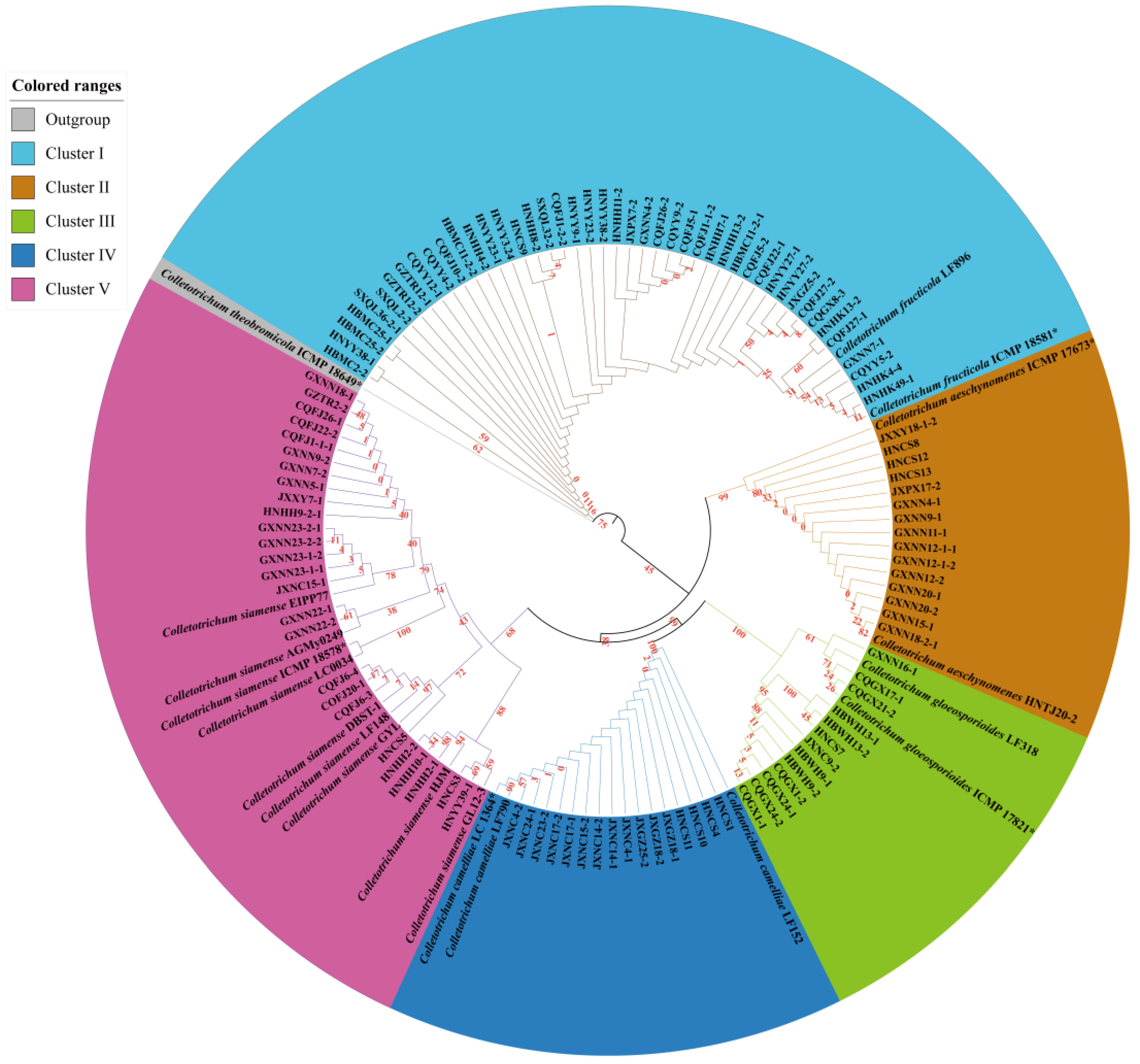
3.1. Phylogenetic Analyses
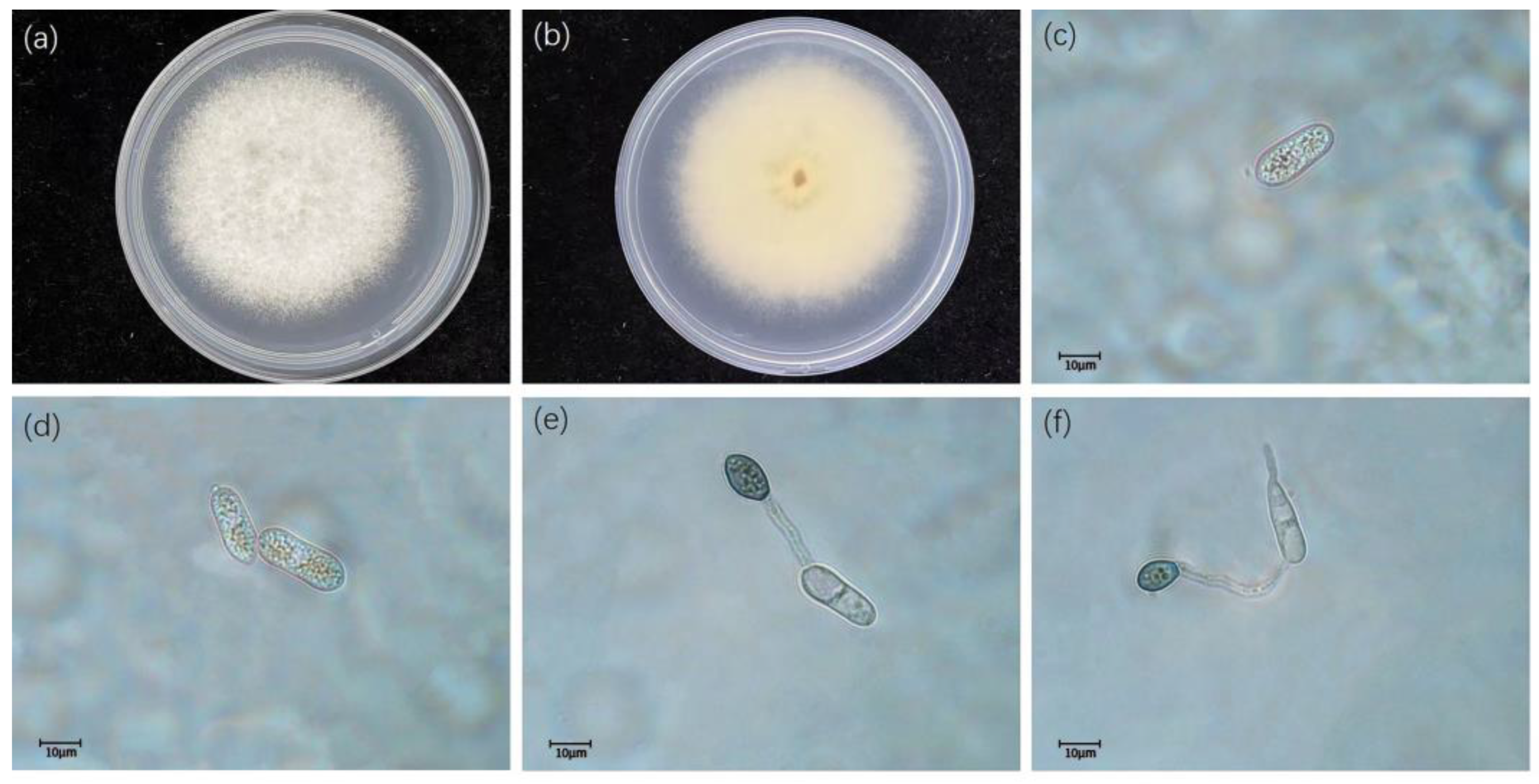
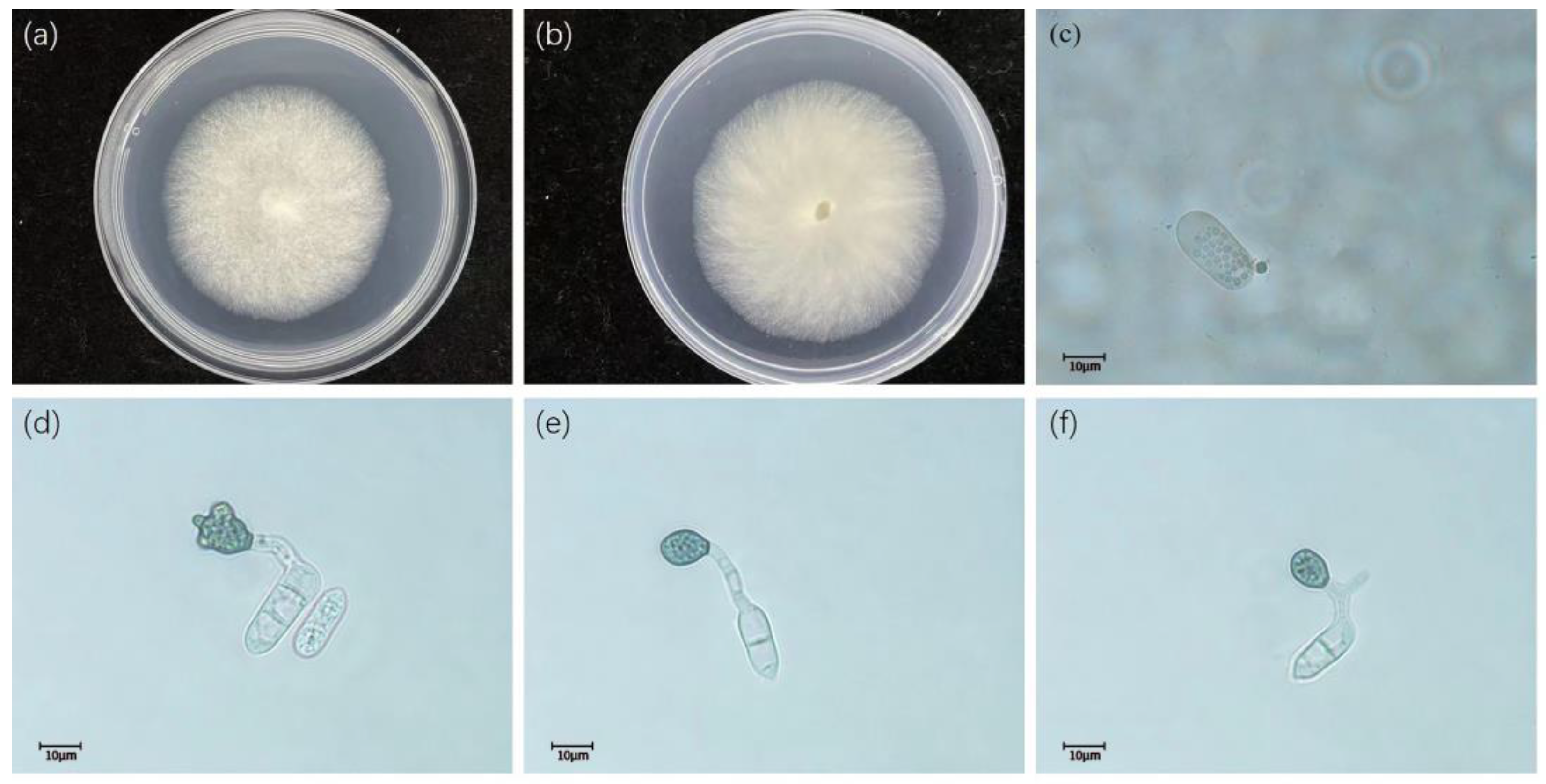
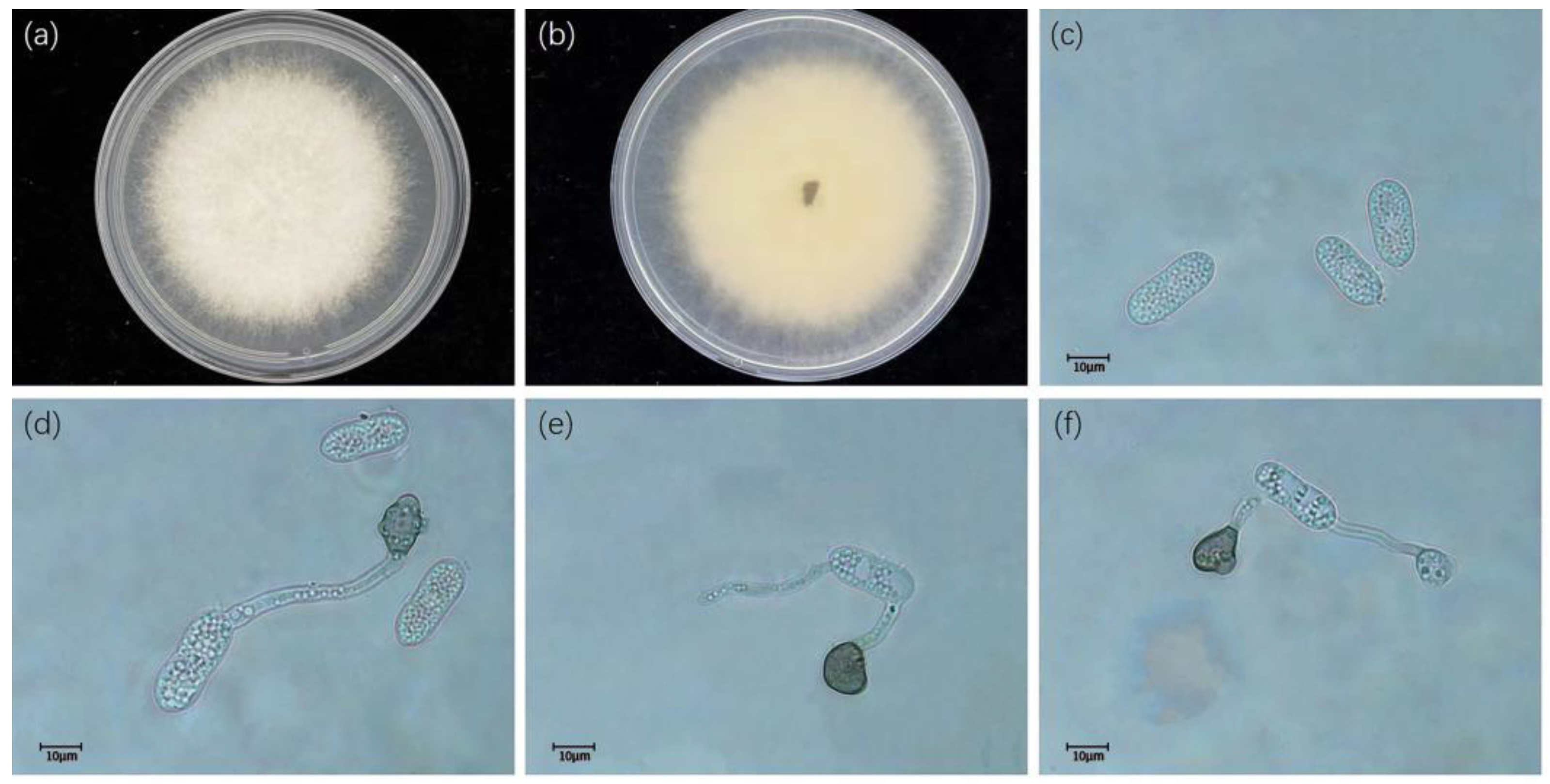
3.2. Taxonomy
- Colletotrichum gloeosporioides (Penz.) Penz. and Sacc. 1884
- Colletotrichum fructicola Priastuti, L. Cai. and K.D. Hyde. 2009.
- Colletotrichum siamense Priastuti, L. Cai. and K.D. Hyde. 2009.
- Colletotrichum camelliae Massee. 2012
- Colletotrichum aeschynomenes B.S. Weir, P.R. Johnston, and U. Damm. 2012
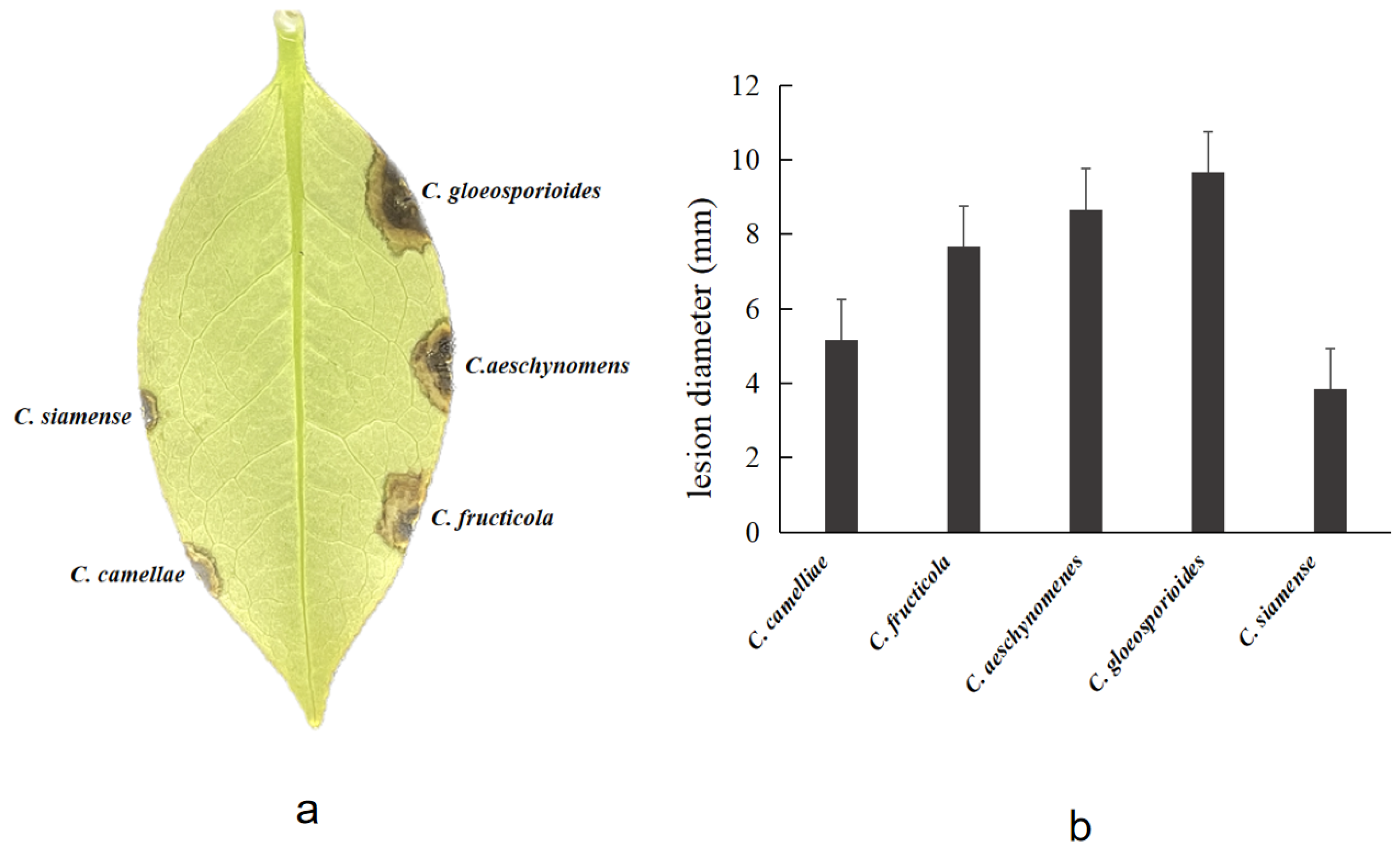
3.3. Pathogenicity Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wei, Y.H. Research progress on phenotypic genetic diversity of Camellia germplasm resources in China. South China Agric. 2020, 14, 66–68. [Google Scholar]
- Li, X.L. Analysis of nutritional components and poisonous elements in flowers of four Camellia species. For. Res. 2010, 23, 298–301. [Google Scholar]
- Ana, M.T.; Clara, S. A review on the biological activity of Camellia species. Molecules 2021, 26, 2178. [Google Scholar] [CrossRef]
- Wang, J.W.; Qian, X.M.; Zhang, B.X. Camellia diseases in Zhejiang Province. Acta Phytopathol. Sin. 1990, 20, 2. [Google Scholar]
- Luo, H.S. The Control of Camellia japonica diseases and pests. China Flowers Hortic. 2016, 4, 32–34. [Google Scholar]
- Lv, Y.F. Diseases and Insect Pests of Camellia japonica L. and Their Control Techniques. J. Agric. Catastropholgy 2021, 11, 18–19. [Google Scholar]
- Liu, L.P.; Gao, J.; Li, Y. Advances in knowledge of the fungi referred to the genus Colletotrichum. J. Fungal Res. 2020, 18, 266–281. [Google Scholar]
- Li, L.L.; Yang, Q.; Li, H. Morphology, phylogeny, and pathogenicity of pestalotioid species on Camellia oleifera in China. J. Fungi 2021, 7, 1080. [Google Scholar] [CrossRef]
- Wang, Y.C.; Hao, X.Y.; Wang, L.; Xiao, B.; Wang, X.C.; Yang, Y.J. Diverse Colletotrichum species cause anthracnose of tea plants (Camellia sinensis (L.) O. Kuntze) in China. Sci. Rep. 2016, 6, 35387. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, R.S.; Hyde, K.D.; Damm, U.; Cai, L.; Liu, M.; Li, X.H.; Zhang, W.; Zhao, W.S.; Yan, J.Y. Notes on currently accepted spcies of Colletotrichum. Mycosphere 2016, 7, 1192–1260. [Google Scholar] [CrossRef]
- Liu, F.; Ma, Z.Y.; Hou, L.W.; Diao, Y.Z.; Wu, W.P.; Damm, U.; Song, S.; Cai, L. Updating species diversity of Colletotrichum, with a phylogenomic overview. Stud. Mycol. 2022, 101, 1–56. [Google Scholar] [CrossRef]
- Hou, S.F.; Hua, Z.Y.; Liu, J.J.; Dong, Z.F.; Feng, Z.K.; Yan, J.Q.; Wang, H.Q. Multi-gene joint identification and pathogenicity analysis of Colletotrichun gloeosporioides complex of strawberry in China. J. China Agric. Univ. 2022, 27, 82–94. [Google Scholar]
- Sun, W.; Lei, T.Y.; Yuan, H.Z.; Chen, S.N. First Report of Anthracnose Caused by Colletotrichum kahawae and Colletotrichum horri on Tea-oil Tree in China. Plant Dis. 2022. [Google Scholar] [CrossRef]
- Wang, Y.X.; Chen, J.Y.; Xu, X.W.; Cheng, J.Y.; Zheng, L.; Huang, J.B.; Li, D.W. Identification and characterization of Colletotrichum species associated with anthracnose disease of Camellia oleifera in China. Plant Dis. 2020, 104, 474–482. [Google Scholar] [CrossRef]
- Lu, Y.; He, C.P.; Wu, W.H.; Zheng, J.L.; Huang, X.; Xi, J.G.; Tan, S.B.; Xian, Y.K. Pathogenicity differentiation of Coffee anthracnose in China. Southwest China J. Agric. Sci. 2021, 34, 1008–1014. [Google Scholar]
- Silva, D.N.; Talhinhas, P.; Várzea, V.; Cai, L.; Paulo, S.O.; Batista, D. Application of the Apn2/MAT locus to improve the systematics of the Colletotrichum gloeosporioides complex: An example from coffee (Coffea spp.) hosts. Mycologia 2012, 104, 396–409. [Google Scholar] [CrossRef]
- Li, L.L.; Li, H. First report of Colletotrichum aeschynomenes causing anthracnose on Camellia oleifera in China. For. Pathol. 2022, 52, e12770. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Kumar, G.S.N.; Weir, B.S.; Hyde, K.D.; Shenoy, B.D. The ApMat market can resolve Colletotrichum species: A case study with Mangifera indica. Fungal Divers. 2013, 61, 117–138. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Liu, F.; Weir, B.S.; Damm, U.; Crous, P.W.; Wang, Y.; Liu, B.; Wang, M.; Zhangm, M.; Cai, L. Unravelling Colletotrichum species associated with Camellia: Employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Persoonia 2015, 35, 63–86. [Google Scholar] [CrossRef]
- Douanla-Meli, C.; Unger, J.G. Phylogenetic study of the Colletotrichum species on imported citrus fruits uncovers a low diversity and a new species in the Colletotrichum gigasporum complex. Fungal Biol. 2017, 121, 858–868. [Google Scholar] [CrossRef]
- Gong, C.Y.; Liu, J.J.; Deng, Q.; Zhang, L.X. Identification and Pathogenicity of Colletotrichum species causing anthracnose on Camellia sinensis. Acta Hortic. Sin. 2022, 49, 1092–1101. [Google Scholar] [CrossRef]
- Li, H.; Li, S.Z.; Wang, Y.C.; Liu, J.A.; Xu, J.P.; Zhou, G.Y. Identification of the pathogens causing anthracnose of Camellia oleifera in nursery and their resistence to fungcides. Sci. Silvae Sin. 2019, 55, 85–94. [Google Scholar]
- Sharma, G.; Maymon, M.; Freeman, S. Epidemiology, pathology and identification of Colletotrichum including a novel species associated with avocado (Persea americana) anthracnose in Israel. Sci. Rep. 2017, 7, 15839. [Google Scholar] [CrossRef]
- Huang, Y.N.; Zhu, T.H. A Preliminary Study of anthracnose on the leaf of Camellia. J. Sichuan For. Sci. Technol. 2005, 26, 10–13. [Google Scholar]
- Peng, X.J.; Yuan, Y.X.; Zhang, S.K.; Zhou, X.Z. First Report of Anthracnose on Camellia japonica Caused by Colletotrichum siamense in Zhejiang Province, China. Am. Phytopathol. Soc. 2022, 106, 768. [Google Scholar] [CrossRef]
- Han, C.Z.; Zhao, H.Y. Pathogen identification of Camellia anthracnose disease. For. Pest Dis. 2016, 35, 5–6. [Google Scholar]
- Chen, X.; Wang, T.; Guo, H.; Zhu, P.K. First Report of Anthracnose of Camellia sasanqua Caused by Colletotrichum aenigma in China. Plant Dis. 2019, 103, 1423. [Google Scholar] [CrossRef]
- Yang, S.; Wang, H.X.; Yi, Y.J.; Tan, L.L. First Report that Colletotrichum aenigma Causes Leaf Spots on Camellia japonica in China. Plant Dis. 2019, 103, 2127. [Google Scholar] [CrossRef]
- Hou, L.W.; Liu, F.; Duan, W.J.; Cai, L. Colletotrichum aracearum and C. camelliae-japoncae, two holomorphic new species from China and Japan. Mycosphere 2016, 7, 1111–1123. [Google Scholar] [CrossRef]
- Weir, B.S.; Johnston, P.R.; Damm, U. The Colletotrichum gloeosporioides species comple. Stud. Mycol. 2012, 73, 115–180. [Google Scholar] [CrossRef]
- Wang, W.; de Silva, D.D.; Moslemi, A.; Edwards, J.; Ades, P.K.; Crous, P.W.; Taylor, P.W.J. Colletotrichum Species Causing Anthracnose of Citrus in Australia. J. Fungi. 2021, 7, 47. [Google Scholar] [CrossRef]
| Taxon | Isolate Designation | Host | Geographic Location | ApMat GenBank | References |
|---|---|---|---|---|---|
| C. camelliae | LF152 | Camellia sp. | China | KJ954506.1 | [21] |
| LF790 | Cinamomum zeylanicum | India | KU239747.1 | Direct Submission | |
| LC1364 * | Camellia sinensis | China | KJ954497.1 | [21] | |
| HNCS1 | Camellia japonica | China | OQ198468 | In this study | |
| HNCS4 | Camellia japonica | China | OQ198469 | In this study | |
| HNCS10 | Camellia japonica | China | OQ198470 | In this study | |
| HNCS11 | Camellia japonica | China | OQ198471 | In this study | |
| JXGZ18-1 | Camellia japonica | China | OQ198472 | In this study | |
| JXGZ18-2 | Camellia japonica | China | OQ198473 | In this study | |
| JXGZ25-2 | Camellia japonica | China | OQ198474 | In this study | |
| JXNC4-1 | Camellia japonica | China | OQ198475 | In this study | |
| JXNC4-2 | Camellia japonica | China | OQ198476 | In this study | |
| JXNC14-1 | Camellia japonica | China | OQ198477 | In this study | |
| JXNC14-2 | Camellia japonica | China | OQ198478 | In this study | |
| JXNC15-2 | Camellia japonica | China | OQ198479 | In this study | |
| JXNC17-1 | Camellia japonica | China | OQ198480 | In this study | |
| JXNC17-2 | Camellia japonica | China | OQ198481 | In this study | |
| JXNC23-2 | Camellia japonica | China | OQ198482 | In this study | |
| JXNC24-1 | Camellia japonica | China | OQ198483 | In this study | |
| C. siamense | AGMy0249 | Citrus pennivesiculata | Bangladesh | KX578769.1 | [22] |
| LC0034 | Coffee berry | Thailand | JQ899288.1 | Direct Submission | |
| HJM | Loropetalum chinense | China | MG717312.1 | Direct Submission | |
| GL12-3 | Plum | China | OM816816.1 | Direct Submission | |
| GYL | Magnolia grandiflora | China | MG717298.1 | Direct Submission | |
| LF148 | Camellia sp. | China | KJ954504.1 | [21] | |
| DBST-1 | Cycas debaoensis | China | MT786728.1 | Direct Submission | |
| EIPP77 | Coffee | China | MK344209.1 | Direct Submission | |
| ICMP18649 * | Coffea arabica | China | JQ899289 | [21] | |
| HNCS3 | Camellia japonica | China | OQ198484 | In this study | |
| HNCS5 | Camellia japonica | China | OQ198485 | In this study | |
| HNYY39-1 | Camellia japonica | China | OQ198486 | In this study | |
| HNHH2-1 | Camellia japonica | China | OQ198487 | In this study | |
| HNHH2-2 | Camellia japonica | China | OQ198488 | In this study | |
| HNHH9-2-1 | Camellia japonica | China | OQ198489 | In this study | |
| HNHH10-1 | Camellia japonica | China | OQ198490 | In this study | |
| JXXY7-1 | Camellia japonica | China | OQ198491 | In this study | |
| JXNC15-1 | Camellia japonica | China | OQ198492 | In this study | |
| GXNN5-1 | Camellia japonica | China | OQ198493 | In this study | |
| GXNN7-2 | Camellia japonica | China | OQ198494 | In this study | |
| GXNN9-2 | Camellia japonica | China | OQ198495 | In this study | |
| GXNN18-1 | Camellia japonica | China | OQ198496 | In this study | |
| GXNN22-1 | Camellia japonica | China | OQ198497 | In this study | |
| GXNN22-2 | Camellia japonica | China | OQ198498 | In this study | |
| GXNN23-1-1 | Camellia japonica | China | OQ198499 | In this study | |
| GXNN23-1-2 | Camellia japonica | China | OQ198500 | In this study | |
| GXNN23-2-1 | Camellia japonica | China | OQ198501 | In this study | |
| GXNN23-2-2 | Camellia japonica | China | OQ198502 | In this study | |
| CQFJ1-1-1 | Camellia japonica | China | OQ198503 | In this study | |
| CQFJ6-3 | Camellia japonica | China | OQ198504 | In this study | |
| CQFJ6-4 | Camellia japonica | China | OQ198505 | In this study | |
| CQFJ20-1 | Camellia japonica | China | OQ198506 | In this study | |
| CQFJ22-2 | Camellia japonica | China | OQ198507 | In this study | |
| CQFJ26-1 | Camellia japonica | China | OQ198508 | In this study | |
| GZTR2-2 | Camellia japonica | China | OQ198509 | In this study | |
| C. gloeosporioides | LF318 | Ca. sinensis | China | KJ954541.1 | [21] |
| ICMP1782 * | Citrus sinensis | Italy | JQ807843 | [21] | |
| HNCS7 | Camellia japonica | China | OQ198510 | In this study | |
| JXNC9-2 | Camellia japonica | China | OQ198511 | In this study | |
| HBWH9-1 | Camellia japonica | China | OQ198512 | In this study | |
| HBWH9-2 | Camellia japonica | China | OQ198513 | In this study | |
| HBWH13-1 | Camellia japonica | China | OQ198514 | In this study | |
| HBWH13-2 | Camellia japonica | China | OQ198515 | In this study | |
| GXNN16-1 | Camellia japonica | China | OQ198516 | In this study | |
| CQGX1-1 | Camellia japonica | China | OQ198517 | In this study | |
| CQGX1-2 | Camellia japonica | China | OQ198518 | In this study | |
| CQGX17-1 | Camellia japonica | China | OQ198519 | In this study | |
| CQGX21-2 | Camellia japonica | China | OQ198520 | In this study | |
| CQGX24-1 | Camellia japonica | China | OQ198521 | In this study | |
| CQGX24-2 | Camellia japonica | China | OQ198522 | In this study | |
| C. aeschynomenes | ICMP1767 * | Aeschynomene viginica | China | KM360145.1 | [21] |
| HNTJ20-1 | Camellia oleifera | China | MZ8321172.1 | [17] | |
| HNCS8 | Camellia japonica | China | OQ198523 | In this study | |
| HNCS12 | Camellia japonica | China | OQ198524 | In this study | |
| HNCS13 | Camellia japonica | China | OQ198525 | In this study | |
| JXPX17-2 | Camellia japonica | China | OQ198526 | In this study | |
| JXXY18-1-2 | Camellia japonica | China | OQ198527 | In this study | |
| GXNN4-1 | Camellia japonica | China | OQ198528 | In this study | |
| GXNN9-1 | Camellia japonica | China | OQ198529 | In this study | |
| GXNN11-1 | Camellia japonica | China | OQ198530 | In this study | |
| GXNN12-1-1 | Camellia japonica | China | OQ198531 | In this study | |
| GXNN12-1-2 | Camellia japonica | China | OQ198532 | In this study | |
| GXNN12-2 | Camellia japonica | China | OQ198533 | In this study | |
| GXNN15-1 | Camellia japonica | China | OQ198534 | In this study | |
| GXNN18-2-1 | Camellia japonica | China | OQ198535 | In this study | |
| GXNN20-1 | Camellia japonica | China | OQ198536 | In this study | |
| GXNN20-2 | Camellia japonica | China | OQ198537 | In this study | |
| C. theobromicola | ICMP18649 * | Theobroma cacao | Panama | KC790726 | [21] |
| C. fructicola | LF896 | Ca. sinensis | China | KJ954624.1 | [21] |
| LC0033 | Coffea arabica | India | JQ807838.1 | Direct Submission | |
| ICMP18581 * | Coffea arabica | Thailand | JQ07838 | [21] | |
| HNCS9 | Camellia japonica | China | OQ198538 | In this study | |
| HNYY3.24 | Camellia japonica | China | OQ198539 | In this study | |
| HNYY9-1 | Camellia japonica | China | OQ198540 | In this study | |
| HNYY23-1 | Camellia japonica | China | OQ198541 | In this study | |
| HNYY23-2 | Camellia japonica | China | OQ198542 | In this study | |
| HNYY27-1 | Camellia japonica | China | OQ198543 | In this study | |
| HNYY27-2 | Camellia japonica | China | OQ198544 | In this study | |
| HNYY38-1 | Camellia japonica | China | OQ198545 | In this study | |
| HNYY38-2 | Camellia japonica | China | OQ198546 | In this study | |
| HNHH4-2 | Camellia japonica | China | OQ198547 | In this study | |
| HNHH7-1 | Camellia japonica | China | OQ198548 | In this study | |
| HNHH8-2 | Camellia japonica | China | OQ198549 | In this study | |
| HNHH11-2 | Camellia japonica | China | OQ198550 | In this study | |
| HNHH13-2 | Camellia japonica | China | OQ198551 | In this study | |
| JXPX7-2 | Camellia japonica | China | OQ198552 | In this study | |
| JXGZ5-2 | Camellia japonica | China | OQ198553 | In this study | |
| HBMC2-2 | Camellia japonica | China | OQ198554 | In this study | |
| HBMC11-2-1 | Camellia japonica | China | OQ198555 | In this study | |
| HBMC11-2-2 | Camellia japonica | China | OQ198556 | In this study | |
| HBMC25-1 | Camellia japonica | China | OQ198557 | In this study | |
| HBMC25-2 | Camellia japonica | China | OQ198558 | In this study | |
| GXNN4-2 | Camellia japonica | China | OQ198559 | In this study | |
| GXNN7-1 | Camellia japonica | China | OQ198560 | In this study | |
| CQGX8-3 | Camellia japonica | China | OQ198561 | In this study | |
| CQFJ1-1-2 | Camellia japonica | China | OQ198562 | In this study | |
| CQFJ1-2-2 | Camellia japonica | China | OQ198563 | In this study | |
| CQFJ5-1 | Camellia japonica | China | OQ198564 | In this study | |
| CQFJ5-2 | Camellia japonica | China | OQ198565 | In this study | |
| CQFJ10-2 | Camellia japonica | China | OQ198566 | In this study | |
| CQFJ22-1 | Camellia japonica | China | OQ198567 | In this study | |
| CQFJ26-2 | Camellia japonica | China | OQ198568 | In this study | |
| CQFJ27-1 | Camellia japonica | China | OQ198569 | In this study | |
| CQFJ27-2 | Camellia japonica | China | OQ198570 | In this study | |
| CQYY4-2 | Camellia japonica | China | OQ198571 | In this study | |
| CQYY5-2 | Camellia japonica | China | OQ198572 | In this study | |
| CQYY9-2 | Camellia japonica | China | OQ198573 | In this study | |
| CQYY12-1 | Camellia japonica | China | OQ198574 | In this study | |
| HNHK4-4 | Camellia japonica | China | OQ198575 | In this study | |
| HNHK13-2 | Camellia japonica | China | OQ198576 | In this study | |
| HNHK49-1 | Camellia japonica | China | OQ198577 | In this study | |
| GZTR12-1 | Camellia japonica | China | OQ198578 | In this study | |
| GZTR12-2 | Camellia japonica | China | OQ198579 | In this study | |
| SXQL2-2 | Camellia japonica | China | OQ198580 | In this study | |
| SXQL32-2 | Camellia japonica | China | OQ198581 | In this study | |
| SXQL36-2-1 | Camellia japonica | China | OQ198582 | In this study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, L.; Li, H. Morphology, Phylogeny, and Pathogenicity of Colletotrichum Species Causing Anthracnose in Camellia japonica in China. Diversity 2023, 15, 516. https://doi.org/10.3390/d15040516
Wen L, Li H. Morphology, Phylogeny, and Pathogenicity of Colletotrichum Species Causing Anthracnose in Camellia japonica in China. Diversity. 2023; 15(4):516. https://doi.org/10.3390/d15040516
Chicago/Turabian StyleWen, Lixia, and He Li. 2023. "Morphology, Phylogeny, and Pathogenicity of Colletotrichum Species Causing Anthracnose in Camellia japonica in China" Diversity 15, no. 4: 516. https://doi.org/10.3390/d15040516
APA StyleWen, L., & Li, H. (2023). Morphology, Phylogeny, and Pathogenicity of Colletotrichum Species Causing Anthracnose in Camellia japonica in China. Diversity, 15(4), 516. https://doi.org/10.3390/d15040516






