Genetic Evidence for Indo-Western Pacific Olive Ridley Sea Turtles in Mexican Waters
Abstract
1. Introduction
2. Materials and Methods
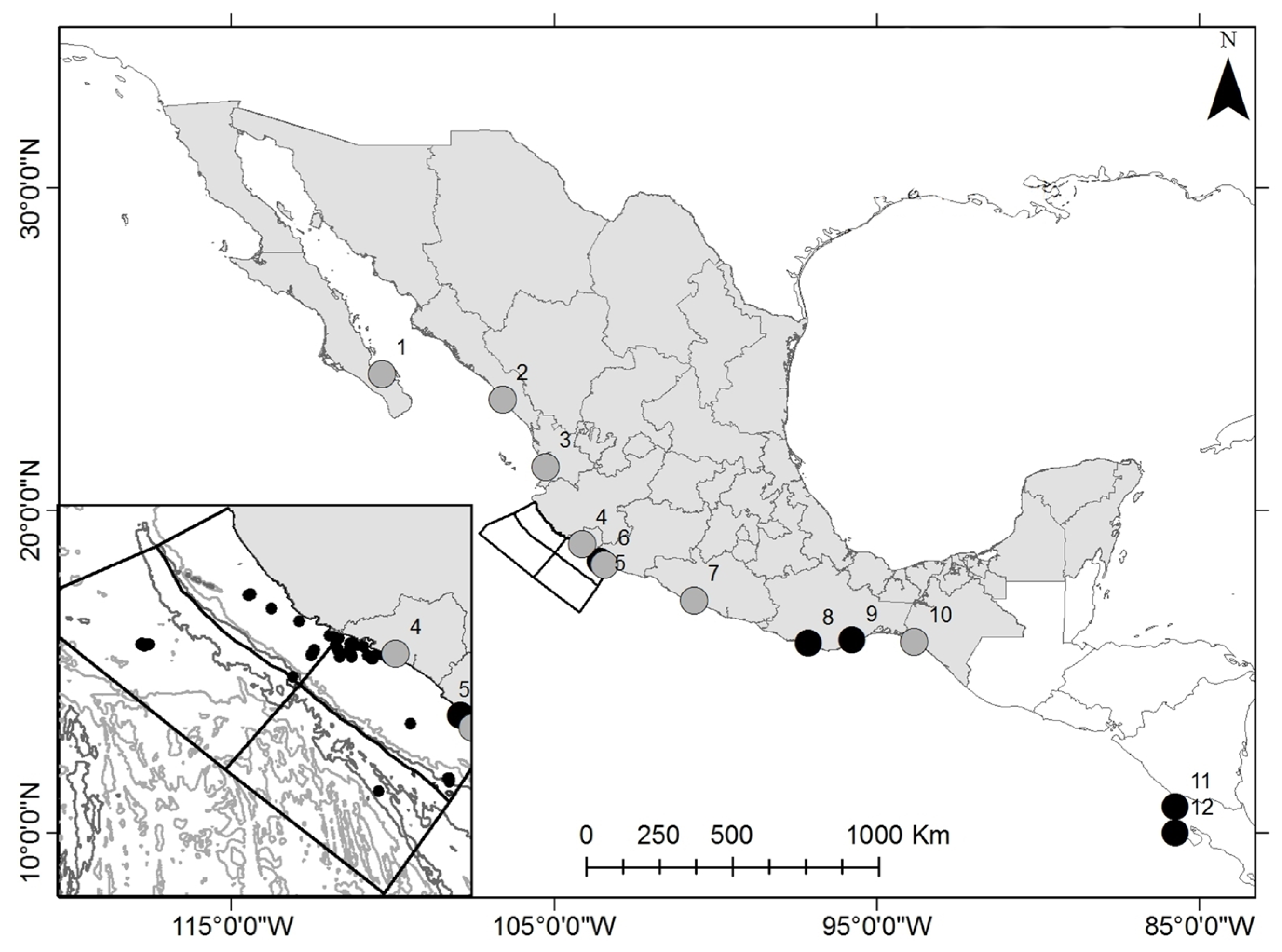
2.1. Study Area and Sampling
2.2. Characterization of mtDNA Haplotypes
2.3. Molecular Analysis
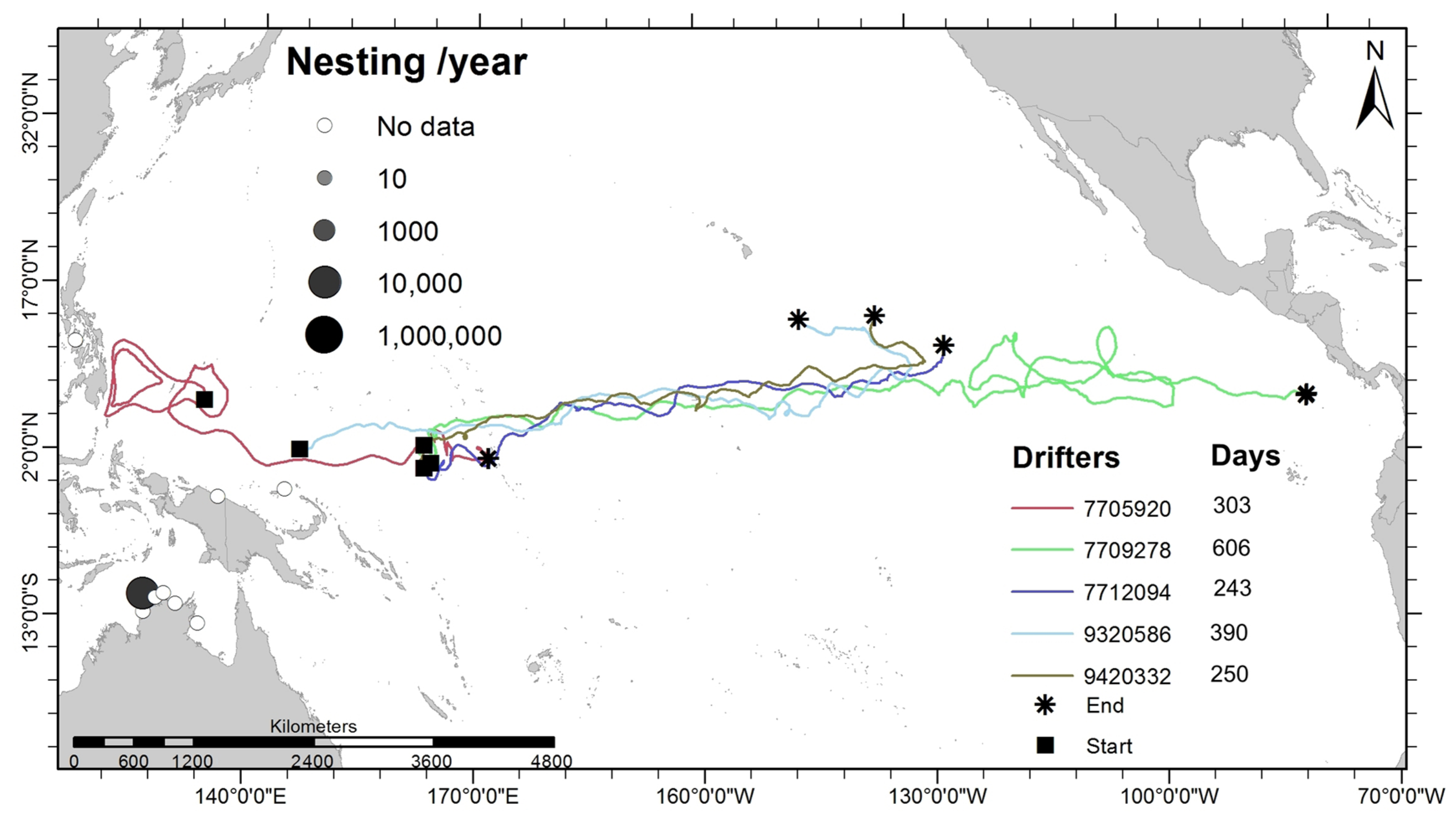
2.4. Trajectories of Satellite-Tracked Lagrangian Drifter Buoys
3. Results
3.1. Haplotype Composition
3.2. Genetic Diversity and Lack of Genetic Differentiation
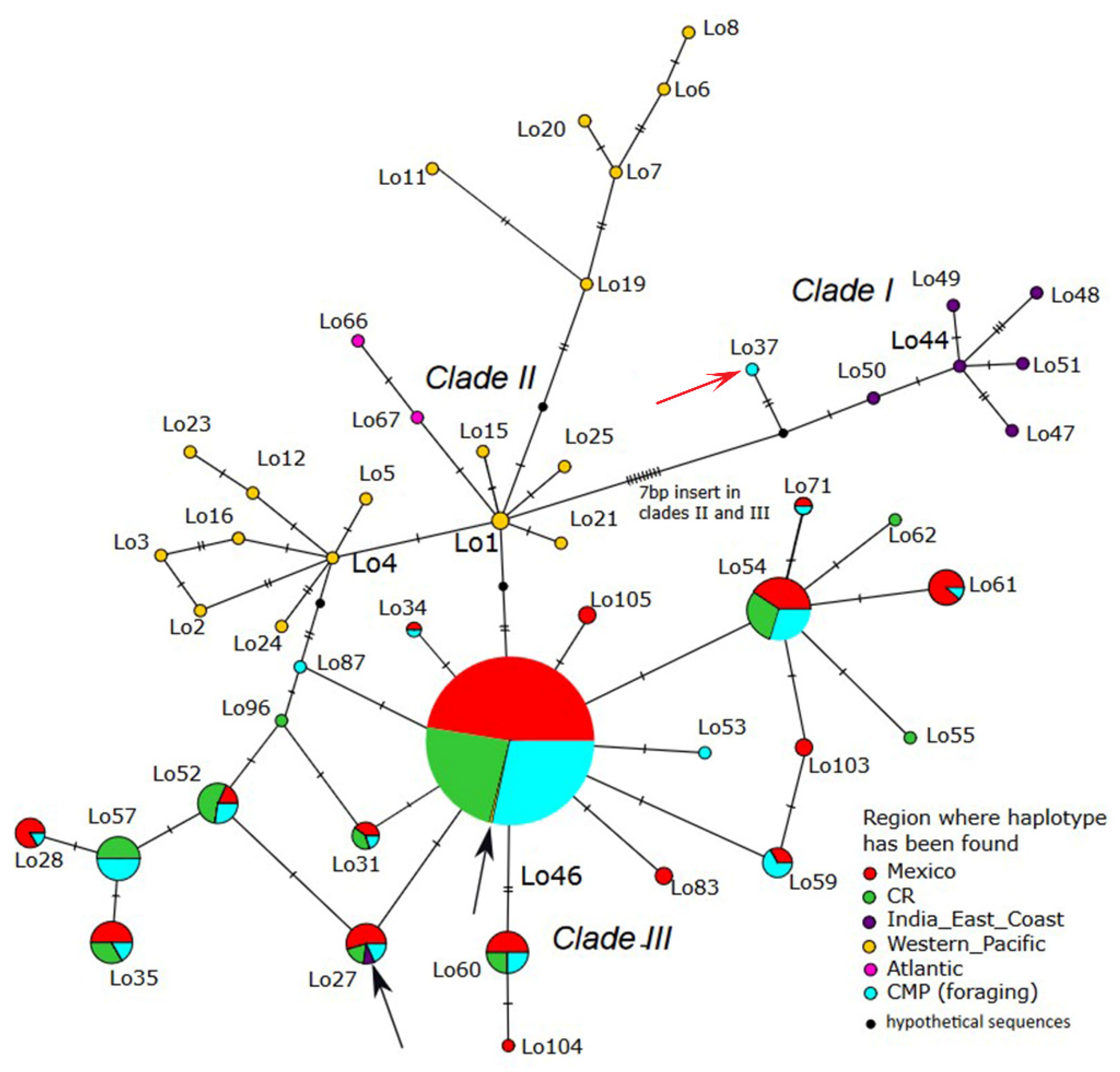
3.3. Phylogenetic Relationships
3.4. Potential Current-Driven Trans-Oceanic Displacement
4. Discussion
4.1. Genetic Diversity and Haplotype Composition
4.2. Absence of Genetic Spatial Structure
4.3. Phylogenetic Relationships
4.4. Role of Ocean Currents
4.5. Implications for Conservation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morreale, S.J.; Plotkin, P.T.; Shaver, D.J.; Kalb, H.J. Adult migration and habitat utilization. In Biology and Conservation of Ridley Sea Turtles; Plotkin, P.T., Ed.; Johns Hopkins University Press: Baltimore, MD, USA, 2007; pp. 213–229. [Google Scholar]
- Pritchard, P.C. Studies of the systematics and reproductive cycles of the genus Lepidochelys. Ph.D. Thesis, University of Florida, Gainesville, FL, USA, 1969. Available online: http://ufdcimages.uflib.ufl.edu/UF/00/09/77/74/00001/studiesofsystema00pritrich.pdf (accessed on 7 November 2022).
- Shanker, K.; Abreu-Grobois, A.; Bezy, V.; Briseño, R.; Colman, L.; Girard, A.; Girondot, M.; Jensen, M.; Manoharakrishnan, M.; Rguez-Baron, J.M.; et al. Olive ridleys: The Quirky Turtles that Conquered the World. SWOT 2021, 16, 24–31. Available online: https://www.seaturtlestatus.org/articles/olive-ridleys-the-quirky-turtles-that-conquered-the-world (accessed on 21 January 2023).
- Abreu-Grobois, A.; Plotkin, P.; IUCN SSC Marine Turtle Specialist Group. Olive Ridley Turtle Lepidochelys olivacea. In IUCN Red List of Threatened Species; Version 2022.2; IUCN: Cambridge, UK, 2008. [Google Scholar] [CrossRef]
- Bowen, B.; Clark, A.; Abreu-Grobois, A.; Chaves, A.; Reichart, H.; Ferl, R. Global phylogeography of the ridley sea turtles (Lepidochelys spp.) as inferred from mitochondrial DNA sequences. Genetica 1997, 101, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Shanker, K.; Ramadevi, J.; Choudhury, B.; Singh, L.; Aggarwal, R. Phylogeography of olive ridley turtles (Lepidochelys olivacea) on the east coast of India: Implications for conservation theory. Mol. Ecol. 2004, 137, 1899–1909. [Google Scholar] [CrossRef]
- Vilaça, S.T.; Hahn, A.T.; Naro-Maciel, E.; Abreu-Grobois, A.; Bowen, B.W.; Castilhos, J.C.; Ciofi, C.; FitzSimmons, N.N.; Jensen, M.P.; Formia, A.; et al. Global phylogeography of ridley sea turtles (Lepidochelys spp.): Evolution, demography, connectivity, and conservation. Conserv. Genet. 2022, 23, 995–1010. [Google Scholar] [CrossRef]
- Bowen, B.W.; Karl, S.A. Population genetics and phylogeography of sea turtles. Mol. Ecol. 2007, 16, 4886–4907. [Google Scholar] [CrossRef]
- Silver-Gorges, I.; Koval, J.; Rodriguez-Zarate, C.J.; Paladino, F.V.; Jordan, M. Large-scale connectivity, cryptic population structure, and relatedness in Eastern Pacific Olive ridley sea turtles (Lepidochelys olivacea). Ecol. Evol. 2020, 10, 8688–8704. [Google Scholar] [CrossRef]
- López-Castro, M.C.; Rocha-Olivares, A. The panmixia paradigm of eastern Pacific olive ridley turtles revised: Consequences for their conservation and evolutionary biology. Mol. Ecol. 2005, 14, 3325–3334. [Google Scholar] [CrossRef]
- Plotkin, P.T. Nomadic behaviour of the highly migratory olive ridley sea turtle Lepidochelys olivacea in the eastern tropical Pacific Ocean. Endanger. Species Res. 2010, 13, 33–40. [Google Scholar] [CrossRef]
- Shanker, K.; Choudhury, B.C.; Pandav, B.; Tripathy, B.; Kar, C.S.; Kar, S.K.; Gupta, N.K.; Frazier, J.G. Tracking olive ridley turtles from Orissa. In Proceedings of the Twenty-Second Annual Symposium on Sea Turtle Biology and Conservation, Miami, FL, USA, 4–7 April 2002; Seminoff, J.A., Ed.; NOAA Technical Memorandum NMFS-SEFSC–503. NOAA: Miami, FL, USA, 2003; pp. 50–51. Available online: https://seaturtlesofindia.org/wp-content/uploads/2017/04/Shanker-K.-B.C.-Choudhury-B.-Pandav-B.-Tripathy-C.S.-Kar-S.K.-Kar-N.K.-Gupta-J.G.-Frazier.-2003.-Tracking-olive-ridley-turtles-from-Orissa.-22nd-Symposium.pdf (accessed on 7 November 2022).
- Whiting, S.D.; Long, J.L.; Coyne, M. Migration routes and foraging behaviour of olive ridley turtles Lepidochelys olivacea in northern Australia. Endanger. Species Res. 2007, 3, 1–9. [Google Scholar] [CrossRef]
- Jensen, M.P.; Limpus, C.J.; Whiting, S.D.; Guinea, M.; Prince, R.I.; Dethmers, K.E.; Adnyana, I.B.; Kennett, R.; FitzSimmons, N.N. Defining olive ridley turtle Lepidochelys olivacea management units in Australia and assessing the potential impact of mortality in ghost nets. Endanger. Species Res. 2013, 21, 241–253. [Google Scholar] [CrossRef]
- Bowen, B.W.; Abreu-Grobois, A.; Balazs, G.; Kamezaki, N.; Limpus, C.; Ferl, R. Trans-Pacific migrations of the loggerhead turtle (Caretta caretta) demonstrated with mitochondrial DNA markers. Proc. Natl. Acad. Sci. USA 1995, 92, 3731–3734. [Google Scholar] [CrossRef]
- Amorocho, D.F.; Abreu-Grobois, F.A.; Dutton, P.H.; Reina, R.D. 2012. Multiple distant origins for green sea turtles aggregating off Gorgona Island in the Colombian eastern Pacific. PloS ONE 2012, 7, e31486. [Google Scholar] [CrossRef]
- Benson, S.R.; Eguchi, T.; Foley, D.G.; Forney, K.A.; Bailey, H.; Hitipeuw, C.; Samber, B.P.; Tapilatu, R.F.; Rei, V.; Ramohia, P.; et al. Large-scale movements and high-use areas of western Pacific leatherback turtles, Dermochelys coriacea. Ecosphere 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Polovina, J.J.; Balazs, G.H.; Howell, E.A.; Parker, D.M.; Seki, M.P.; Dutton, P.H. Forage and migration habitat of loggerhead (Caretta caretta) and olive ridley (Lepidochelys olivacea) sea turtles in the central North Pacific Ocean. Fish. Oceanogr. 2004, 13, 36–51. [Google Scholar] [CrossRef]
- Wyrtki, K. Equatorial currents in the Pacific 1950 to 1970 and their relations to the trade winds. J. Phys. Oceanogr. 1974, 4, 372–380. [Google Scholar] [CrossRef]
- Davenport, J. Temperature and the life-history strategies of sea turtles. J. Therm. Biol. 1997, 22, 479–488. [Google Scholar] [CrossRef]
- Cummins, P.F.; Freeland, H.J. Variability of the North Pacific Current and its bifurcation. Prog. Oceanogr. 2007, 75, 253–265. [Google Scholar] [CrossRef]
- Wyrtki, K. Teleconnections in the equatorial Pacific Ocean. Science 1973, 180, 66–68. [Google Scholar] [CrossRef] [PubMed]
- Kessler, W.S. The circulation of the eastern tropical Pacific: A review. Prog. Oceanogr. 2006, 69, 181–217. [Google Scholar] [CrossRef]
- Work, T.M.; Dagenais, J.; Stacy, B.A.; Ladner, J.T.; Lorch, J.M.; Balazs, G.H.; Barquero-Calvo, E.; Berlowski-Zier, B.M.; Breeden, R.; Corrales-Gómez, N.; et al. A novel host-adapted strain of Salmonella Thypimurium causes renal disease in olive ridley turtles (Lephdochelys olivacea) in the Pacific. Sci. Rep. 2019, 9, 9313. [Google Scholar] [CrossRef]
- Wyneken, J. The Anatomy of Sea Turtles; U.S. Department of Commerce NOAA Technical Memorandum NMFS-SEFSC-470: Miami, FL, USA, 2001; p. 172. Available online: https://www.dnr.sc.gov/seaturtle/Literature/TM_470_Wyneken.pdf (accessed on 10 March 2012).
- Márquez, M.R. FAO Species Catalogue. Vol. 11: Sea Turtles of the world. An Annotated and Illustrated Catalogue of Sea Turtle Species Known to Date; FAO: Rome, Italy, 1990; p. 81. Available online: https://www.fao.org/3/t0244e/t0244e00.htm (accessed on 21 January 2023).
- Amos, B.; Hoelzel, A.R. Long-term preservation of whale skin for DNA analysis. Rep. Int. Whal. Comm. Spec. Issue 1991, 13, 99–103. [Google Scholar]
- Gemmell, N.J.; Akiyama, S. An efficient method for the extraction of DNA from vertebrate tissues. Trends Genet. 1996, 12, 338–339. [Google Scholar] [CrossRef]
- Abreu-Grobois, A.; Horrocks, J.; Formia, A.; Dutton, P.; LeRoux, R.; Vélez-Zuazo, X.; Soares, L.; Meylan, P. New mtDNA D-loop primers which work for a variety of marine turtle species may increase the resolution of mixed stock analyses. In Proceedings of the 26th Annual Symposium on Sea Turtle Biology, Island of Crete, Greece, 3–8 April 2006; Book of Abstracts. Frick, M., Panagopoulou, A., Rees, A.F., Williams, K., Eds.; International Sea Turtle Society: Athens, Greece, 2006; p. 179. Available online: https://internationalseaturtlesociety.org/wp-content/uploads/2021/02/26-turtle.pdf (accessed on 7 November 2022).
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1998, 16, 10881–10890. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987; p. 512. [Google Scholar]
- Excoffier, L.; Lischer, H. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. The genetical structure of populations. Ann. Eugen. 1949, 15, 323–354. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Campista-León, S.; Beltrán-Espinoza, J.B.; Sosa-Cornejo, I.; Castillo-Ureta, H.; Martín-del-Campo, R.; Sánchez-Zazueta, J.G.; Peinado-Guevara, L.I. Haplotypic characterization of the olive ridley turtle (Lepidochelys olivacea) in northwest Mexico: The northernmost limit of its distribution. Anim. Biodivers. Conserv. 2019, 42, 113–126. [Google Scholar] [CrossRef]
- Pinou, T.; Prunier, R.; Bresson, M.; Enciso-Padilla, I.; Javier-Perez, J.F.; Trejo-Robles, J.A.; DiGiovanni, R.A.; Robinson, N.J. Repeated sampling adds to the genetic diversity of Lepidochelys olivacea (Eschscholtz 1829) olive ridley sea turtle. J. Nat. Hist. 2018, 52, 2899–2917. [Google Scholar] [CrossRef]
- Clement, M.; Snell, Q.; Walker, P.; Posada, D.; Crandall, K. TCS: Estimating gene genealogies. Parallel Distrib. Process. Symp. Int. Proc. 2002, 2, 184. [Google Scholar] [CrossRef]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy; Freeman: San Francisco, CA, USA, 1973; p. 573. [Google Scholar]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Duchene, S.; Frey, A.; Alfaro-Núñez, A.; Dutton, P.H.; Gilbert, M.T.P.; Morin, P.A. Marine turtle mitogenome phylogenetics and evolution. Mol. Phylogenet. Evol. 2012, 65, 241–250. [Google Scholar] [CrossRef]
- Adnyana, W.; Jayaratha, M.; Purwanasari, H.N.; Wandia, I.N.; Dethmers, K.; Limpus, C.J. Post-Nesting Migration and Mitochondrial DNA Structures of Olive Ridley Turtles (Lepidochelys olivacea) Nested on Beaches of the Bird’s Head of Papua and the Lesser Sunda Regions, Indonesia. Int. J. Sci. Res. Publ. 2020, 10, 809–817. [Google Scholar] [CrossRef]
- Avise, J.C. Molecular Markers, Natural History, and Evolution; Chapman & Hall: New York, NY, USA, 1994; p. 511. [Google Scholar]
- Briseño-Dueñas, R. Variación genética de la región control del ADN mitocondrial de poblaciones de la tortuga golfina (Lepidochelys olivacea) en el Pacífico oriental e implicaciones para su conservación. Master’s Thesis, Universidad Autónoma de Sinaloa, Mazatlán, Mexico, 1998. [Google Scholar]
- Tripathy, B.; Pandav, B. Beach fidelity and internesting movements of olive ridley turtles (Lepidochelys olivacea) at Rushikulya, India. Herpetol. Conserv. Biol. 2008, 3, 40–45. Available online: https://www.herpconbio.org/Volume_3/Issue_1/Tripathy_Pandav_2008.pdf (accessed on 7 November 2022).
- Wallace, B.P.; DiMatteo, A.D.; Bolten, A.B.; Chaloupka, M.Y.; Hutchinson, B.J.; Abreu-Grobois, A.; Mortimer, J.A.; Seminoff, J.A.; Amorocho, D.; Bjorndal, K.A.; et al. Global conservation priorities for marine turtles. PLoS ONE 2011, 6, e24510. [Google Scholar] [CrossRef]
- Behera, S.; Choudhury, B.C.; Dutta, S.K. Spatial Dynamics of Olive Ridley Turtles (Lepidochelys olivacea) Density in the Tropical Sea Water of India. Proc. Zool. Soc. 2019, 72, 364–371. [Google Scholar] [CrossRef]
- Carr, A. New perspectives on the pelagic stage of sea turtle development. Conserv. Biol. 1987, 1, 103–121. [Google Scholar] [CrossRef]
- Luschi, P.; Hays, G.C.; Papi, F. A review of long-distance movements by marine turtles, and the possible role of ocean currents. Oikos 2003, 103, 293–302. [Google Scholar] [CrossRef]
- Lambardi, P.; Lutjeharms, J.R.E.; Menacci, R.; Hays, G.C.; Luschi, P. Influence of ocean currents on long distance movements of leatherback sea turtles in the southwest Indian ocean. Mar. Ecol. Prog. Ser. 2008, 353, 289–301. [Google Scholar] [CrossRef]
- Hays, G.C.; Fossette, S.; Katselidis, K.A.; Mariani, P.; Schofield, G. Ontogenetic development of migration: Lagrangian drift trajectories suggest a new paradigm for sea turtles. J. R. Soc. Interface 2010, 7, 1319–1327. [Google Scholar] [CrossRef] [PubMed]
- Carpena-Catoira, C.; Ortega-Ortiz, C.D.; Liñán-Cabello, M.A.; Olivos-Ortiz, A.; Elorriaga-Verplancken, F.R. Foraging ecology of the olive ridley sea turtle (Lepidochelys olivacea) from the Mexican Central Pacific based on stable isotopes. Reg. Stud. Mar. Sci. 2022, 52, 102296. [Google Scholar] [CrossRef]
- Zepeda-Borja, K.M.; Ortega-Ortiz, C.D.; Torres-Orozco, E.; Olivos-Ortiz, A. Spatial and temporal distribution of sea turtles related to sea surface temperature and chlorophyll-a in Mexican Central Pacific waters. Rev. Biol. Mar. Oceanogr. 2017, 52, 375–385. [Google Scholar] [CrossRef]
- Peavey, L.E.; Popp, B.N.; Pitman, R.L.; Gaines, S.D.; Arthur, K.E.; Kelez, S.; Seminoff, J.A. Opportunism on the high seas: Foraging ecology of olive ridley turtles in the eastern Pacific Ocean. Front. Mar. Sci. 2017, 4, 348. [Google Scholar] [CrossRef]
- Collins, M.; Sutherland, M.; Bouwer, L.; Cheong, S.-M.; Frölicher, T.; Jacot Des Combes, H.; Koll Roxy, M.; Losada, I.; McInnes, K.; Ratter, B.; et al. Extremes, Abrupt Changes and Managing Risk. In IPCC Special Report on the Ocean and Cryosphere in a Changing Climate; Pörtner, H.-O., Roberts, D.C., Masson-Delmotte, V., Zhai, P., Tignor, M., Poloczanska, E., Mintenbeck, K., Alegría, A., Nicolai, M., Okem, A., et al., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2019; pp. 589–655. [Google Scholar] [CrossRef]
- Boyle, M.C.; Fitz-Simmons, N.N.; Limpus, C.J.; Kelez, S.; Velez-Zuazo, X.; Waycott, M. Evidence for transoceanic migrations by loggerhead sea turtles in the southern Pacific Ocean. Proc. R. Soc. B: Biol. Sci. 2009, 276, 1993–1999. [Google Scholar] [CrossRef]
- Brongersma, L.D.; Carr, A.F. Lepidochelys kempi, (Garman) from Malta. Proc. K. Ned. Akad. Wet. C 1983, 86, 445–454. [Google Scholar]
- Tomás, J.; Formia, A.; Fernández, M.; Raga, J.A. Occurrence and genetic analyses of a Kemp’s ridley sea turtle (Lepidochelys kempii) in the Mediterranean Sea. Sci. Mar. 2003, 67, 367–369. [Google Scholar] [CrossRef]
- Oliver, G.; Pigno, A. Première observation d’une Tortue de Kemp, Lepidochelys kempii (Garman, 1880), (Reptilia, Chelonii, Cheloniidae) sur les côtes françaises de Méditerranée. Bull. Soc. Herp. Fr. 2005, 116, 31–38. [Google Scholar]
- Tomás, J.; Raga, J.A. Occurrence of Kemp’s Ridley sea turtle (Lepidochelys kempii) in the Mediterranean. Mar. Biodivers. Rec. 2008, 1, e58. [Google Scholar] [CrossRef]
- Insacco, G.; Spadola, F. First record of Kemp’s ridley sea turtle, Lepidochelys kempii (Garman 1880) (Cheloniidae), from the italian waters (Mediterranean Sea). Acta Herpetol. 2010, 5, 113–117. [Google Scholar] [CrossRef]
- Carreras, C.; Monzon-Arguello, C.; López-Jurado, L.F.; Calabuig, P.; Bellido, J.J.; Castillo, J.J.; Sanchez, P.; Medina, P.; Tomas, J.; Gozalbes, P.; et al. Origin and dispersal routes of foreign green and Kemp’s ridley turtles in Spanish Atlantic and Mediterranean waters. Amphib. Reptil. 2014, 35, 73–86. [Google Scholar] [CrossRef]
- Lahanas, P.N.; Bjorndal, K.A.; Bolten, A.B.; Encalada, S.E.; Miyamoto, M.M.; Valverde, R.A.; Bowen, B.W. Genetic composition of a green turtle (Chelonia mydas) feeding ground population: Evidence for multiple origins. Mar. Biol. 1998, 130, 345–352. [Google Scholar] [CrossRef]
- Luke, K.; Horrocks, J.A.; LeRoux, R.A.; Dutton, P.H. Origins of green turtle (Chelonia mydas) feeding aggregations around Barbados, West Indies. Mar.Biol. 2004, 144, 799–805. [Google Scholar] [CrossRef]
- Prosdocimi, L.; Dutton, P.; Albareda, D.; Remis, M.I. Origin and genetic diversity of leatherbacks (Dermochelys coriacea) at Argentine foraging grounds. J. Exp. Mar. Biol. Ecol. 2014, 458, 13–19. [Google Scholar] [CrossRef]
- Doyle, T.K.; Houghton, J.D.R.; O’Súilleabháin, P.F.; Hobson, V.J.; Marnell, F.; Davenport, J.; Hays, G.C. Leatherback turtles satellite-tagged in European waters. Endanger. Species Res. 2008, 23, 23–31. [Google Scholar] [CrossRef]
- Vargas, S.M.; Molfetti, E.; Vilaça, S.T.; Monteiro, D.S.; Estima, S.C.; Soares, L.S.; Almeida, A.P.; Thoisy, B.; Naro-Maciel, E.; Santos, F.R. Mixed stock analysis of leatherback turtles feeding in Brazil: Records over four years. In Proceedings of the 33rd Symposium on Sea Turtle Biology and Conservation, Baltimore, MD, USA, 2–8 February 2013; NOAA technical memorandum NMFS-SEFSC–645. p. 246. [Google Scholar]
- Fossette, S.; Witt, M.J.; Miller, P.; Nalovic, M.A.; Albareda, D.; Almeida, A.P.; Broderick, A.C.; Chacón-Chaverri, D.; Coyne, M.S.; Domingo, A.; et al. Pan-Atlantic analysis of the overlap of a highly migratory species, the leatherback turtle, with pelagic longline fisheries. Proc. Royal Soc. B 2014, 281, 20133065. [Google Scholar] [CrossRef]
- Bolten, A.B.; Bjorndal, K.A.; Martins, H.R.; Dellinger, T.; Biscoito, M.J.; Encalada, S.E.; Bowen, B.W. Transatlantic developmental migrations of loggerhead sea turtles demonstrated by mtDNA sequence analysis. Ecol. Appl. 1998, 8, 1–7. [Google Scholar] [CrossRef]
- Laurent, L.; Casale, P.; Bradai, M.N.; Godley, B.J.; Gerosa, G.; Broderick, A.C.; Schroth, W.; Shierwater, B.; Levy, A.M.; Freggi, D.; et al. Molecular resolution of marine turtle stock composition in fishery bycatch: A case study in the Mediterranean. Mol. Ecol. 1998, 7, 1529–1542. [Google Scholar] [CrossRef]
- Hahn, A.T. Filogeografia global da tartaruga oliva (Lepidochelys olivacea). Ph.D. Thesis, Pontifícia Universidade Católica do Rio Grande do Sul, Porto Alegre, Rio Grande do Sul, Brasil, 2011. [Google Scholar]
- Bonjean, F.; Lagerloef, G.S.E. Diagnostic model and analysis of the surface currents in the tropical Pacific Ocean. J. Phys. Oceanogr. 2002, 32, 2938–2954. [Google Scholar] [CrossRef]
| Maturity Stages | ||||
|---|---|---|---|---|
| Haplotype | Females | Males | Immatures | Total |
| Lo46 | 18 (8-3-5-2) | 19 (4-14-0-1) | 17 (6-6-3-2) | 54 (18-23-8-5) |
| Lo54 | 4 (0-3-0-1) | 4 (1-2-1-0) | 8 (1-5-1-1) | |
| Lo59 | 2 (1-1-0-0) | 2 (1-0-0-1) | 4 (2-1-0-1) | |
| Lo52 | 1 (0-1-0-0) | 1 (0-1-0-0) | 1 (1-0-0-0) | 3 (1-2-0-0) |
| Lo60 | 2 (1-0-0-1) | 1 (0-1-0-0) | 3 (1-1-0-1) | |
| Lo27 | 1 (0-1-0-0) | 1 (0-1-0-0) | 2 (0-2-0-0) | |
| Lo28 | 1 (1-0-0-0) | 1 (1-0-0-0) | ||
| Lo31 | 1 (0-0-1-0) | 1 (0-0-1-0) | ||
| Lo57 | 1 (0-0-1-0) | 1 (0-0-1-0) | ||
| Lo61 | 1 (0-1-0-0) | 1 (0-1-0-0) | ||
| Lo62 | 1 (0-1-0-0) | 1 (0-1-0-0) | ||
| Lo71 | 1 (0-1-0-0) | 1 (0-1-0-0) | ||
| Lo34 | 1 (1-0-0-0) | 1 (1-0-0-0) | ||
| Lo35 | 1 (0-1-0-0) | 1 (0-1-0-0) | ||
| Lo37 * | 1 (0-1-0-0) | 1 (0-1-0-0) | ||
| Lo53 * | 1 (0-1-0-0) | 1 (0-1-0-0) | ||
| Lo87 * | 1 (0-0-1-0) | 1 (0-0-1-0) | ||
| 27 (11-6-7-3) | 31 (4-24-1-2) | 27 (10-10-4-3) | 85 (25-40-12-8) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martín-del-Campo, R.; Ortega-Ortiz, C.D.; Abreu-Grobois, A.; Enríquez-Paredes, L.M.; Petatán-Ramírez, D.; García-Gasca, A.; Quijano-Scheggia, S.I. Genetic Evidence for Indo-Western Pacific Olive Ridley Sea Turtles in Mexican Waters. Diversity 2023, 15, 430. https://doi.org/10.3390/d15030430
Martín-del-Campo R, Ortega-Ortiz CD, Abreu-Grobois A, Enríquez-Paredes LM, Petatán-Ramírez D, García-Gasca A, Quijano-Scheggia SI. Genetic Evidence for Indo-Western Pacific Olive Ridley Sea Turtles in Mexican Waters. Diversity. 2023; 15(3):430. https://doi.org/10.3390/d15030430
Chicago/Turabian StyleMartín-del-Campo, Rodolfo, Christian D. Ortega-Ortiz, Alberto Abreu-Grobois, Luis M. Enríquez-Paredes, David Petatán-Ramírez, Alejandra García-Gasca, and Sonia I. Quijano-Scheggia. 2023. "Genetic Evidence for Indo-Western Pacific Olive Ridley Sea Turtles in Mexican Waters" Diversity 15, no. 3: 430. https://doi.org/10.3390/d15030430
APA StyleMartín-del-Campo, R., Ortega-Ortiz, C. D., Abreu-Grobois, A., Enríquez-Paredes, L. M., Petatán-Ramírez, D., García-Gasca, A., & Quijano-Scheggia, S. I. (2023). Genetic Evidence for Indo-Western Pacific Olive Ridley Sea Turtles in Mexican Waters. Diversity, 15(3), 430. https://doi.org/10.3390/d15030430







