Microbial Communities of Peaty Permafrost Tundra Soils along the Gradient of Environmental Conditions and Anthropogenic Disturbance in Pechora River Delta in the Eastern European Arctic
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Region
2.2. Study Sites and Objects
2.3. Sample Collection and Soil Characterization
2.4. DNA Extraction, qPCR Assays, and Sequencing
2.5. Sequence Data Processing and Statistical Analysis
3. Results
3.1. The Characteristics of the Sampling Sites and Soil Physicochemical Properties
3.2. Results of qPCR Estimates of Bacterial and Fungal Abundance
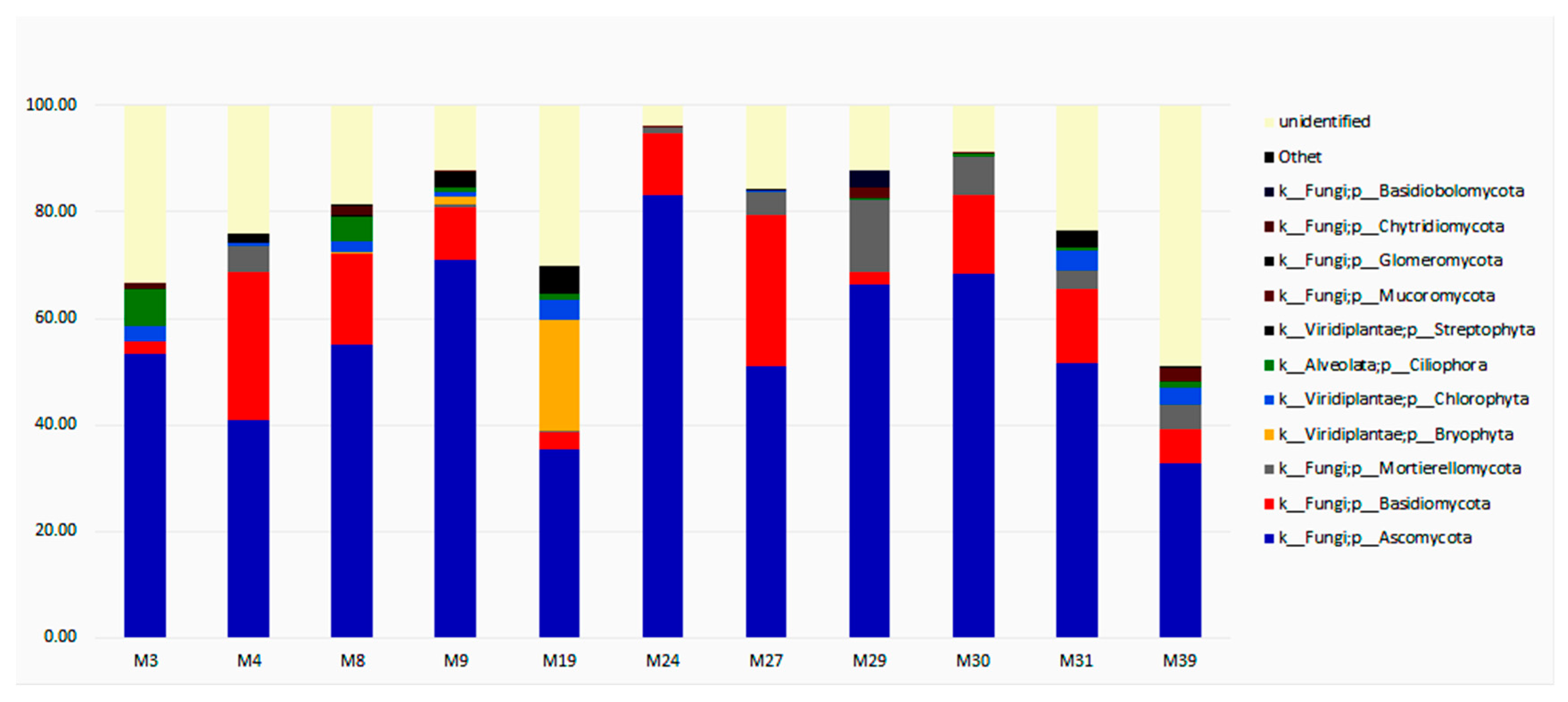
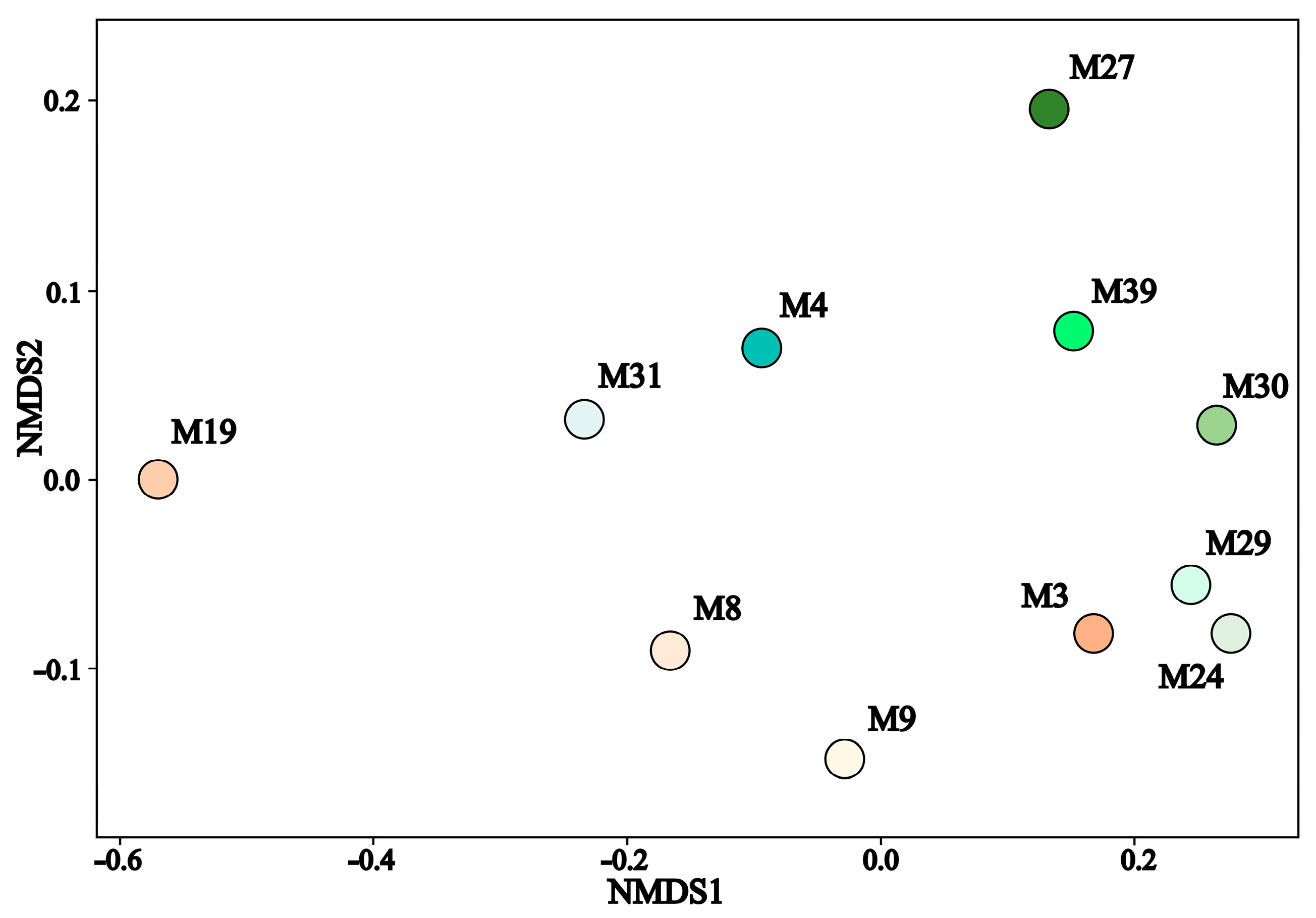
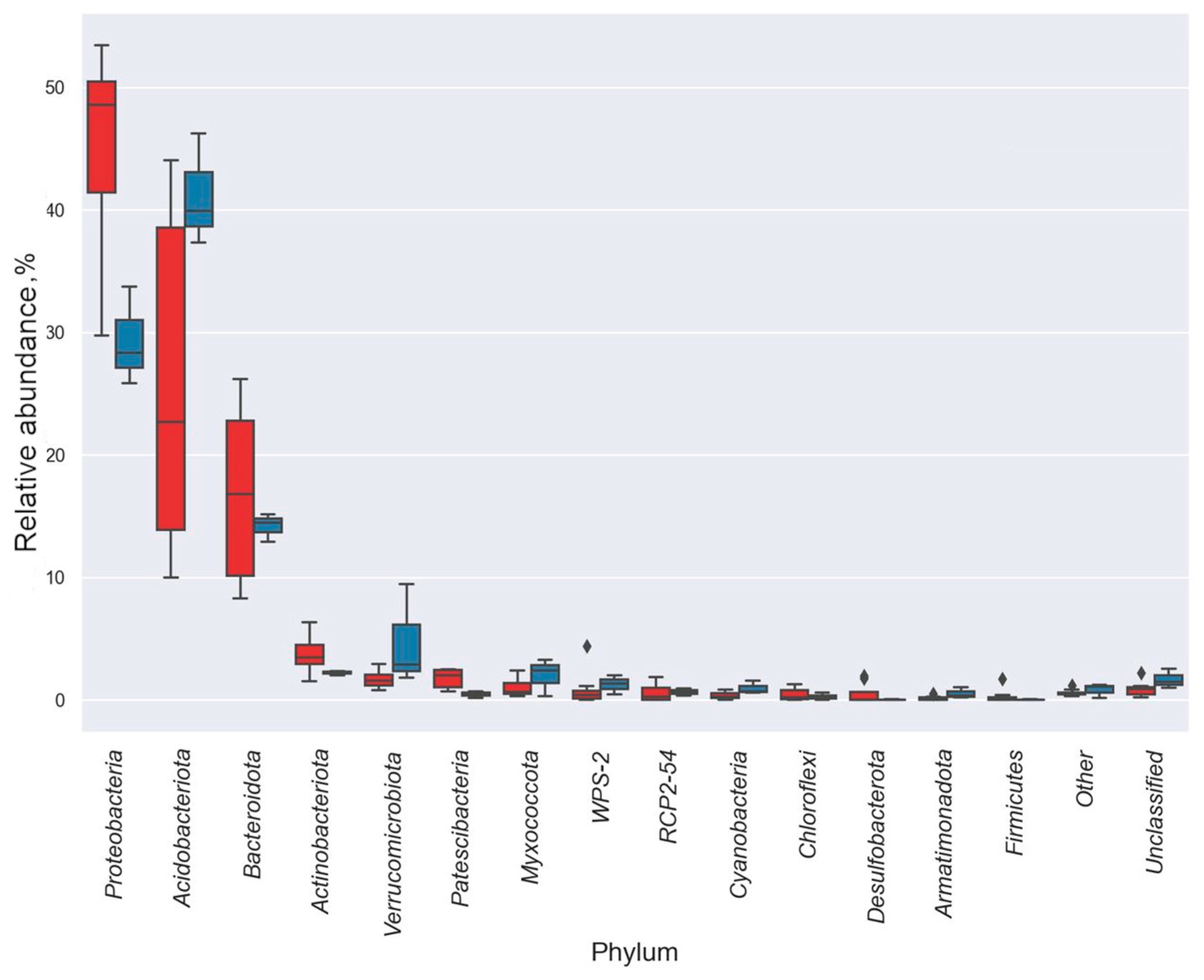
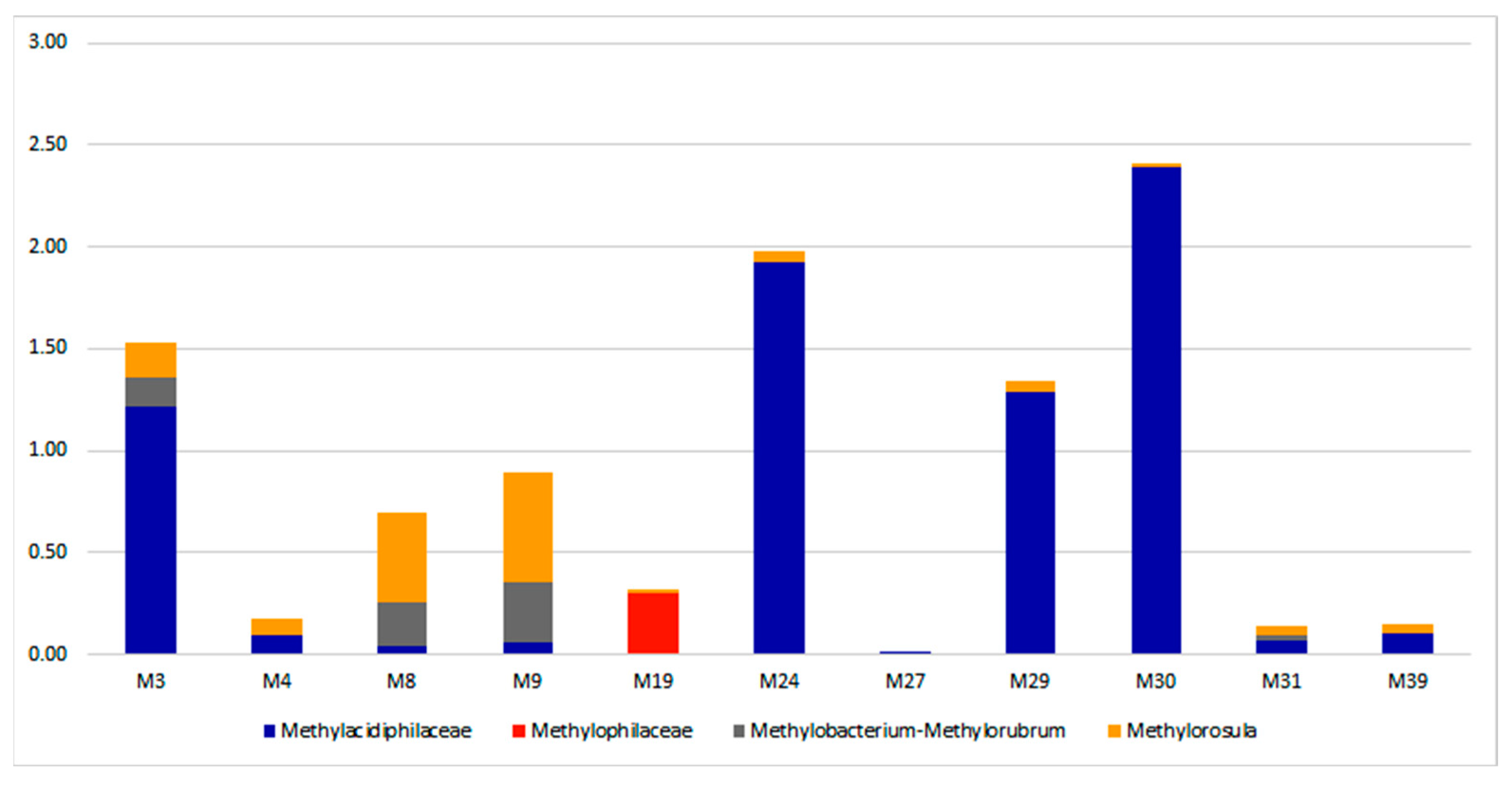
3.3. Taxonomic Diversity and Microbial Community’s Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Masson-Delmotte, V.; Zhai, P.; Pirani, A.; Connors, S.; Péan, C.; Berger, S.; Caud, N.; Chen, Y.; Goldfarb, L.; Gomis, M.; et al. (Eds.) IPCC, 2021: Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2021; p. 2391. [Google Scholar] [CrossRef]
- Shukla, P.R.; Skea, J.; Calvo Buendia, E.; Masson-Delmotte, V.; Pörtner, H.-O.; Roberts, D.C.; Zhai, P.; Slade, R.; Connors, S.; van Diemen, R.; et al. (Eds.) IPCC, 2019. Climate Change and Land: An IPCC Special Report on Climate Change, Desertification, Land Degradation, Sustainable Land Management, Food Security, and Greenhouse Gas Fluxes in Terrestrial Ecosystems; IPCC: Geneva, Switzerland, 2019; Available online: https://www.ipcc.ch/srccl/ (accessed on 20 November 2022).
- Parish, F.; Sirin, A.; Charman, D.; Joosten, H.; Minayeva, T.; Silvius, M.; Stringer, L. (Eds.) Assessment on Peatlands, Biodiversity and Climate Change: Main Report; Global Environment Centre: Kuala Lumpur, Malaysia; Wetlands International: Wageningen, The Netherlands, 2008; pp. 118–138. [Google Scholar]
- Joosten, H.; Sirin, A.; Couwenberg, J.; Laine, J.; Smith, P. The role of peatlands in climate regulation. In Peatland Restoration and Ecosystem Services: Science, Policy and Practice; Bonn, A., Allott, T., Evans, M., Joosten, H., Stoneman, R., Eds.; Cambridge University Press: Cambridge, UK, 2016; pp. 66–79. [Google Scholar] [CrossRef]
- Minayeva, T.Y.; Sirin, A.A. Peatland biodiversity and climate change. Biol. Bull. Rev. 2012, 2, 164–175. [Google Scholar]
- Minayeva, T.Y.; Bragg, O.M.; Sirin, A.A. Towards ecosystem-based restoration of peatland biodiversity. Mires Peat 2017, 19, 1–36. [Google Scholar] [CrossRef]
- Minayeva, T.; Sirin, A.; Kershaw, P.; Bragg, O. Arctic Peatlands. In The Wetland Book II: Distribution, Description, and Conservation; Springer: Dordrecht, The Netherlands, 2018; pp. 275–288. [Google Scholar] [CrossRef]
- Minayeva, T.; Sirin, A. Arctic peatlands. In Arctic Biodiversity Trends 2010–Selected Indicators of Change; CAFF International Secretariat: Akureyri, Iceland, 2010; pp. 71–74. Available online: http://www.arcticbiodiversity.is/images/stories/report/pdf/Arctic_Biodiversity_Trends_Report_2010.pdf (accessed on 20 November 2022).
- Sirin, A.A. Bolota. In Rossijskaya Arktika: Prostranstvo. Vremya. Resursy. Atlas; Agafonova, S.A., Ajbulatov, D.N., Baburin, V.L., Eds.; Feoriya: Moscow, Russia, 2019; pp. 344–345. (In Russian) [Google Scholar]
- Naylor, D.; Sadler, N.; Bhattacharjee, A.; Graham, E.B.; Anderton, C.R.; McClure, R.; Lipton, M.; Hofmockel, K.S.; Jansson, J.K. Soil Microbiomes Under Climate Change and Implications for Carbon Cycling. Annu. Rev. Environ. Resour. 2020, 45, 29–59. [Google Scholar] [CrossRef]
- Grosse, G.; Harden, J.W.; Turetsky, M.; McGuire, A.D.; Camill, P.; Tarnocai, C.; Frolking, S.; Schuur, E.A.G.; Jorgenson, T.; Marchenko, S.; et al. Vulnerability of high-latitude soil organic carbon in North America to disturbance. J. Geophys. Res. Atmos. 2011, 116, G00K06. [Google Scholar] [CrossRef]
- Muhammad, I.; Lv, J.Z.; Wang, J.; Ahmad, S.; Farooq, S.; Ali, S.; Zhou, X.B. Regulation of Soil Microbial Community Structure and Biomass to Mitigate Soil Greenhouse Gas Emission. Front. Microbiol. 2022, 13, 868862. [Google Scholar] [CrossRef]
- Andersen, R.; Chapman, S.; Artz, R. Microbial communities in natural and disturbed peatlands: A review. Soil Biol. Biochem. 2012, 57, 979–994. [Google Scholar] [CrossRef]
- Yu, S.; Ehrenfeld, J.G. Relationships among plants, soils and microbial communities along a hydrological gradient in the New Jersey Pinelands, USA. Ann. Bot. 2010, 105, 185–196. [Google Scholar] [CrossRef]
- Ji, M.; Kong, W.; Liang, C.; Zhou, T.; Jia, H.; Dong, X. Permafrost thawing exhibits a greater influence on bacterial richness and community structure than permafrost age in Arctic permafrost soils. Cryosphere 2020, 14, 3907–3916. [Google Scholar] [CrossRef]
- Aksenov, A.; Shirokova, L.; Kisil, O.; Kolesova, S.; Lim, A.; Kuzmina, D.; Pouillé, S.; Alexis, M.; Castrec-Rouelle, M.; Loiko, S.; et al. Bacterial Number and Genetic Diversity in a Permafrost Peatland (Western Siberia): Testing a Link with Organic Matter Quality and Elementary Composition of a Peat Soil Profile. Diversity 2021, 13, 328. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Z.; Ma, D.; Zhang, M.; Fu, L. Diversity and Distribution Characteristics of Soil Microbes across Forest–Peatland Ecotones in the Permafrost Regions. Int. J. Environ. Res. Public Health 2022, 19, 14782. [Google Scholar] [CrossRef]
- Vitorino, L.C.; Bessa, L.A. Microbial Diversity: The Gap between the Estimated and the Known. Diversity 2018, 10, 46. [Google Scholar] [CrossRef]
- Liu, S.; Moon, C.D.; Zheng, N.; Huws, S.; Zhao, S.; Wang, J. Opportunities and challenges of using metagenomic data to bring uncultured microbes into cultivation. Microbiome 2022, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Tanneberger, F.; Moen, A.; Barthelmes, A.; Lewis, E.; Miles, L.; Sirin, A.; Tegetmeyer, C.; Joosten, H. Mires in Europe—Regional Diversity, Condition and Protection. Diversity 2021, 13, 381. [Google Scholar] [CrossRef]
- Moen, A.; Joosten, H.; Tanneberger, F. Mire diversity in Europe: Mire regionality. In Mires and Peatlands of Europe. Status, Distribution and Conservation; Joosten, H., Tanneberger, F., Moen, A., Eds.; Schweizerbart: Stuttgart, Germany, 2017; pp. 97–150. [Google Scholar]
- Sirin, A.; Minayeva, T.; Yurkovskaya, T.; Kuznetsov, O.; Smagin, V.; Fedotov, Y.U. Russian Federation (European Part). In Mires and Peatlands of Europe: Status, Distribution and Conservation; Joosten, H., Tanneberger, F., Moen, A., Eds.; Schweizerbart Science Publishers: Stuttgart, Germany, 2017; pp. 589–616. [Google Scholar] [CrossRef]
- Minaevoj, T.Y. E`Kologicheskaya Restavracziya v Arktike: Obzor Mezhdunarodnogo i Rossijskogo opy`ta. [Ecological Restoration in the Arctic: A Review of International and Russian Experience]; Sy`kty`vkar-Nar`yan-Mar; Triada Publ.: Sewickley, PA, USA, 2016; p. 287. ISBN 978-5-94789-751-7. (In Russian) [Google Scholar]
- Sudakova, M.; Sadurtdinov, M.; Skvortsov, A.; Tsarev, A.; Malkova, G.; Molokitina, N.; Romanovsky, V. Using Ground Penetrating Radar for Permafrost Monitoring from 2015–2017 at CALM Sites in the Pechora River Delta. Remote Sens. 2021, 13, 3271. [Google Scholar] [CrossRef]
- Nikonova, A.N. Transformation of Floodplain Ecosystems in the Pechora Delta within the Kumzhinsk Gas Condensate Field (Nenets Autonomous Okrug). Izvestiya Rossiiskoi Akademii Nauk. Seriya Geogr. 2015, 5, 117–129. (In Russian) [Google Scholar] [CrossRef]
- Korchagin, A.A. Structure of plant communities. In Field Geobotany; Nauka: Moscow, Russia, 1976; Volume 5, 313p. [Google Scholar]
- Fierer, N.; Jackson, J.A.; Vilgalys, R.; Jackson, R.B. Assessment of Soil Microbial Community Structure by Use of Taxon-Specific Quantitative PCR Assays. Appl. Environ. Microbiol. 2005, 71, 4117–4120. [Google Scholar] [CrossRef]
- Martin, K.J.; Rygiewicz, P.T. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 2005, 5, 28. [Google Scholar] [CrossRef]
- Banos, S.; Lentendu, G.; Kopf, A.; Wubet, T.; Glöckner, F.O.; Reich, M. A comprehensive fungi-specific 18S rRNA gene sequence primer toolkit suited for diverse research issues and sequencing platforms. BMC Microbiol. 2018, 18, 190. [Google Scholar] [CrossRef]
- Martin, V. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Vieira, S.; Sikorski, J.; Dietz, S.; Herz, K.; Schrumpf, M.; Bruelheide, H.; Scheel, D.; Friedrich, M.W.; Overmann, J. Drivers of the composition of active rhizosphere bacterial communities in temperate grasslands. ISME J. 2020, 14, 463–475. [Google Scholar] [CrossRef]
- Strauss, J.; Schirrmeister, L.; Mangelsdorf, K.; Eichhorn, L.; Wetterich, S.; Herzschuh, U. Organic-matter quality of deep permafrost carbon—A study from Arctic Siberia. Biogeosciences 2015, 12, 2227–2245. [Google Scholar] [CrossRef]
- Wu, Y.; Ulrich, C.; Kneafsey, T.; Lopez, R.; Chou, C.; Geller, J.; McKnight, K.; Dafflon, B.; Soom, F.; Peterson, J.; et al. Depth-Resolved Physicochemical Characteristics of Active Layer and Permafrost Soils in an Arctic Polygonal Tundra Region. J. Geophys. Res. Biogeosci. 2018, 123, 1366–1386. [Google Scholar] [CrossRef]
- Matyshak, G.V.; Bogatyrev, L.G.; Goncharova, O.Y.; Bobrik, A.A. Specific features of the development of soils of hydromorphic ecosystems in the Northern Taiga of Western Siberia under conditions of cryogenesis. Eurasian Soil Sci. 2017, 50, 1115–1124. [Google Scholar]
- Pastukhov, A.; Kovaleva, V.; Kaverin, D. Microbial community structure in ancient European Arctic Peatlands. Plants 2022, 11, 2704. [Google Scholar] [CrossRef]
- Alekseev, I.; Shamilishvilly, G.; Abakumov, E. Content of trace elements in selected permafrost-affected soils of Yamal region with different functional load. Polarforschung 2014, 88, 125–133. [Google Scholar] [CrossRef]
- Kitrsidelli, I.Y.; Vlasov, D.Y.; Barantsevich, E.P.; Krylenkov, V.A.; Sokolov, V.T. Kompleksy mikroskopicheskih gribov v pochvah i gruntah poljarnogo ostrova Izvestija Cik (Karskoje more). (Microfungi from soil of polar Island Izvestia tsik [Kara Sea]). Mikol. Fitopatol. 2014, 48, 365–371. [Google Scholar]
- Vlasov, D.; Abakumov, E.; Tomashunas, V.; Krylenkov, V.; Zelenskaya, M. Micobiota pochv i antropogennyh substratov poluostrova Jamal. (Mycobiota of soil and anthropogenic substrates of the Yamal Peninsula). Gig. Sanit. 2014, 5, 49–51. [Google Scholar]
- Alekseev, I.; Zverev, A.; Abakumov, E. Organic carbon and microbiome in tundra and forest–tundra permafrost soils, southern Yamal, Russia. Polar Res. 2021, 40, 5283. [Google Scholar] [CrossRef]
- Malard, L.; Anwar, M.Z.; Jacobsen, C.S.; Pearce, D. Biogeographical patterns in soil bacterial communities across the Arctic region. FEMS Microbiol. Ecol. 2019, 95, fiz128. [Google Scholar] [CrossRef] [PubMed]

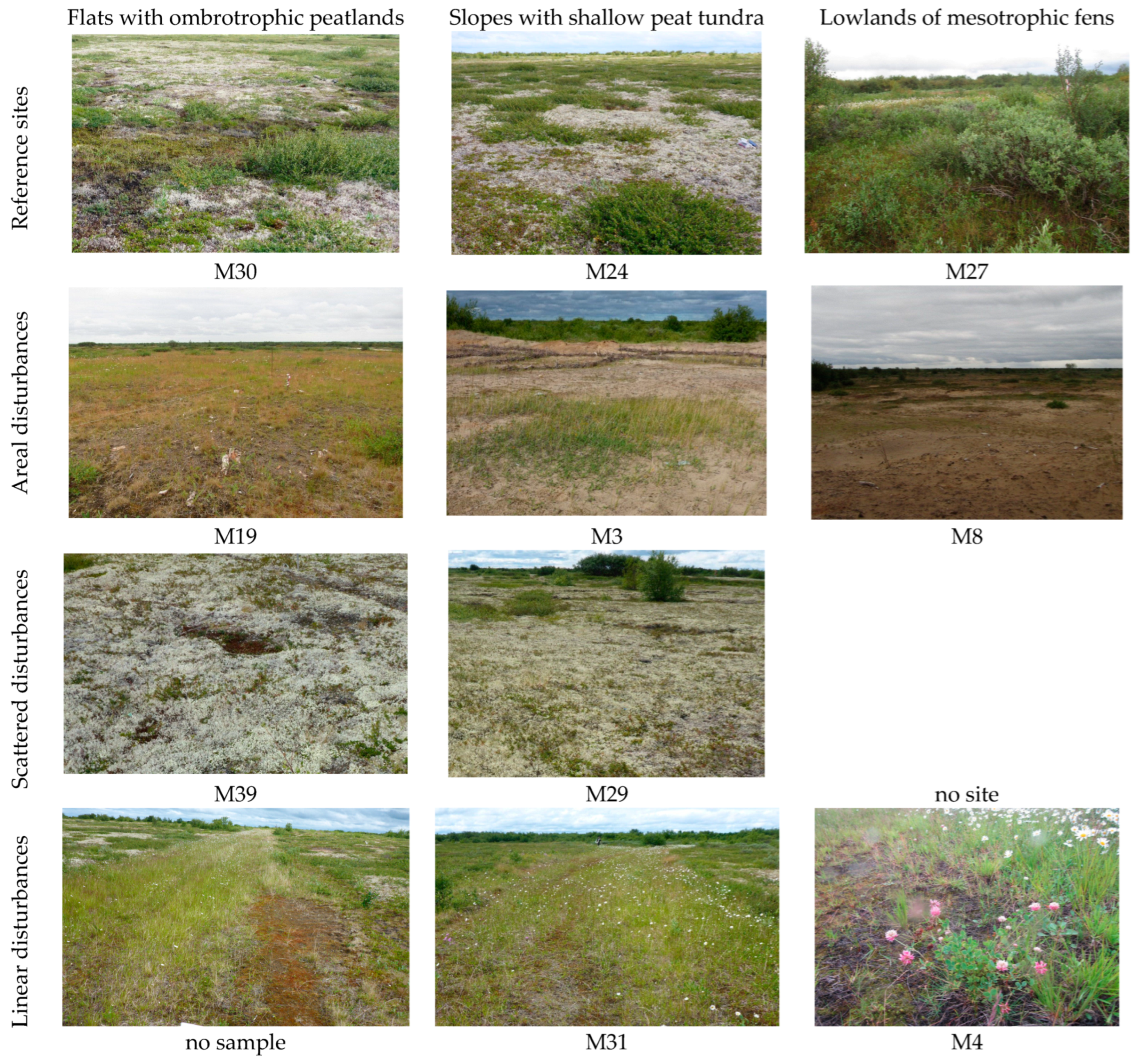

| Site ID | Coordinates | Elevation, m | Initial Micro-Landscape | Dominating Current Vegetation | Disturbance Type | Restoration Action |
|---|---|---|---|---|---|---|
| M30 | N 68°11′31.25″; E 53°46′41.29″ | 4.0 | Ombrotrophic mires of flat landscapes | Sedge, lichen, sphagnum hummocks within fen | Reference | No |
| M24 | N 68°11′36.51″; E 53°46′46.35″ | 6.1 | Shallow peat tundra on the slope | Lichen, dwarf shrubs with dwarf birch | Reference | No |
| M27 | N 68°11′38.58″; E 53°46′47.18″ | 4.0 | Fens on the lowlands and riverine habitats | Brown moss forbs, willow dwarf birch | Reference | No |
| M19 | N 68°11′35.70″; E 53°47′11.92″ | 6.5 | Ombrotrophic mires of flat landscapes | Grass, moss mesophilic with shrubs | Areal | Self-regeneration |
| M3 | N 68°11′37.78″; E 53°47′15.74″ | 6.1 | Shallow peat tundra on the slope | Grass, forbs xerophilic with crust | Areal | Self-regeneration |
| M8 | N 68°11′37.04″; E 53°47′15.68″ | 5.4 | Fens on the lowlands and riverine habitats | Grass, forbs xerophilic with crust | Areal | Self-regeneration |
| M9 | N 68°11′38.27″; E 53°47′15.68″ | 5.4 | Fens on the lowlands and riverine habitats | Grass, forbs xerophilic with lichens | Areal | Ecological restoration |
| M39 | N 68°11′35.55″; E 53°46′50.42″ | 5.6 | Ombrotrophic mires of flat landscapes | Lichen, dwarf birch | Scattered | Self-regeneration |
| M29 | N 68°11′35.81″; E 53°46′50.50″ | 5.6 | Shallow peat tundra on the slope | Lichen, dwarf shrubs | Scattered | Self-regeneration |
| M31 | N 68°11′37.49″; E 53°46′54.36″ | 5.2 | Shallow peat tundra on the slope | Moss, forbs, willow dwarf birch | Linear | Self-regeneration |
| M4 | N 68°11′38.94″; E 53°46′47.45″ | 4.0 | Fens on the lowlands and riverine habitats | Brown moss, forbs with willow | Linear | Self-regeneration |
| Site ID | Organic Soil Layer | Underlying Bedrock | Vegetation Cover, % | Litter Cover, % | Number of High Plants Species | Average Vegetation Height, cm | Soil Water Level, cm | Active Layer Depth, m | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | Cover, % | Depth, cm | Overall | Shrub | Herb | Moss, Lichen | Shrub | Herb | ||||||
| Reference sites | ||||||||||||||
| M30 | peat | 100 | 12 | sandy loess | 95 | 40 | 50 | 90 | 15 | 19 | 60 | 25 | −3 | 1.25 |
| M24 | peat and litter | 100 | 10 | sandy loess | 90 | 3 | 40 | 85 | 0 | 23 | 100 | 40 | −5 | 2.2 |
| M27 | moss, litter, turf, peat | 100 | 25 | sand | 90 | 30 | 70 | 30 | 0 | 15 | 120 | 40 | −7 | >1.80 |
| Areal disturbance | ||||||||||||||
| M19 | turf, moss, litter | 80 | 3 | sand and technogenic loam | 90 | 5 | 70 | 20 | 20 | 19 | 15 | 35 | >30 | >1.80 |
| M3 | litter, crust, and lichen | 50 | 2 | sand | 30 | 0 | 20 | 10 | 10 | 6 | 0 | 30 | >30 | >1.80 |
| M8 | crust and lichen | 30 | 2 | sand | 35 | 0 | 30 | 30 | 5 | 11 | 0 | 30 | >30 | >1.80 |
| M9 | crust | 5 | 1 | sand | 20 | 0 | 20 | 5 | 5 | 11 | 0 | 30 | >30 | 2.2 |
| Scattered disturbance | ||||||||||||||
| M39 | peat and litter | 100 | 6 | sand | 100 | 40 | 60 | 50 | 20 | 14 | 80 | 25 | >30 | 2 |
| M29 | moss, lichen, turf, peat | 80 | 6 | sand | 90 | 0 | 75 | 60 | 0 | 13 | 0 | 40 | >30 | >1.80 |
| Linear disturbance | ||||||||||||||
| M31 | turf and peat | 90 | 6 | sand | 70 | 0 | 70 | 20 | 20 | 11 | 0 | 30 | >30 | >1.80 |
| M4 | litter and crust | 65 | 2 | sand | 90 | 0 | 80 | 60 | 10 | 14 | 0 | 15 | −15 | >1.80 |
| Site ID | Material | pH | Bulk Density, g/cm3 | Moisture Content, % | Ash Content, % | Organic Matter, % | TC, % | TN, % | C:N | TS, % |
|---|---|---|---|---|---|---|---|---|---|---|
| Reference sites | ||||||||||
| M30 | peat and moss | 4.94 | 0.54 | 64.00 | 64.2 | 35.8 | n.d. | n.d. | n.d. | n.d. |
| M24 | peat, moss, lichen | 4.55 | 0.72 | 38.00 | 72.3 | 27.7 | n.d. | n.d. | n.d. | n.d. |
| M27 | moss, turf and peat with sand | 5.03 | 0.49 | 54.00 | 76.8 | 23.16 | 16.2 | 0.85 | 19 | 0.00 |
| Areal disturbance | ||||||||||
| M19 | turf with sand | 6.25 | 1.15 | 12.90 | 89.5 | 10.55 | 5.2 | 0.08 | 65 | 1.07 |
| M3 | sand and humified sand | 5.37 | 1.35 | 10.38 | 98.1 | 1.88 | 7.4 | 0.23 | 32 | 0.00 |
| M8 | sand and humified sand | 5.30 | 1.34 | 5.70 | 98.9 | 1.09 | 1.4 | 0.05 | 28 | 0.00 |
| M9 | sand and humified sand | 5.34 | 1.44 | 10.44 | 99.1 | 0.91 | 1.4 | 0.05 | 28 | 0.00 |
| Scattered disturbance | ||||||||||
| M39 | lichen and peat | 5.32 | 0.26 | 39.00 | 68.87 | 31.13 | 23.4 | 0.61 | 38 | 0.00 |
| M29 | moss, turf with sand | 4.63 | 0.94 | 12.80 | 93.8 | 6.23 | 6.5 | 0.24 | 27 | 0.00 |
| Linear disturbance | ||||||||||
| M31 | litter and turf with sand | 5.32 | 1.26 | 6.70 | 98.8 | 1.20 | 2.2 | 0.07 | 31 | 0.00 |
| M4 | moss, crust and turf with sand | 4.90 | 1.01 | 16.60 | 96.4 | 3.56 | 4.3 | 0.19 | 23 | 0.00 |
| Sample ID | Bacteria, 16S rRNA Copies g−1 | Fungi, ITS Copies g−1 |
|---|---|---|
| Reference sites | ||
| M30 | 10.50 ± 2.1 × 109 | 14.56 ± 4.2 × 106 |
| M24 | 1.30 ± 0.8 × 1010 | 7.20 ± 2.2 × 107 |
| M27 | 8.32 ± 1.8 × 109 | 6.94 ± 2.2 × 107 |
| Areal disturbance | ||
| M19 | 3.22 ± 1.2 × 1010 | 4.42 ± 0.4 × 107 |
| M3 | 2.42 ± 0.8 × 109 | 15.72 ± 4.4 × 105 |
| M8 | 4.98 ± 0.8 × 109 | 7.98 ± 2.4 × 106 |
| Scattered disturbance | ||
| M39 | 3.24 ± 0.8 × 1010 | 5.52 ± 1.1 × 106 |
| M29 | 9.46 ± 2.2 × 109 | 14.52 ± 5.4 × 106 |
| M9 | 6.20 ± 2.3 × 109 | 2.14 ± 0.6 × 107 |
| Linear disturbance | ||
| M31 | 4.36 ± 1.1 × 1010 | 5.72 ± 1.7 × 107 |
| M4 | 2.78 ± 0.6 × 1010 | 2.70 ± 0.3 × 107 |
| Site ID | OTU Number | Chao1 | Shannon | Simpson | Richness | Raw |
|---|---|---|---|---|---|---|
| Bacteria | ||||||
| M30 | 1290 ± 27 | 1850.4 ± 123.1 | 8.58 ± 0.15 | 0.9916 ± 0.002 | 0.6971± 0.004 | 17,138 |
| M24 | 1102 ± 42 | 1590.6 ± 111.5 | 7.82 ± 0.31 | 0.9809± 0.003 | 0.6928 ± 0.007 | 16,413 |
| M27 | 1594 ± 21 | 2205.0 ± 126.1 | 8.97 ± 0.17 | 0.9935± 0.001 | 0.7229 ± 0.009 | 19,948 |
| M19 | 1910 ± 24 | 2149.3 ± 118.4 | 9.32 ± 0.23 | 0.9941± 0.004 | 0.8887± 0.012 | 23,801 |
| M3 | 1149 ± 15 | 1766.4 ± 117.3 | 8.56 ± 0.30 | 0.9932± 0.003 | 0.6504 ± 0.011 | 12,281 |
| M8 | 1483 ± 23 | 2313.3 ± 124.4 | 9.2 ± 0.27 | 0.9959± 0.001 | 0.6411± 0.022 | 12,929 |
| M9 | 1498 ± 19 | 2120.3 ± 119.8 | 8.86 ± 0.16 | 0.9947± 0.004 | 0.7065 ± 0.016 | 17,177 |
| M39 | 1207 ± 11 | 1604.9 ± 111.2 | 8.83 ± 0.17 | 0.9935± 0.003 | 0,7521 ± 0.011 | 13,580 |
| M29 | 1287 ± 16 | 1775.2 ± 121.1 | 8.90 ± 0.22 | 0.9952± 0.004 | 0.7250 ± 0.014 | 16,899 |
| M31 | 1936 ± 22 | 2789.9 ± 125.4 | 9.93 ± 0.24 | 0.9976± 0.002 | 0.6939 ± 0.023 | 13,521 |
| M4 | 2030 ± 26 | 3055.0 ± 128.6 | 9.96 ± 0.19 | 0.9978± 0.004 | 0.6645 ± 0.019 | 15,428 |
| Fungi | ||||||
| M30 | 407 ± 3 | 444.0 ± 47.4 | 6.17 ± 0.11 | 0.9694 ± 0.004 | 0.9167 ± 0.006 | 19,346 |
| M24 | 435 ± 5 | 476.6 ± 39.5 | 3.98 ± 0.23 | 0.7591 ± 0.002 | 0.9128 ± 0.008 | 20,937 |
| M27 | 116 ± 2 | 139.0 ± 29.3 | 4.21 ± 0.24 | 0.8632 ± 0.001 | 0.8348 ± 0.005 | 60,281 |
| M19 | 410 ± 5 | 422.2 ± 22.1 | 6.14 ± 0.33 | 0.9635 ± 0.003 | 0.9711 ± 0.007 | 23,848 |
| M3 | 413 ± 2 | 438.4 ± 14.2 | 5.15 ± 0.18 | 0.9341± 0.002 | 0.9420 ± 0.005 | 13,908 |
| M8 | 670 ± 6 | 705.6 ± 31.1 | 6.72 ± 0.17 | 0.9724 ± 0.004 | 0.9495 ± 0.008 | 16,541 |
| M9 | 594 ± 6 | 651.0 ± 23.2 | 5.63 ± 0.19 | 0.9121 ± 0.001 | 0.9124 ± 0.007 | 23,872 |
| M39 | 403 ± 7 | 414.1 ± 22.2 | 5.77 ± 0.27 | 0.9122 ± 0.003 | 0.9731 ± 0.007 | 14,049 |
| M29 | 378 ± 2 | 417.1 ± 13.1 | 6.12 ± 0.31 | 0.9647 ± 0.003 | 0.9062 ± 0.008 | 10,035 |
| M31 | 639 ± 9 | 705.1 ± 29.2 | 7.35 ± 0.25 | 0.9861 ± 0.003 | 0.9063 ± 0.005 | 16,505 |
| M4 | 511 ± 4 | 575.1 ± 18.4 | 6.16 ± 0.23 | 0.9552 ± 0.005 | 0.8886 ± 0.004 | 29,912 |
| Initial Microlandscape Type | Disturbances | Reference | |||
|---|---|---|---|---|---|
| Areal—Self-Regeneration | Areal—Restoration Activities | Linear | Scattered | ||
| Shallow peat tundra on slopes | M3 | No sample | M31 | M29 | M24 |
| Ombrotrophic shallow peatlands of flat landscapes | M19 | No sample | No sample | M39 | M30 |
| Rich and transition fens in the lowlands and riverine habitats | M8 | M9 | M4 | No sample | M27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kravchenko, I.; Grouzdev, D.; Sukhacheva, M.; Minayeva, T.; Sirin, A. Microbial Communities of Peaty Permafrost Tundra Soils along the Gradient of Environmental Conditions and Anthropogenic Disturbance in Pechora River Delta in the Eastern European Arctic. Diversity 2023, 15, 251. https://doi.org/10.3390/d15020251
Kravchenko I, Grouzdev D, Sukhacheva M, Minayeva T, Sirin A. Microbial Communities of Peaty Permafrost Tundra Soils along the Gradient of Environmental Conditions and Anthropogenic Disturbance in Pechora River Delta in the Eastern European Arctic. Diversity. 2023; 15(2):251. https://doi.org/10.3390/d15020251
Chicago/Turabian StyleKravchenko, Irina, Denis Grouzdev, Marina Sukhacheva, Tatyana Minayeva, and Andrey Sirin. 2023. "Microbial Communities of Peaty Permafrost Tundra Soils along the Gradient of Environmental Conditions and Anthropogenic Disturbance in Pechora River Delta in the Eastern European Arctic" Diversity 15, no. 2: 251. https://doi.org/10.3390/d15020251
APA StyleKravchenko, I., Grouzdev, D., Sukhacheva, M., Minayeva, T., & Sirin, A. (2023). Microbial Communities of Peaty Permafrost Tundra Soils along the Gradient of Environmental Conditions and Anthropogenic Disturbance in Pechora River Delta in the Eastern European Arctic. Diversity, 15(2), 251. https://doi.org/10.3390/d15020251









