Abstract
This paper addresses the genetic delimitation of narrowly distributed Scapania magadanica and broadly circumpolar S. kaurinii. The phylogenetic trees based on maximum likelihood and Bayesian inference constructed for one of the most informative loci (ITS1–2) showed that Scapania magadanica was deeply nested within the clade of S. kaurinii. The comparison of the obtained topologies with known strong morphological dissimilarities of two taxa has led to the understanding that this approach does not work. The latter may be due to a widespread variable tentatively ‘ancestral’ species (S. kaurinii) having no joint molecular synapomorphies that would delimit it from the locally distributed derived taxon (S. magadanica). Therefore, the relationships of these two species were evaluated using molecular genetic distances with the Neighbor Net split network and TCS haplotype network. The obtained data have confirmed the speculation above, and it is possible to assume that when the S. magadanica lineage split, S. kaurinii already occupied a rather wide range, which could limit further gene flow among its remote populations.
1. Introduction
The speciation of a new species from local populations within the ranges of the widely distributed species is apparently a common and inevitable process. However, the question of how a newly formed species with a complex of specific morphological and genetic traits should be treated in terms of taxonomy if the widespread variable ancestral species has no molecular synapomorphies that would delimit it from the newly formed taxon remains unanswered. Even if speciation is proven by a shift in ecology and the achievement of distinct morphological traits, treating such derived populations as representing a separate species is forbidden from the cladistic point of view since an ancestral taxon appears paraphyletic, although an evolutionary taxonomic approach allows for an alternative decision. We encountered one such case while studying Scapania magadanica S.S. Choi, Bakalin and B.Y. Sun, a species with, apparently, a Beringian range, and the problem of its taxonomic treatment and delimitation from a widely distributed S. kaurinii Ryan.
Scapania magadanica was described by Choi et al. [1] and placed in sect. Curtae based on morphological features, including strongly conduplicate leaves, relatively narrow leaf lobes with rounded apices, and bicellular colorless gemmae. This placement was also inspired by the refinements of the concept of this section by Heinrichs et al. [2], who made the circumscription of the section considerably broader. Based on a molecular phylogenetic study, this section was shown to include Scapania irrigua (Nees) Nees and S. obcordata (Berggr.) S.W. Arnell, in addition to the taxa traditionally placed on it [2]. That action changed the morphological boundaries of the section. In particular, it turned out that plants of a lophozioid appearance, with almost equal, widely diverging lobes (S. obcordata) or distinctly (although shortly) decurrent leaves on the ventral side and oblong cells in the middle part of the ventral lobe of the leaf (S. irrigua), are not alien to sect. Curtae (see [3]). While working in the Hemiarctic environments of the Magadan Province of Russia in 2011, Choi and Bakalin collected remarkable plants in the crevices between stones in rock fields. These plants were characterized by strongly conduplicate leaves with rounded lobe apices and resembled a large modification of mainly Korean–Japanese S. diplophylloides Amakawa and S. Hatt. or Mediterranean S. helvetica Gottsche. Aside from its remarkably distinct distribution and minor morphological traits, the plants were characterized by paroicous inflorescence—a rare case in Scapania that has not been known in sect. Curtae before. Therefore, Choi et al. [1] did not doubt the need for a description of collected plants as a new species (such as S. magadanica) and suggested its placement in sect. Curtae.
Within several years of the original description by Choi et al. [1], the species was found in other areas of the Magadan Province as well as in the high mountain heathlands and mountain tundras of the central part of the Kamchatka Peninsula. These records and phytogeographical speculations on the distribution of S. magadanica were published by Klimova and Bakalin [4]. In addition, the cited authors provided an updated morphological description and figures of the taxon, taking into account the newly obtained data [4]. Thus, it turned out that in the mountains of northeast Asia, characterized by a continental or subcontinental climate, this species occurs, if not sporadically, then surely not exceptionally rarely. The morphological distinctions along with the current preparation of the Scapania integrative revision for Asia have inspired us to check the phylogenetic position of S. magadanica using a molecular phylogenetic approach. The first unpublished attempt by Klimova to recognize the phylogenetic position of the species by a traditionally used tree-based approach to the visualization of the inferred phylogenetic relationships among the included accessions revealed specimens assigned to this species in a clade deeply nested within the clade of S. kaurinii, a species morphologically very different from S. magadanica due to having loosely conduplicate leaves concave and dorsally turned, commonly as wide as long lobes with acute to widely obtuse and rarer rounded apices (versus strongly conduplicate leaves with convex lobes that are distinctly longer than wide with rounded apices), although noticeably shared with the latter paroicous inflorescence. Thus, we faced a dilemma: either to refer S. magadanica to S. kaurinii or to look for an approach that could justify its segregation. The presentation of the obtained assessments of the phylogenetic relationships of S. magadanica is the goal of the present account. Since the state of liverwort diversity knowledge in Russian Asia is still fragmentary, there were no similar works in other genera of hepatics in Russian Asia, and this is the first attempt to resolve such kind of problem. At the same, molecular phylogenetic studies of Asian mosses revealed several cases of such “problematic” topologies [5,6,7].
2. Materials and Methods
2.1. Taxon Sampling
To compile the dataset for molecular phylogenetic analysis, we studied six specimens of S. magadanica from the Kamchatka Territory and Magadan Province, and four specimens of S. kaurinii from Magadan Province. The same specimens were the base for the morphological comparison of these taxa. Sequences of S. kaurinii obtained from specimens from Chitinskaya Province available in GenBank were added to our dataset. Based on the data available in GenBank, we also included three accessions of Scapania compacta (Roth) Dumort. from Germany, Spain, and the United Kingdom and two accessions of Scapania spitsbergensis from Norway and Buryatiya Republic (originally studied by Vilnet et al. [8], Heinrichs et al. [2]); two specimens of S. crassiretis from Primorsky Territory were used as an outgroup for tree rooting. Specimen voucher details, including GenBank accession numbers, are listed in Table 1.

Table 1.
The list of voucher details and GenBank accession numbers for the specimens used in phylogenetic analysis in the present paper. The newly obtained sequences are marked in bold.
2.2. DNA Isolation, Amplification, and Sequencing
Three markers, the nuclear ITS1–2 plastid trnL–F region and trnG-intron, were used for a phylogenetic study. DNA was extracted from dried liverwort tissues using the NucleoSpin Plant II Kit (Macherey-Nagel, Germany) and DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Amplification of ITS1–2, trnL–F, and trnG-intron was performed using an Encyclo Plus PCR kit (Evrogen, Moscow, Russia) with the primers listed in Table 2.

Table 2.
Primers used in polymerase chain reaction (PCR) and cycle sequencing.
The polymerase chain reaction was performed in a total volume of 20 µL, including 1 µL of template DNA, 0.4 µL of Encyclo polymerase, 5 µL of Encyclo buffer, 0.4 µL of dNTP-mixture (included in Encyclo Plus PCR Kit), 13.4 µL (for trnL–F, trnG-intron)/12.4 µL (for ITS1–2) of double-distilled water (Evrogen, Moscow, Russia), 1 µL of dimethylsulfoxide/DMSO (for ITS1–2), and 0.4 µL of each primer (forward and reverse, at a concentration of 5 pmol/µL). Polymerase chain reactions were carried out using the following program: 180 sec. initial denaturation at 95 °C, followed by 30–40 cycles of 30 sec. denaturation at 94 °C, 20 (for trnL–F)—30 s. (for ITS1–2, trnG-intron) annealing at 56 °C (trnG-intron), 58 °C (trnL–F and ITS1–2), and 30 sec. elongation at 72 °C. Final elongation was carried out in one 5-min. step at 72 °C. Amplified fragments were visualized on 1% agarose TAE gels by EthBr staining and purified using the Cleanup Mini Kit (Evrogen, Moscow, Russia). The DNA was sequenced using the ABI PRISM® BigDye™ Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Waltham, MA, USA) with further analysis of the reaction products following the standard protocol on an automatic sequencer 3730 DNA Analyzer (Applied Biosystems, Waltham, MA, USA) in the Genome Center (Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow).
2.3. Phylogenetic Analyses
The independent alignments were compiled for the ITS1–2, trnL–F, and trnG-intron loci and aligned using MAFFT [15] with standard settings and then edited manually in BioEdit ver. 7.2.5 [16]. All positions of the final alignment were included in the phylogenetic analyses. Absent data at the ends of regions and missing loci were coded as missing data.
Phylogenetic trees were reconstructed using two approaches: maximum likelihood (ML) with IQ-tree ver. 1.6.12 [17] and Bayesian inference (BA) with MrBayes ver. 3.2.7 [18].
For the ML analysis, the best fitting evolutionary model of nucleotide substitutions according to the BIC value was TIM+F+G4 determined by IQ-tree. Consensus trees were constructed with 1000 bootstrap replicates.
Bayesian analyses were performed by running two parallel analyses using the GTR+I+G model. The analysis consisted of four Markov chains. Chains were run for five million generations, and trees were sampled every 500th generation. The first 2500 trees in each run were discarded as burn-in; thereafter, 15,000 trees were sampled from both runs to produce a resulting tree. Bayesian posterior probabilities were calculated from the trees sampled after burn-in. The average standard deviation of split frequencies between two runs reached 0.0028 before the analysis was stopped.
As phylogenetic trees often incompletely reflect molecular affinities between closely related species, we used Splits tree and Neighbor Net methods implemented in SplitsTree v.4.14.2 [19] and TCS to visualize molecular relationships within the S. magadanica—S. kaurinii complex neglected within the tree-like phylogeny interface inferred from ML and Bayesian analyses. A haplotype network was constructed by the TCS network inference method [20] using the PopART package (http://popart.otago.ac.nz/), accessed on 8 November 2019 [21]. The PopART program automatically removes positions having at least one N or a gap value from the consideration.
3. Results
In total, we sequenced twelve specimens; newly obtained ITS1–2 sequences from S. magadanica, S. kaurinii, and S. crassiretis specimens were deposited in GenBank. The trnL–F and trnG-intron sequences were non variable; therefore, all further inferences were based solely on ITS data. ITS1–2 alignment of the 17 specimens consisted of 808 character sites: conservative sites—712 (88.12%); variable sites—90 (11.14%); and parsimony-informative sites—47 (5.82%).
The ML criterion recovered a bootstrap consensus tree with a log-likelihood = −1701.327. The arithmetic means of the log likelihoods in Bayesian analysis for each sampling run were −1715.42 and −1714.90.
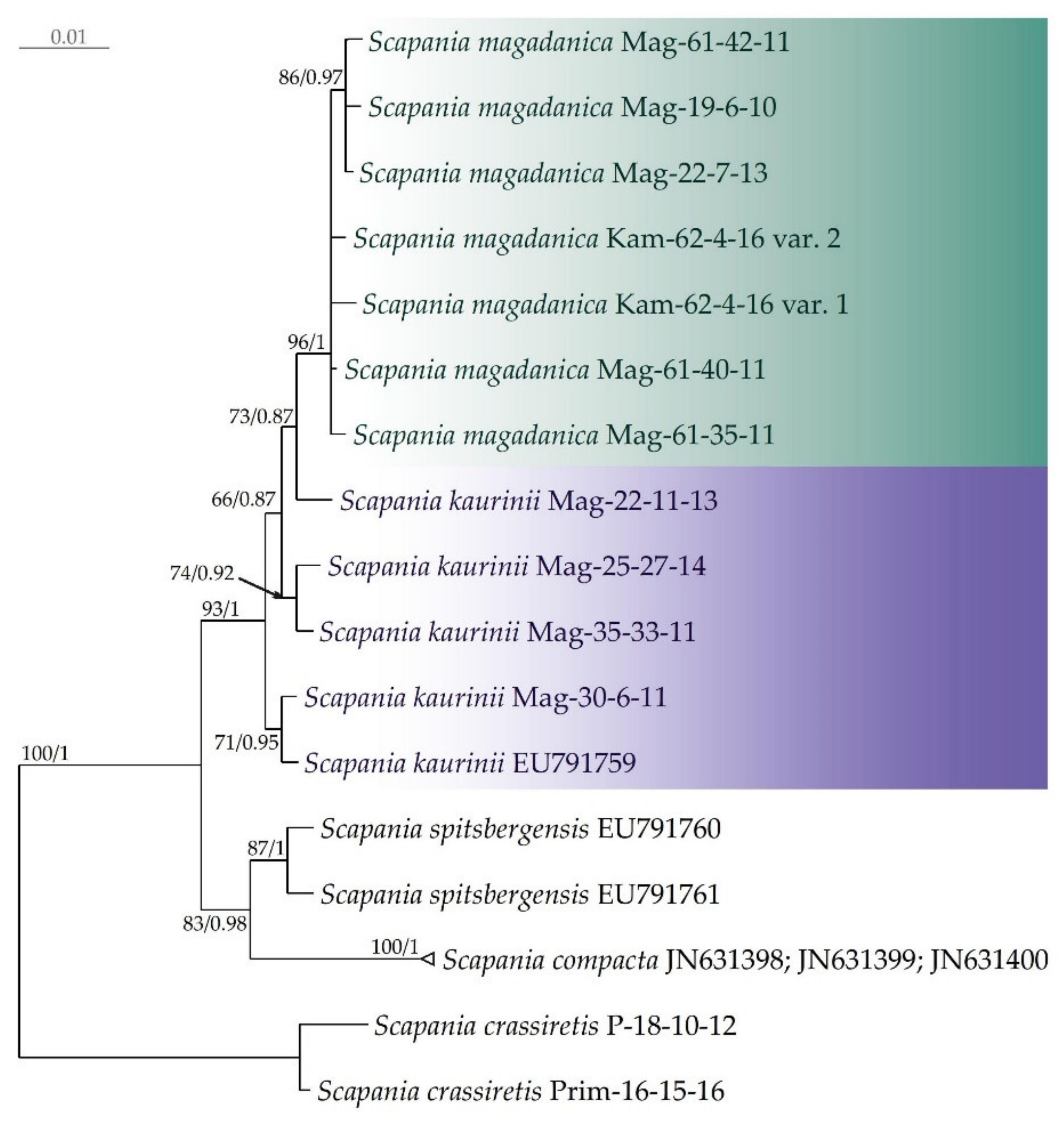
In the Bayesian phylogenetic tree, inferred from ITS (Figure 1), the moderately supported S. magadanica clade is nested within the S. kaurinii clade, i.e., accessions, assigned to S. kaurinii, do not form a separate clade but rather a grade crowned with the S. magadanica clade. The split tree shows that the molecular polymorphism of S. kaurinii is partly shared by S. magadanica, which might result in a weakly resolved and supported phylogenetic tree. Accessions, representing two other species of sect. Compactae (S. compacta and S. spitsbergensis), form a clade, sister to the clade, corresponding to the S. kaurinii—S. magadanica complex.
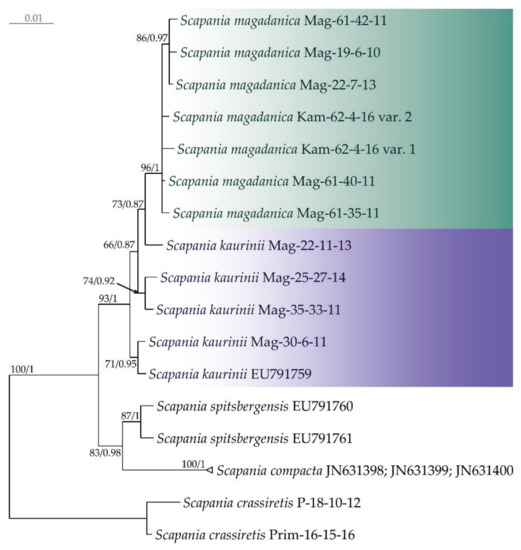
Figure 1.
Phylogram obtained in a Bayesian analysis for the Scapania magadanica and S. kaurinii taxa based on the ITS1–2 dataset. The values of bootstrap support from the ML analysis and Bayesian posterior probabilities greater than 0.50 (50%) are indicated. Three sequences of S. compacta from GenBank were identical, so we combined them. Taxon names, vouchers, and GenBank accession numbers are provided.
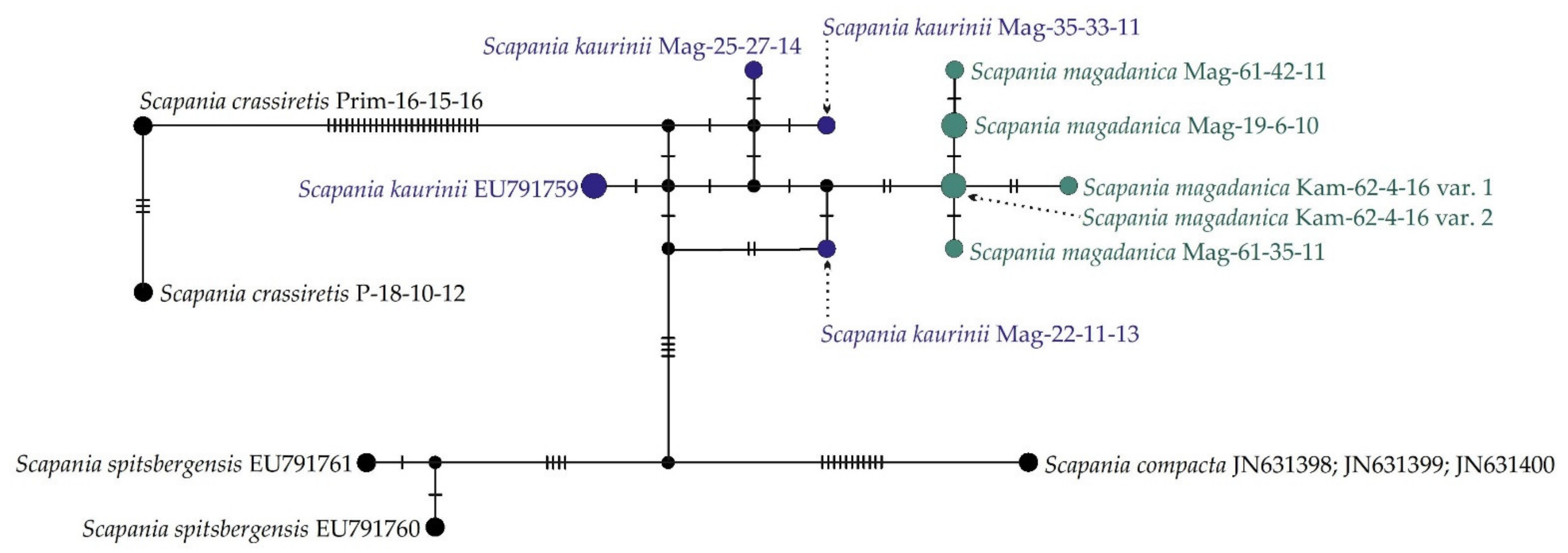
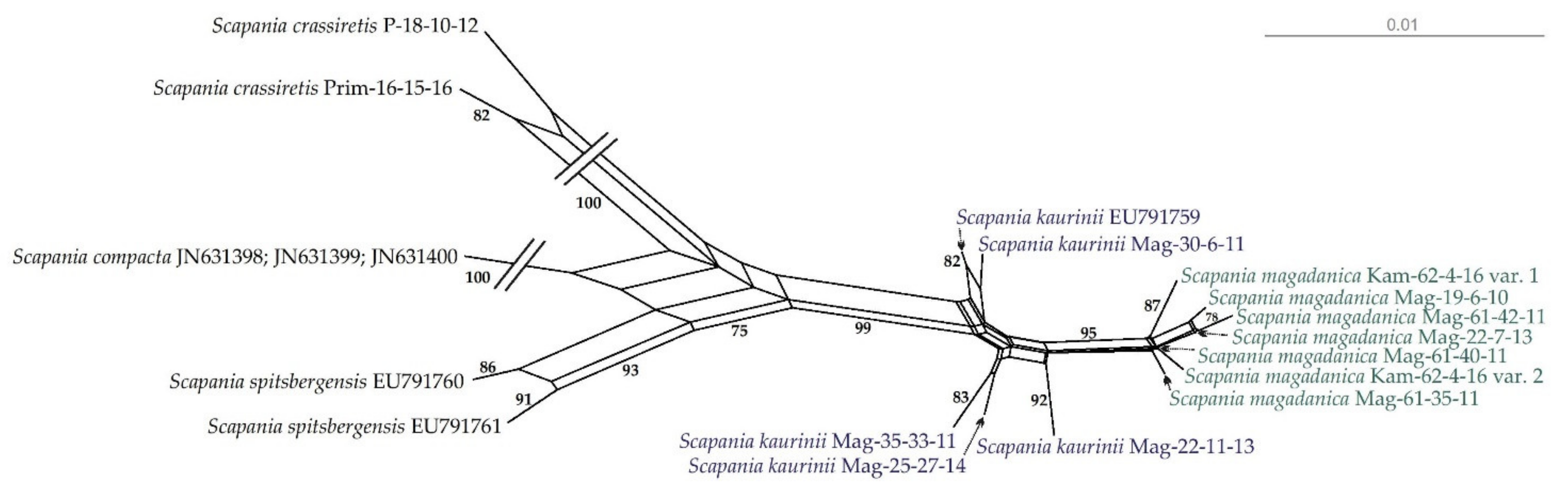
At the same time, the haplotype network (Figure 2) provides a clearer illustration of molecular relationships among specimens of S. magadanica and S. kaurinii. The NN split network (Figure 3) revealed six haplotypes forming a rather compact group, separated from the “S. kaurinii” group by at least three nucleotide substitutions and from the closest of the revealed S. kaurinii haplotypes by five substitutions. All five of the involved S. kaurinii specimens represent different haplotypes, which form a lax group. Scapania spitsbergensis and S. compacta appeared reasonable genetically distant from the S. kaurinii plus S. magadanica group.
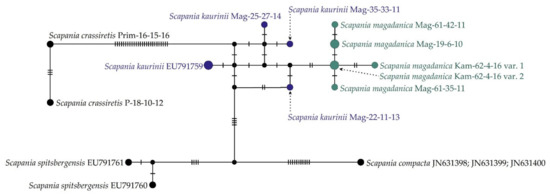
Figure 2.
TCS haplotype network of ITS1–2 sequences. Dashes indicate the number of nucleotide substitutions.
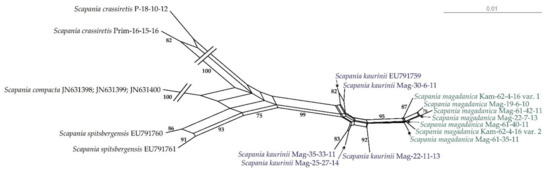
Figure 3.
NeighborNet split network for S. kaurinii and S. magadanica, based on the nuclear ITS1–2 dataset. Support values of 75 or higher are indicated.
The morphological comparison of S. magadanica and S. kaurinii has revealed several differences in
(1) Leaf shape in microscope slide, which is as wide as long to wider than long and commonly having acute to obtuse leaf apex in S. kaurinii versus distinctly longer than wide with inevitable rounded apices leaves in S. magadanica.
(2) Leaf shape in ‘living’ condition that is concave and turned to ventral side and loosely conduplicate in S. kaurinii versus laterally spreading, tightly conduplicate, and slightly convex leaves in S. magadanica.
Moreover, S. magadanica is meso-xerophyte growing in commonly drying-up crevices in stony fields, whereas S. kaurinii is hygro-hydrophyte growing near running water or in snowbed habitats always remained moistened.
4. Discussion
To facilitate the visualization of the results obtained using SplitsTree software, five species were selected: undoubtedly evolutionarily distant Scapania crassiretis; two closely related entities treated in this account, S. magadanica and S. kaurinii; and two species considered within the same section (Compactae), S. compacta and S. spitsbergensis. Among these five species, S. kaurinii occupies the middle (conditionally ‘transitional’) position, while S. magadanica differs more strongly from other involved taxa due to possessing additional molecular synapomorphies. Taking into account the wide arctic-hypoarctic circumpolar distribution of S. kaurinii and, conversely, the rather narrow distribution of S. magadanica in Beringia (currently, it is known to occur in NE Asia but may also be expected in the Alaskan highlands), it can be assumed that S. magadanica arose from an isolated population of S. kaurinii in northeast Asia. The distinctive morphological features of S. magadanica, compared to S. kaurinii (such as strongly conduplicate leaves with convex leaf lobes), likely appeared as an adaptation to growth in the xeric habitats typically occupied by S. magadanica (contrary to the rather wet habitats of S. kaurinii). Furthermore, S. magadanica occurs in dry crevices in areas with relatively xeric climatic conditions (e.g., in the Annachag Range in the Magadan region, see Pisarenko, 2015 [22]) or uneven distribution of precipitation caused by mesorelief (Kamchatka Territory, see Klimova and Bakalin, 2017 [4]), where S. kaurinii occurs near streams only. Counting the generally low variability of the studied molecular markers, we suggest that S. magadanica formed rather recently, probably under aridization of the climate in NE Asia in the Pliocene or Pleistocene, when the Beringian sector was occupied by cryo-xeric communities (so-called tundra-steppe) and became a specific element of this cryo-xeric species complex. The preliminary character of this and some other estimation is also determined by the fact of involvement of S. kaurinii vouchers from Asia only, while the original of the species is from Norway. Therefore, future research may find additional genetic diversity within S. kaurinii s.l.
Based on the available molecular data, which show a remarkable divergence of remote populations of S. kaurinii, we suggest that at the moment of S. magadanica lineage split, S. kaurinii already occupied a rather wide range that might have limited further gene flow between its remote populations; thus, molecular synapomorphies, which would distinguish it from the derived S. magadanica, simply could not be yielded, which is the reason why S. kaurinii s.str. (i.e., without S. magadanica) does not form a clade on the phylogenetic tree. Notably, in such cases, the cladistics approach is obviously prone to underrecognition of the derived taxa that, in our opinion, should be assigned to the “method limitation”. Thus, we suggest relying on other species criteria, such as morphological distinctiveness, ecology, distribution, and molecular divergence, while judging the taxonomic status of the particular derived lineage.
It is worth mentioning that the monoicous sex distribution is a rare case inside Scapania, where it is the only one typically present in sect. Compactae. All four of the currently known species of the section (S. compacta, S. kaurinii, S. magadanica, and S. spitsbergensis) are clearly paroicous [1,23]. The sister position of the clade S. kaurinii (plus S. magadanica embedded in it, as we found here) to the other two species of sect. Compactae (S. spitsbergensis—S. compacta) is revealed in the general topologies [2]. Taking this into account, the treatment of the pair S. magadanica—S kaurinii within a separate infrageneric taxon, as proposed by Potemkin [24], who separated S. kaurinii into a monotypic sect. Incurvae (a point of view currently not widely accepted), may be supported. This group probably required acceptance, if not in the rank of a section then in the rank of a subsection within a sect. Compactae.
Author Contributions
Conceptualization, V.A.B. and V.E.F.; methodology, V.E.F. and Y.D.M.; validation, V.E.F., V.A.B., S.S.C. and Y.D.M.; formal analysis, V.E.F. and Y.D.M.; investigation, all coauthors; resources, all coauthors; data curation, V.A.B., V.E.F., Y.D.M., K.G.K. and S.S.C.; writing—original draft preparation, V.A.B., V.E.F. and Y.D.M.; writing—review and editing, V.A.B., V.E.F., Y.D.M., K.G.K. and S.S.C.; and visualization, Y.D.M. All authors have read and agreed to the published version of the manuscript.
Funding
The work of Y.D.M., V.A.B., and K.G.K. was partially supported by the Russian Foundation for Basic Research (grant no. 20-04-00278) and is within the frame of the institutional research project “Cryptogamic Biota of Pacific Asia” (no. 122040800088-5). The work of V.E.F. was performed within the framework of the Development Program of the Interdisciplinary Scientific and Educational School of M.V. Lomonosov Moscow State University “The future of the planet and global environmental change” and is within the frame of the institutional research project “Cryptogamic Biota of Pacific Asia” (no. 122040800088-5). The work of S.S.C. was supported by the Korean National Institute of Ecology, grant number NIE-A-2023-01.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are very grateful to the persons who helped V.A.B. and K.G.K. in field expeditions in wild environments in Magadan Province and Kamchatka Territory. K.G.K. is sincerely grateful to Max Nitzsche, Vitalina Lobanova, and Thomas Linss, members of the Kamchatka expedition in 2016, without whom a record of Scapania magadanica would be impossible. K.G.K. also thanks I.G. Kokorin, P.P. Sychyov, A.P. Adukanov, and A.L. Pavlyuchenkov for transfer support in the hardly accessible studied area.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Choi, S.S.; Bakalin, V.A.; Sun, B.-Y. Scapania and Macrodiplophyllum in the Russian Far East. Bot. Pacifica 2012, 1, 31–95. [Google Scholar] [CrossRef]
- Heinrichs, J.; Bombosch, A.; Feldberg, K.; Kreier, H.-P.; Hentschel, J.; Eckstein, J.; Long, D.; Zhu, R.-L.; Schäfer-Verwimp, A.; Schmidt, A.R.; et al. A phylogeny of the northern temperate leafy liverwort genus Scapania (Scapaniaceae, Jungermanniales). Mol. Phylogenetics Evol. 2012, 62, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Schljakov, R.N. Hepatics of the North USSR; Nauka: Leningrad, Russia, 1981; Volume 4, p. 221. [Google Scholar]
- Klimova, K.G.; Bakalin, V.A. Two Scapania Species (Scapaniaceae) Newly Recorded from Kamchatka. Arctoa 2017, 26, 125–131. [Google Scholar] [CrossRef]
- Ignatov, M.S.; Maksimov, A.I.; Fedorova, A.V.; Ignatova, E.A. On the Taxonomy of Fontinalis Gracilis (Fontinalaceae, Bryophyta) and Superficially Similar Species. Nova Hedwig. Beih. Beih. 2020, 150, 243–264. [Google Scholar] [CrossRef] [PubMed]
- Ignatova, E.A.; Kuznetsova, O.I.; Shafigullina, N.R.; Fedosov, V.E.; Ignatov, M.S. The Genus Pylaisia (Pylaisiaceae, Bryophyta) in Russia. Arctoa 2020, 29, 135–178. [Google Scholar] [CrossRef]
- Ignatova, E.A.; Czernyadjeva, I.V.; Fedorova, A.V.; Ignatov, M.S. A Morphologocal and Molecular Phylogengetic Study of the Genus Calliergon (Calliergonaceae, Bryophyta) in Russia. Arctoa 2021, 30, 8–24. [Google Scholar] [CrossRef]
- Vilnet, A.A.; Konstantinova, N.A.; Troitsky, A.V. Molecular Insight on Phylogeny and Systematics of the Lophoziaceae, Scapaniaceae, Gymnomitriaceae and Jungermanniaceae. Arctoa 2010, 19, 31–50. [Google Scholar] [CrossRef]
- Feldberg, K.; Váňa, J.; Krusche, J.; Kretschmann, J.; Patzak, S.D.F.; Pérez-Escobar, O.A.; Rudolf, N.R.; Seefelder, N.; Schäfer-Verwimp, A.; Long, D.G.; et al. A Phylogeny of Cephaloziaceae (Jungermanniopsida) Based on Nuclear and Chloroplast DNA Markers. Org. Divers. Evol. 2016, 16, 727–742. [Google Scholar] [CrossRef]
- Friedl, T. Evolution of the Polyphyletic Genus Pleurastrum (Chlorophyta): Inferences from Nuclear-Encoded Ribosomal DNA Sequences and Motile Cell Ultrastructure. Phycologia 1996, 35, 456–469. [Google Scholar] [CrossRef]
- Milyutina, I.A.; Goryunov, D.V.; Ignatov, M.S.; Ignatova, E.A.; Troitsky, A.V. The Phylogeny of Schistidium (Bryophyta, Grimmiaceae) Based on the Primary and Secondary Structure of Nuclear RDNA Internal Transcribed Spacers. Mol. Biol. 2010, 44, 883–897. [Google Scholar] [CrossRef]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal Primers for Amplification of Three Non-Coding Regions of Chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Bakalin, V.; Maltseva, Y.; Vilnet, A.; Choi, S.S. The Transfer of Tritomaria koreana to Lophozia Has Led to Recircumscription of the Genus and Shown Convergence in Lophoziaceae (Hepaticae). Phytotaxa 2021, 512, 41–56. [Google Scholar] [CrossRef]
- Pacak, A.; Szweykowska-Kulińska, Z. Molecular Data Concerning Alloploid Character and the Origin of Chloroplast and Mitochondrial Genomes in the Liverwort Pellia borealis. Plant Biotechnol. J. 2000, 2, 101–108. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Clement, M.; Snell, Q.; Walke, P.; Posada, D.; Crandall, K. TCS: Estimating Gene Genealogies. In Proceedings of the Proceedings 16th International Parallel and Distributed Processing Symposium, Washington, DC, USA, 15–19 April 2002; IEEE: Fort Lauderdale, FL, USA, 2002; p. 33. [Google Scholar]
- Leigh, J.W.; Bryant, D. Popart: Full-feature Software for Haplotype Network Construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Pisarenko, O.Y. Mosses of the Bolshoi Annachag Range (Magadan Province, Russian Far East). Arctoa 2015, 24, 187–193. [Google Scholar] [CrossRef]
- Potemkin, A.D. Evolution, Phylogeny and Classification of Scapaniaceae Family (Hepaticae). Ph.D. Dissertation, Botanical Institute, Saint-Petersburg, Russia, 2001. [Google Scholar]
- Potemkin, A.D. On the Origin, Evolution and Classification of the Genus Scapania (Dum.) Dum. (Hepaticae). J. Hallori. Bot. Lab. 1998, 85, 33–61. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).