2. Materials and Methods
Fish preservation. Fish were sampled with the permission of the local environment department of Iran and euthanized with an overdose of clove oil, fixed in 10% formalin for 24 h, and preserved in 70% ethanol. The samples used for molecular analyses were fixed in 99% EtOH (whole or a fin clip) after being euthanized. The species were identified following the original description and revisions of the genus [
9,
10].
Morphological analysis. Measurements were made point-to-point with a digital calliper and recorded to 0.1 mm. Counts and measurements were made on the left side of the specimens whenever possible, following Kottelat & Freyhof [
11], except for post-dorsal length, which was measured from the dorsal fin origin to the end of the caudal peduncle. The head length and measurements of body parts are given as proportions of the standard length (SL). The subunits of the head are presented as proportions of the head length (HL). The standard length (SL) was measured from the tip of the snout to the posterior extremity of the hypural complex. The skin fold at the posterior part of the gill cover was included in the measurement of the HL. The length of the caudal peduncle was measured from behind the base of the posterior anal-fin ray to the posterior extremity of the hypural complex at mid-height of the caudal-fin base. The length of the adipose fin was measured from the point where the skin fold elevates to the posteriormost tip. Simple rays of dorsal and anal fins were not counted as they are deeply embedded. The last two branched rays articulating on a single pterygiophore in the dorsal and anal fins are noted as “1½”. The distribution map (
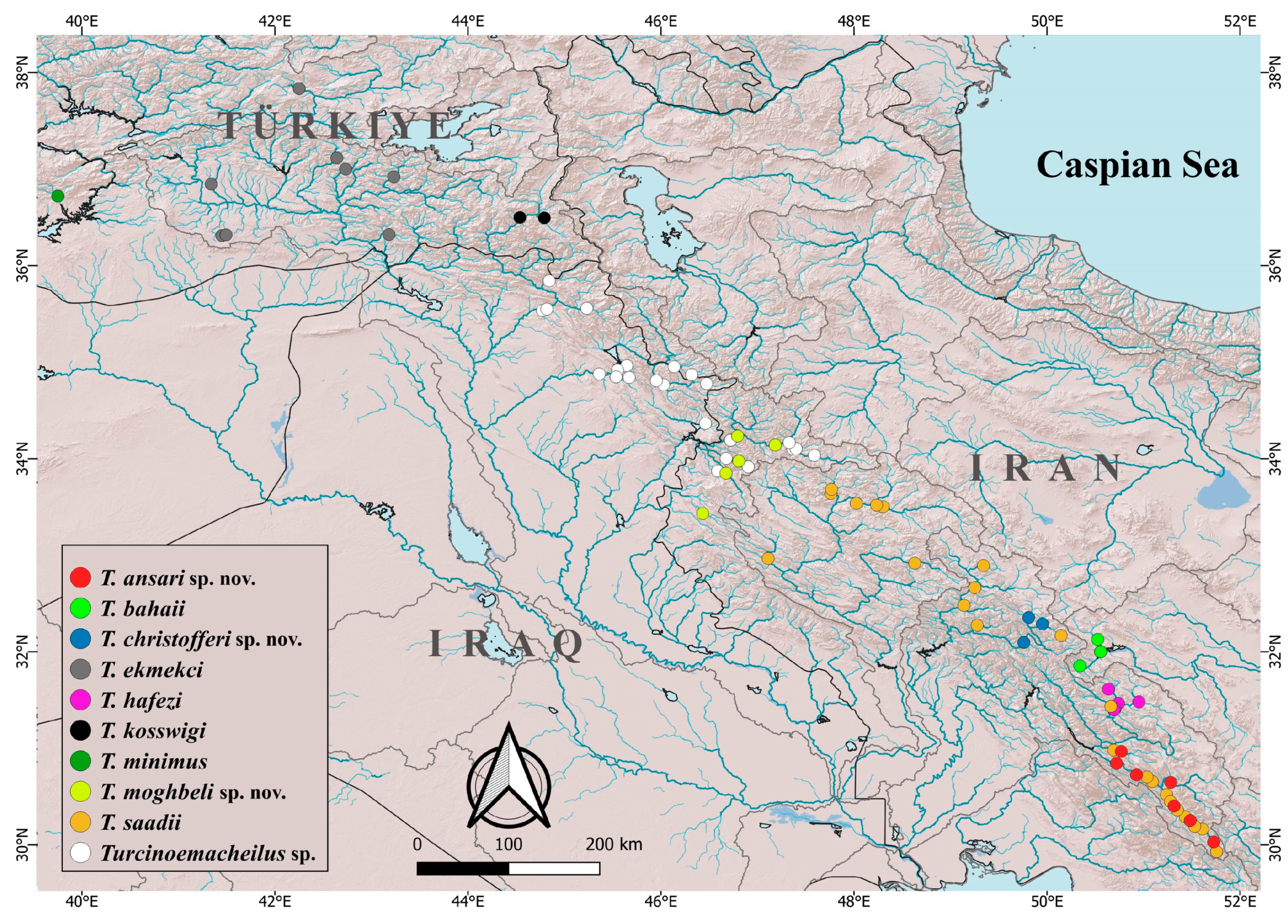
Figure 1) was created with QGIS v.3.18 software (
http://qgis.org, accessed on 10 October 2023). Morphometric data for
T. ekmekciae Kaya, Yoğurtçuoğlu, Aksu, Bayçelebi & Turan, 2023,
T. kosswigi, and
T. minimus Esmaeili, Sayyadzadeh, Özuluğ, Geiger & Freyhof, 2014 [
9] were obtained from Kaya et al. [
10].
Morphometric characteristics were examined through a Correspondence Analysis (CA) and compared using a Non-Parametric Multivariate Analysis of Variance (NPMANOVA). The analysis involved Bonferroni-corrected p-values, determined through a permutation test with 1000 replicates in PAST software (version 2.14).
DNA extraction, PCR amplification, and Sequencing. DNA was extracted using Macherey & Nagel NucleoSpin
® Tissue kit (Düren, Germany) following the provided protocol. The barcode region of the COI (cytochrome c oxidase subunit 1) gene was amplified using FishF1-5′TCAACCAACCACAAAGACATTGGCAC3′ and FishR1-5′TAGACTTCTGGGTGGCCAAAGAATCA3′ [
12]. The T7Promoter (5′TAATACGACTCACTATAGGG3′) and T3 (5′ATTAACCCTCACTAAAGGG3′) standard sequences were added to the sequence of forward and reverse primers, respectively, to simplify the sequencing of different PCR products on the same plate. The sequencing of the PCR products was performed at an external sequencing service provider.
Molecular data analysis. The obtained barcode sequences and the ones downloaded from GenBank (
Table 1), were aligned using MAFFT [
13,
14], as implemented in Geneious v. 10.0.2 (Biomatters,
http://www.geneious.com/, accessed on 10 October 2023). To determine the inter- and intraspecific uncorrected pairwise genetic distances (p-distances) (
Table 2), we employed Mega 6 [
15].
Both the maximum likelihood (ML) and Bayesian (BI) methods have been used to construct the phylogenetic relationships of the group. In the case of the ML approach, IQ-TREE 1.6.12 [
16] was used. In this case, the optimal substitution model and the best partitioning scheme based on the codon information were investigated using ModelFinder [
17] with the Bayesian information criterion (BIC). The bootstrap (-b 500) approximations were used to calculate support values [
18]. FigTree 1.4.4 (
http://tree.bio.ed.ac.uk/software/figtree/, accessed on 10 October 2023) was used to visualize the resulting trees. In the case of the BI approach, MrBayes 3.2.7 [
19] was used with two parallel simultaneous analyses for 2 × 107 generations, each with four MCMC chains, and sampling every 2000 generations. The initial 25% of generations were discarded as burn-in. An rjMCMC [
20] approach was implemented using the nst = mixed command. The proper convergence of the runs was verified using Tracer 1.7 [
21].
Three distance-based molecular species delimitation methods were used: Automatic Barcode Gap Discovery (ABGD) [
22], Assemble Species by Automatic Partitioning (ASAP) [
23], and the Bayesian Poisson Tree Processes model (bPTP) [
24]. The ABGD analysis was performed on its online webserver (
https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html, accessed on 10 October 2023) using the simple distance option and the rest of the options as default. An ASAP analysis was also carried out using simple distances (p-distances) via an online implementation on its web interface (
https://bioinfo.mnhn.fr/abi/public/asap/asapweb.html, accessed on 10 October 2023). The bPTP analysis was run only in the in-group on the online implementation of it (
https://species.h-its.org/, accessed on 10 October 2023) using default settings.
Abbreviations. SL, standard length; HL, lateral head length; BIAUBM, Babol Islamic Azad University Biological Museum, Babol, Iran; AJRPC, A. Jouladeh-Roudbar personal fish collection, Tehran, Iran; VPFC, S. Vatandoust Personal Fish collection, Qaem Shahr, Iran; MZLU, Lund University Biological Museum, Lund, Sweden; MNCN, Museo Nacional de Ciencias Naturales, Madrid, Spain; FSJF, Fischsammlung J. Freyhof, Berlin, Germany.
3. Results
We generated 85 new sequences for seven species of
Turcinoemacheilus from Western Asia and downloaded an additional 40 sequences (+22 outgroups) from NCBI GenBank (
Table 1). The final alignment consisted of 701 base pairs, with 484 positions being constant, 163 being parsimony informative, and 23 being singletons (considering only the in-group). The resulting tree topology is, in general, in agreement with previously published phylogenies that included samples of
Turcinoemacheilus [
9,
10]. Both the BI and ML methods resulted in a similar tree (
Figure 2), recovering 10 well-supported clades corresponding to 10 species in the
Turcinoemacheilus genus:
T. ansari sp. nov.,
T. bahaii,
T. christofferi sp. nov.,
T. ekmekciae,
T. hafezi,
T. kosswigi,
T. minimus,
T. moghbeli sp. nov.,
T. saadii, and
Turcinoemacheilus sp. The barcode COI dataset showed an intraspecific uncorrected p genetic distance of between 3.6% (
T. hafezi and
T. bahaii) and 14.1% (
T. saadii and
T. christofferi sp. nov.). The average interspecific distance for
Turcinoemacheilus species was 0.4%, ranging from 0.0 in
T. christofferi sp. nov. to 1.0% in
T. hafezi.
Table 2 shows the genetic distances between and within the different species of the genus. The resulting distance tree results in two relatively well-supported clades, with
T. hafezi,
T. bahaii,
T. christofferi sp. nov., and
T. ansari sp. nov. on one clade and the rest in the other.
T. hafezi and
T. bahaii are sisters to each other, as are
T. christofferi sp. nov. and
T. ansari sp. nov., though these relationships are not well supported. On the other hand,
T. moghbeli sp. nov.,
T. saadi,
T. minimus,
T. ekmekciae,
T. kosswigi, and
Turcinoemacheilus sp. form a clade, also with poor support for phylogenetic relationships.
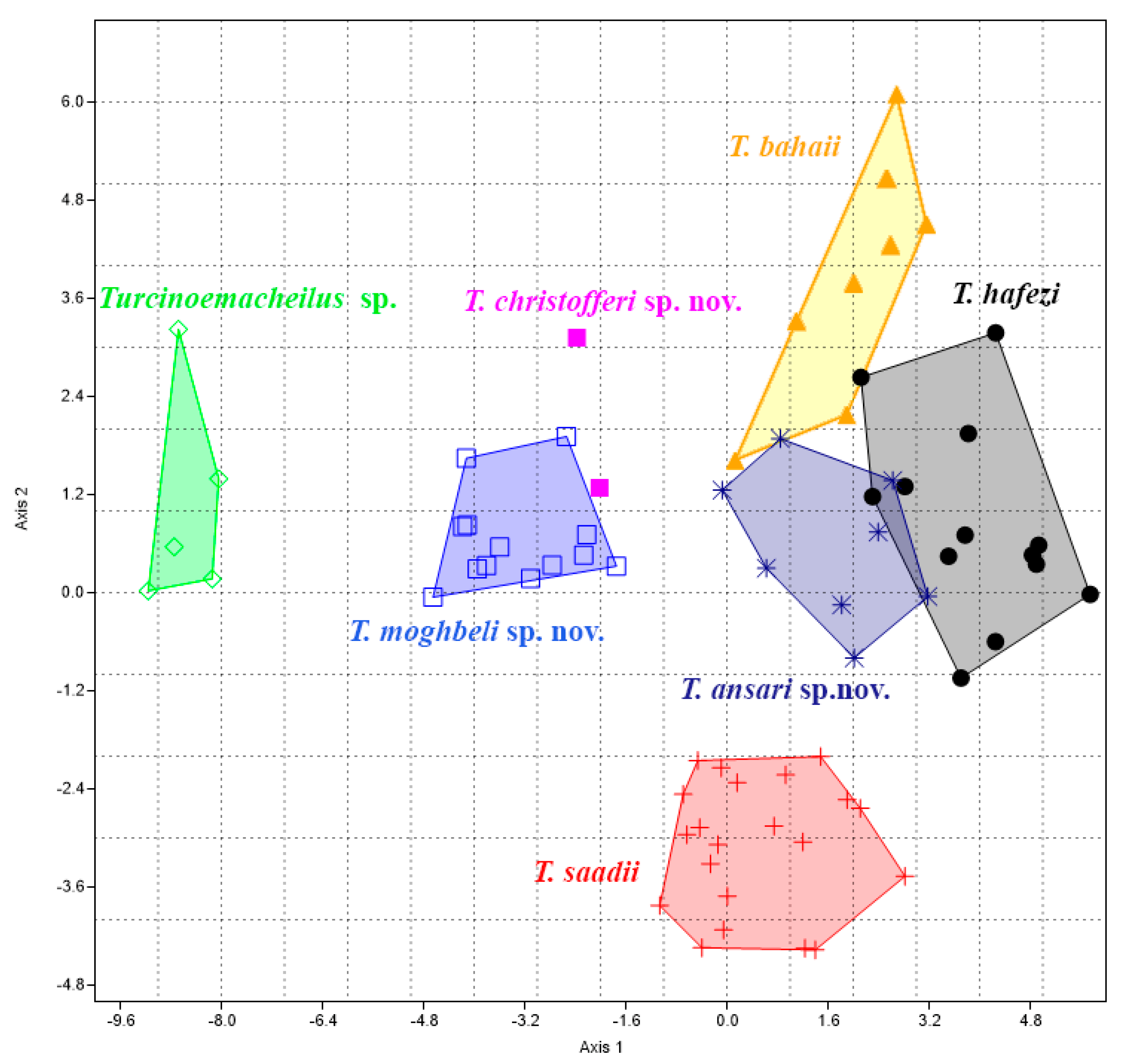
MANOVA/CVA revealed significant separation among the examined species concerning morphometric characteristics (
p < 0.0005). The results indicated that the first two CVA axes (CVA1 = 49.61% and CVA2 = 22.40%) collectively accounted for 72.01% of the morphometric variations across all the specimens. The plotting of the first and second CVAs demonstrated the separation of the seven species (
Figure 3). The “Distance vent and anal-fin origin/Distance pelvic and anal-fin origins” and “Length of caudal peduncle” characters were the most influential in discriminating between the groups across several dimensions.
3.1. Key to Species of Turcinoemacheilus in Western Asia (Expanded from [10])
1a—Anus situated behind midway between pelvic and anal origins.
………………2
1b—Anus situated midway or in front of midpoint between pelvic and anal origins.
………………5
2a—Lateral line complete, reaching to anterior part of caudal fin.
……………… T. christofferi sp. nov.
2a—Lateral line incomplete, reaching to midway between tip of pectoral fin and dorsal fin origin to midway between dorsal-fin insertion and anal-fin origin.
………………3
3a—A dark blotch at base of anal-fin on both sides; anal-fin origin at vertical of tip of dorsal fin when adpressed to body.
……………… T. bahaii
3b—No dark blotch on side of anal-fin base; anal-fin origin behind vertical of tip of dorsal fin when adpressed to body.
………………4
4a—Body depth at dorsal-fin origin: 10–13% SL.
……………… T. ansari
4b—Body depth at dorsal-fin origin: 13–15% SL.
……………… T. hafezi
5a—Lateral stripe or row of blotches absent along lateral midline; 7–9 distinct dark saddles on body.
……………… T. saadii
5b—Prominent row of dark brown blotches along lateral midline, usually fused into a lateral stripe.
………………6
6a—Caudal peduncle length: 6–7% SL; 4–5 mandibular pores.
………………T. minimus
6b—Caudal peduncle length: 7–9% SL; 5–7 mandibular pores.
………………7
7a—A dark stripe narrower than the eye diameter along the lateral midline; pre-pelvic distance: 47–50% SL.
………………T. kosswigi
7b—A dark stripe broader than the eye diameter along the lateral midline; pre-pelvic distance: 50–54% SL.
………………8
8a—Caudal peduncle length to depth: 2.6–3.2.
………………T. ekmekciae
8b—Caudal peduncle length to depth: 1.5–2.3.
……………… T. moghbeli sp. nov/Turcinoemacheilus sp. (identifiable through combination of morphological characters, genetic comparisons, and phylogenetic analyses).
3.2. Taxonomy
3.2.1. Turcinoemacheilus ansari, New Species
Holotype. BIAUBM 3-H, 43.8 mm SL; Iran: Kohgiluyeh and Boyer-Ahmad prov., Beshar River, Allah Abad village, Karun drainage, 30.45029, 51.75346.
Paratypes. AJRPC 1779–1780, 2, MZLU L021/00004-L021/00006 (AJRPC-DNA1782-1784), 3, MNCN-ICTIO 297.327–297.328 (AJRPC-DNA1785-1786), 2, 33–50 mm SL; same data as holotype.
Diagnosis. Turcinoemacheilus ansari is distinguished from any of the species of the T. kosswigi group (T. ekmekciae, T. kosswigi, T. minimus, T. moghbeli, T. saadii, Turcinoemacheilus sp.) by the anus situated behind the middle, between the pelvic and anal origins (vs. front middle).
Turcinoemacheilus ansari is similar to T. bahaii but can be distinguished by no dark blotch at the anal-fin base (vs. an elongated, irregularly shaped blotch on the sides of the anal-fin base) and a shorter anal-fin base length (5–8 vs. 9–12% SL). Turcinoemacheilus ansari can be distinguished from T. christofferi by its incomplete lateral line, reaching anterior to or under the dorsal-fin base (vs. complete, reaching to the anterior part of the caudal fin), and a shorter anal-fin base length (5–8 vs. 8–10% SL). It is distinguished from T. hafezi by a shorter body depth (10–13 vs. 13–15% SL).
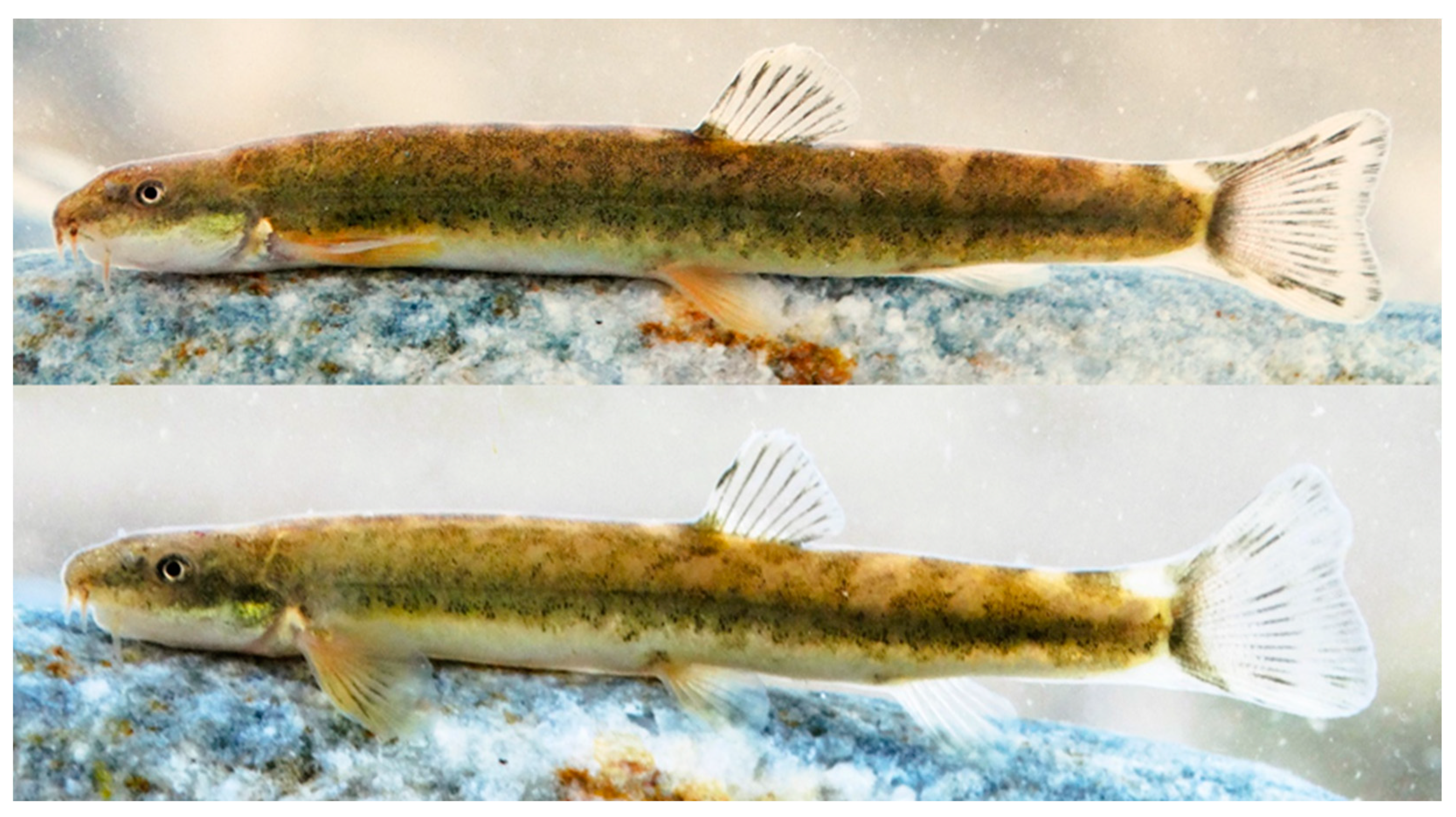
Description. See
Figure 4,
Figure 5 and
Figure 6 for general appearance and
Table 3 for morphometric data. Small, slender, and round-bodied species with short head. Body deepest at tip of pelvic fin, depth slightly decreasing towards caudal-fin base. No hump at nape. Body almost equally wide until dorsal fin origin. Section head round, flattened ventrally. Caudal peduncle compressed laterally, 1.3–2.9 (mean 2.2) times longer than deep. Pelvic axillary lobe absent or present; if present, oval, its tip attached to body. Pelvic-fin origin distinctly in front of dorsal-fin origin. Pectoral fin reaching approximately 35–45% of the distance from pectoral-fin origin to pelvic-fin origin. Pelvic fin not reaching anus. Distance from anus to anal-fin origin 2.4–3.7 times the distance from pelvic-fin to anal-fin origins. Anal-fin origin behind vertical of tip of dorsal fin when adpressed to body. Anal fin reaching or not reaching middle of caudal peduncle. No adipose crest on caudal peduncle. Margin of dorsal fin straight. Caudal fin emarginate. Largest known specimen 50 mm SL.
Three to five supratemporal canals, 10–13 pores in infraorbital canal, and 5–8 pores in suborbital canal. No suborbital flap or groove in male. Dorsal fin with 7½ branched rays. Anal fin with 5½ branched rays. Caudal fin with 8+8 or 8+7 branched rays. Pectoral fin with 9 and pelvic fin with 5–7 branched rays. Body without scales. Lateral line incomplete or complete; if complete, with 12–46 pores, reaches to base of caudal fin. Anterior nostril opening at end of a pointed flap-like tube. Posterior nostril oval, posterior tip of anterior nostril not or just overlapping posterior nostril when folded backwards. Mouth small, slightly arched. Lips moderately thick. A median interruption on lower lip. Upper lip with a shallow median incision. Processus dentiformis small and blunt. No median notch in lower jaw. Barbels short; inner rostral barbel not reaching base of maxillary rostral barbel; outer one reaching to base of maxillary barbel. Maxillary barbel reaching vertical of anterior part of eye. No external sexual dimorphism observed.
Coloration. In fresh and ethanol-fixed specimens, body cream or yellow with olive or dark brown mottled pattern. Lateral midline with irregular blotches, forming irregularly set and shaped saddles in most individuals; in a few individuals, body pattern uniform olive or pale brown without blotch and saddles or few blotches on caudal peduncle. Flank below lateral midline without pigmentation. A semilunar or irregularly shaped, black or dark brown bar at caudal-fin base. A whitish or triangle-shaped patch in front of bar on upper and lower caudal peduncle. Cheeks white or cream, ventral surface of head white or cream, head above cheeks olive or dark brown. Operculum yellowish or golden brown. Pectoral, pelvic, anal fins hyaline, caudal and dorsal fins olive to dark brown. Caudal fin with elongated faint spots on rays, forming a mottled pattern of one vertical row approximately in middle of ray length. Median part of last unbranched ray of dorsal fin with dark brown or black pigments.
Etymology. This species is named after Anousheh Ansari, the first Iranian, as well as the first self-funded woman, to fly to the space station. The naming of this species pays tribute to her, serving as a source of inspiration and motivation for future generations, especially women in Iran. A noun in apposition.
Distribution. The species is known from the Merian, Beshar (
Figure 7), and Khersan rivers in the headwaters of the Karun drainage.
Habitat. Turcinoemacheilus ansari typically inhabits the headwaters of rivers with clear, cold, and well-oxygenated water. The species demonstrates a preference for high-gradient riffles and rapids containing coarse substrata such as gravel, pebbles, and rocks. The species has a low tolerance for sediments and is not found in low-current areas, slack water, or deep water in the lower Khersan or Karun River. This species is found in sympatry with T. saadii.
3.2.2. Turcinoemacheilus christofferi, New Species
Holotype. BIAUBM 4-H, 1, 54.2 mm SL; Iran: Lorestan prov., Gholiyan River, tributary of Bakhtiyari River, Karun drainage, 33.07499, 49.64562.
Paratypes. FSJF 4122 (AJRPC-DNA1713), 1, 41.5 mm SL; same data as holotype.
Diagnosis. Turcinoemacheilus christofferi is distinguished from any of the species of the T. kosswigi group (T. ekmekciae, T. kosswigi, T. minimus, T. moghbeli, T. saadii, Turcinoemacheilus sp.) by the anus situated behind the middle, between the pelvic and anal origins (vs. front middle). Turcinoemacheilus christofferi is similar to T. bahaii and T. hafezi but can be distinguished by a complete lateral line with more than 70 pores, reaching to the anterior part of caudal fin (vs. an incomplete lateral line, reaching to the anterior part of the dorsal fin, with less than 35 pores).
Description. See
Figure 8 and
Figure 9 for general appearance and
Table 4 for morphometric data. Small, slender, and oval-bodied species with short head. Body deepest at tip of pelvic fin, depth slightly decreasing towards caudal-fin base. Small hump at nape. Body almost equally wide until dorsal-fin origin. Section of head triangle, flattened on ventral surface. Caudal peduncle compressed laterally, 1.6–1.8 (mean 1.7) times longer than deep. Pelvic axillary lobe oval, its tip attached to body. Pelvic-fin origin distinctly in front of dorsal-fin origin. Pectoral fin tip reaching approximately 40–55% of the distance from pectoral-fin origin to pelvic-fin origin. Pelvic-fin tip not reaching anus. Distance from anus to anal-fin origin 2.6–2.9 times the distance from pelvic-fin to anal-fin origins. Anal-fin origin behind vertical of tip of dorsal fin when adpressed to body. Anal fin not reaching to middle of caudal peduncle. No adipose crest on caudal peduncle. An indentation in front of dorsal-fin origin. Margin of dorsal fin straight. Caudal fin forked or deeply emarginate. Largest known specimen 54 mm SL.
One central and one lateral pore on each side of supratemporal canal, 6–8 pores in anterior infraorbital canal, 3–4 pores in posterior infraorbital canal, 10 pores in the supraorbital canal, and 5–6 pores in mandibular canal. No suborbital flap or groove in male. Dorsal fin with 7½–8½ branched rays. Anal-fin with 5½ branched rays. Caudal fin with 8+8 or 8+7 branched rays. Pectoral fin 9 and pelvic-fin 6 branched rays. Body without scales. Lateral line complete; 72–74 pores reach base of caudal fin. Anterior nostril opening at end of a pointed flap-like tube. Posterior nostril oval, posterior tip of anterior nostril not or just overlapping posterior nostril when folded backwards. Mouth small, slightly arched. Lips moderately thick. A median interruption in lower lip. Upper lip without median incision. Processus dentiformis small and blunt. No median notch in lower jaw. Barbels: short, inner, and outer rostral barbel not reaching base of maxillary rostral barbel. Maxillary barbel reaching vertical of pupil. No external sexual dimorphism observed.
Coloration. White or cream background in life and ethanol preserved. A row of large irregular, brown, and longitudinally elongated blotches and saddles along lateral midline, often fused into a prominent irregular lateral stripe. Pattern on flank and back more or less faded in anterior part of body, possessing a plain brown on back, darker on lateral midlines. Large lateral blotches behind dorsal fin, connected to lateral blotches along whole body. A semilunar-shaped black or dark brown bar at caudal-fin base. A whitish or triangle-shaped patch in front of bar on upper and lower caudal peduncle. Cheeks dark olive or brown; ventral surface of head white or cream; head above cheeks pale dark brown. Operculum golden brown. Pectoral fin pale brown with elongated black spots on rays; pelvic fins yellowish; anal-fin hyaline; caudal and dorsal fins olive to dark brown. Caudal fins hyaline, with elongated spots on rays, forming two dark vertical rows, one approximately in middle of ray length and one on distal part. Dorsal fin rays with melanophores extending from approximately middle of ray length to distal margin.
Etymology. The species has been named in honor of Christoffer Fägerström (Lund University, Sweden) in recognition of his invaluable contributions to the photography and comprehensive documentation of type specimens of a variety of taxa, including insects, fishes, and other organisms. Most pictures of the type materials in this study have been very kindly contributed by him. A noun in the genitive case.
Distribution. The species appears to have an extremely limited distribution. It is currently only known to exist in the Gholiyan River (
Figure 10), a headwater tributary of the Bakhtiyari River system of the Karun drainage.
Habitat. Turcinoemacheilus christofferi inhabits the fast-flowing parts of river, with a preference for high-gradient riffles and rapids. It is typically found in areas with clear, cold, and well-oxygenated water running over coarse substrata such as gravel, pebbles, and rocks. The species occurs among the interstitial spaces within loosely embedded gravel beds and avoids silt-bedded pools or slow currents. Several rainbow trout fish farms have been recently established around the type locality of this species. Unfortunately, a recent flood released many rainbow trout into the river. As a consequence, the population of this species in the Gholiyan River has significantly diminished.
Remark. Turcinoemacheilus christofferi is restricted to the headwaters of the Bakhtiyari River system. Despite extensive survey efforts encompassing more than 30 sampling stations across the river’s headwaters in different seasons, no additional specimens have been collected. The species’ limited range is likely a result of competitive exclusion by the larger and more widespread Oxynoemacheilus euphraticus, which dominates optimal habitats. Further field studies and extensive sampling of the region are needed to elucidate T. christofferi’s distribution range and biology.
3.2.3. Turcinoemacheilus moghbeli, New Species
Holotype. BIAUBM 5-H, 54.2 mm SL; Iran: Kermanshah prov., Leyleh River at Sepidbarg, tributary of Sirvan drainage, 34.87334, 46.35020.
Paratypes. AJRPC 1635–1636, 2, 40–47 mm SL; MNCN-ICTIO 297.329–297.331, 3, 38–51 mm SL; Kermanshah prov., Goleyn River at Goleyn Village, 34.25603, 45.96339. FSJF 4123, 1, 48 mm SL; Kermanshah prov., Alvand River near Shirinab Village, 34.48136, 45.75623. MZLU L021/00007-L021/00009, 3, 44–50 mm SL; AJRPC 15-P, 2, 56–59 mm SL; data same as holotype. FSJF 4124, 1, 44 mm SL; Kermanshah prov., Zemkan River near Siyah Taher Village, 34.73312, 46.20455.
Diagnosis. Turcinoemacheilus moghbeli is distinguished from any of the species of the
T. hafezi group (
T. hafezi,
T. bahaii,
T. cristtofferi,
T. ansari) by the anus situated at or in front of the midpoint of the pelvic-fin and anal-fin origins (vs. behind the middle)(
Table 5 and
Table 6).
This new species is distinguished from T. kosswigi by having the dark stripe broader than the eye diameter along the lateral midline (vs. narrower) and a greater pre-pelvic distance (50–55 vs. 47–50% SL). It is further distinguished from T. minimus by possessing a deeper caudal peduncle (6–7 vs. 9–10% SL), a deeper head (47–58 vs. 37–47% HL), and 5–6 mandibular pores in the mandibular canal (vs. 4–5). This species can be distinguished from T. ekmekciae by often having 7½ branched rays (vs. 6½) and a smaller caudal peduncle length to its depth (1.5–2.3 vs. 2.6–3.2). Turcinoemacheilus moghbeli can be distinguished from T. saadii by a prominent row of dark brown blotches along the lateral midline, usually fused into a lateral stripe (vs. 7–9 distinct dark saddles or blotches on the body) and is superficially similar to Turcinoemacheilus sp. and can be distinguished by the caudal fin deeply emarginate (vs. truncate or slightly emarginate).
Description. For general appearance, see
Figure 11,
Figure 12 and
Figure 13; morphometric data are provided in
Table 3. Small-sized and slender species. Head short, 1.2–1.8 times body depth at dorsal-fin origin. Pre-dorsal profile convex, pre-pelvic profile slightly convex. Body deepest and widest at mid-point of pre-dorsal distance, depth decreasing towards caudal-fin base. A small hump at nape. Section of head roundish, flattened ventrally, interorbital straight or slightly convex, straight on snout. Snout pointed. Caudal peduncle compressed laterally, 1.5–2.3 times longer than deep. Pelvic axillary lobe present, oval shape; in specimens larger than 40 mm SL, its tip not attached to body. Pelvic-fin origin in front of dorsal-fin origin. Pectoral fin reaching 32–48% of the distance from pectoral-fin origin to pelvic-fin origin. Tip of pelvic-fin reaching beyond anus. Distance from anus to anal-fin origin 1.8–2.7 times the distance from pelvic-fin to anal-fin origins. Anal-fin origin behind vertical of tip of dorsal fin when adpressed to body. Anal fin not reaching to middle of caudal peduncle. A shallow and incomplete adipose crest under and below caudal peduncle. Margin of dorsal fin straight. Caudal fin emarginate. Largest known specimen: 59 mm SL.
Three to five pores in supratemporal canal, 10–11 pores in anterior infraorbital canal and 7–10 supraorbital canal. No suborbital flap or groove in male. Dorsal fin with 6½–7½ (usually 7½) branched rays. Anal-fin with 5½ branched rays. Caudal fin with 8+8 or 8+7 branched rays. Pectoral fin with 7–9 and pelvic-fin with 6–7 branched rays. Body without scales. Lateral line incomplete, 16–32 pores, often interrupted in posterior part, reaching midpoint between tip of pectoral fin and dorsal-fin origin to midpoint between dorsal-fin insertion and anal-fin origin. Anterior nostril opening at end of a pointed flap-like tube. Posterior nostril oval, posterior tip of anterior nostril not or just overlapping posterior nostril when folded backwards. Mouth small, slightly arched. Lips moderately thick. A median interruption in lower lip. Upper lip with a shallow median incision. Processus dentiformis small and blunt. No median notch in lower jaw. Barbels short, inner rostral barbel not reaching base of maxillary rostral barbel, outer one reaching to base of maxillary barbel. Maxillary barbel reaching vertical of anterior part or middle of eye. No external sexual dimorphism observed.
Coloration. In fresh and ethanol-fixed specimens, body yellowish or cream. In specimens from the Alvand and Zemkan drainages, a distinct dark stripe runs from the head to the caudal peduncle; some individuals change to a marmorated pattern on posterior part of body. In contrast, in specimens from the Sirvan drainage, midlateral blotches on their flanks tend to fuse into a stripe. Additionally, a series of saddles or blotches on their backs and a marmorated pattern on the upper flank, positioned between the saddles and the midlateral stripe. Lateral blotches behind the dorsal and anal fin extend below the midlateral stripe. A distinct, irregularly shaped dark brown or black at the caudal-fin base. In front of this bar, a whitish or yellowish triangular patch on upper and lower caudal peduncles. The cheeks dark olive or dark brown; the ventral surface of the head white or cream. The area above the cheeks dark brown. The pectoral and pelvic fins yellowish, anal-fin hyaline, and the caudal and dorsal fins’ shades ranging from olive to dark brown. Black spots on caudal fin rays, creating a mottled pattern, with a single dark brown vertical row in approximately the middle of the rays. Dark brown pigments on the rays of the distal and median parts of the dorsal fin and the anterior half of the pectoral fin.
Etymology. This species is named after Jasmin Moghbeli, a NASA astronaut of Iranian descent, in recognition of her contributions to space exploration and her achievements in the field of aerospace. Her accomplishments and dedication serve as a beacon of hope and encouragement for Iranian women. A noun in apposition.
Distribution. This species is distributed in the Goleyn, Alvand, Zemkan, and Leyley Rivers in the Sirvan drainage and potentially in other headwaters of the Sirvan drainage (
Figure 14).
Habitat. These species are typically found in fast-flowing, clear, and cold-water parts of rivers and streams, frequently occupying areas with rapids and riffles containing coarse gravel or rocks. They tend to inhabit the interstices within gravel substrates. In the proximity of their native habitat, such as the Leyleh River (
Figure 14), there are rainbow trout farms. Escaped fish from these farms predate on
T. moghbeli, exerting a detrimental impact.
4. Discussion
Our extensive investigation examined samples from the Karkheh, Karun, Lesser Zab, Sirvan, and Alvand drainages, with the last three being shared between Iran and Iraq. We also considered previously collected data from other valid species available. We used the p-uncorrelated distance model to calculate the genetic distances between species. Our dataset encompasses a diverse range of interspecific distances, ranging from 3.6% to 14.1%. Based on our result, we clearly see the usefulness of the barcode region of the COI gene in species delimitation and differentiation for the studied group of organisms. Even if a phylogenetic framework has been used, this result is not a true investigation of the evolutionary history of the group, and the obtained tree is only used as a clustering method to delimit and differentiate species. In general, our dataset shows the poor phylogenetic signal of the marker used, precluding meaningful phylogenetic discussion, and shows the need for more markers for an in-depth phylogenetic analysis of the group.
Conway et al. [
2] described
T. himalaya from Nepal, establishing a substantially increased distribution range for the genus spanning approximately 3500 km. Although this study lacked specimens of
T. himalaya, we speculate that this species belongs to a different genus. The
T. hafezi and
T. kosswigi groups, displaying a genetic distance exceeding 12% in the COI marker, approach the acceptable threshold for differentiating genera in Nemacheilidae, especially considering their presence in neighboring basins.
Turcinoemacheilus also demonstrates a limited ability to migrate along rivers. Taken together, these factors strongly support placing
T. himalaya in a different genus.
Golzarianpour et al. [
3] initially recorded
T. kosswigi from the Sezar drainage in Iran. However, based on the photos they provided, it appears that the fish corresponds most probably to
T. saadii, a species abundant in the Sezar drainage. Our sampling from the same locality found both
Sasanidus kermanshahensis and
T. saadii. Golzarianpour et al. [
8] described
T. hafezi from the Shalamzar stream based on its morphology, but no DNA sequences from the type materials were provided. The original paper included a photo of live specimens of
T. ansari (their
Figure 5, Beshar River), assigned to
T. hafezi. These discrepancies prompted an investigation into the species’ type locality, which found that
T. hefezi is restricted to Shalamzar and Beheshtabad and absent from other drainage areas previously reported.
Esmaeili et al. [
9] conducted a review of
Turcinoemacheilus using both morphological and molecular data. They referenced two sequences of
T. saadii (KJ179257, KJ179250) from the Leyleh River, which were found to be almost identical to our samples of
T. saadii from Karkheh. We sampled the Leyleh River six times from Sepidbarg to Kalash Bakhan, but we were unable to locate any
T. saadii specimens. Therefore, it is highly probable that their claim is due to mislabeling in their dataset. Even if they examined the type materials of
T. hafezi, they did not include any COI sequences from the type material or the type localities of
T. hafezi and
T. kosswigi in their molecular dataset. They assigned populations from the Lesser Zab, Great Zab, and Sirvan as
T. kosswigi and the Beshar River as
T. hafezi. However, the results of the morphometric and phylogenetic tree analyses indicate that specimens from the Great and Lesser Zab, Sirvan, and Alvand belong to both
T. moghbeli and an undescribed
Turcinoemacheilus, and the populations of the Beshar River belong to
T. saadii and
T. ansari.
A note on the status of Paracobitis basharensis Freyhof, Esmaeili, Sayyadzadeh, Geiger 2014.
Based on the original description and pictures, Paracobitis basharensis might be wrongly placed in Paracobitis and possibly be within Turcinoemacheilus. We contacted Hamidreza Esmaeili, the contact person of the Zoological Museum of Shiraz University, Collection of Biology Department (ZM-CBSU), on several occasions to access the type material deposited in their collection, but without success. Even when we offered to send someone to examine the material and/or obtain higher-quality pictures of the deposited samples to include them in our study, our request was rejected. The type locality of this species (Beshar River) has been sampled on numerous occasions by our team and others, but no other species of loaches have been obtained other than the Turcinoemacheilus species. The habitat at the type locality is well preserved, and the fishes inhabiting it are caught in relatively high numbers. We believe we would have caught them if any Paracobitis species had been there. In addition, Saber Vatandoust, a collaborator of Omid Tabiee, is recognized in the original description as the collector of the type material of this species, and he cannot confirm catching any Paracobitis species on that occasion. A morphological character which makes us believe it might be in the wrong genus is the position of the pelvic fin in front of the dorsal-fin origin, which is diagnostic for Turcinoemacheilus. The only morphological difference we observed between the P. basharensis and Turcinoemacheilus species, based on the original description pictures, is the presence of an adipose keel on the caudal peduncle. This might be an abnormal specimen, observed sometimes in other specimens, or it might have a seasonal character. This might be a lipid reserve developed during the nutrient-rich season. Due to these complications, we suggest to our colleagues that they consider depositing the type material used in taxonomic works in serious public collections, preferably in multiple institutions, where access to the samples is not limited for other colleagues working in the field.
5. Conclusions
Using an integrative taxonomical approach, with extensive field work and morphologic, genetic, and phylogenetic analyses, we studied the diversity of an understudied and often neglected group of loaches. This resulted in the description of three new species, showing the importance of such integrative approaches for modern taxonomy. This study also shows the need for more effective conservation policies for the often remote and inaccessible freshwater habitats of the region. This is especially important for the newly described species, as their habitats are under serious threat. The presence of many unregulated or poorly regulated fish farms is an example of where a more serious conservation policy could benefit the conservation of such habitats. Overall, we would like to point out the importance of dedicated public institutions to preserve and protect biological materials as a valuable resource for future generations of scientists, especially in a world where many undescribed and unstudied organisms are disappearing at an accelerated and alarming rate.
Material examined
Turcinoemacheilus saadii. (
Figure 15)—VPFC TangGhir 1400.9., 5, 39−46 mm SL; Iran: Ilam prov.: Seymareh River at Tang-e Ghir, 33.75044, 46.6837.—VPFC Atishgah 1400.8., 1, 41 mm SL; Chaharmahal and Bakhtiari prov.: Khersan River at Atishgah, 31.24365, 50.99066.—AJRPC Kangavarkohne 1402.1., 9, 29−46 mm SL; Kermanshah prov., Kangavar stream at Kangavar Kohne, 34.34849, 47.98972.—AJRPC Beheshtabad 1400.8., 1, 43 mm SL; Chaharmahal and Bakhtiari prov.: Behesht Abad River at Liro (Beheshtabad), 32.04038, 50.64484.
Turcinoemacheilus hafezi. (
Figure 16)—VPFC Filabad 1400.8., 13, 36–51 mm SL; Chaharmahal and Bakhtiari prov.: Filabad stream at Filabad, 32.29427, 50.50578.
Turcinoemacheilus bahaii. (
Figure 17)—VPFC Khorbeh 1402.7., 8, 30–51 mm SL; Esfahan prov.: Khorbeh stream at Khorbeh, 32.51877, 50.22835.
Turcinoemacheilus sp. (
Figure 18)—MZLU L021/00002-3, 2, 48–55 mm SL; Iran: Kurdistan prov., Choman at Cherush, a tributary of Lesser Zab, 35.95400, 45.69216.—AJRPC 1758–17599, 40–42 mm SL; Iran: Kurdistan prov., Choman at Cherush, a tributary of Lesser Zab, 35.95400, 45.69216.—AJRPC 1637, 1, 43 mm SL; Iran: Kurdistan prov., Lesser Zab at Armardeh, 35.91506, 45.72798.
New material used in molecular genetic analysis
Turcinoemacheilus ansari.—AJRPC-DNA 925, OR800668, AJRPC-DNA 927, OR800669, AJRPC-DNA 929, OR800670, AJRPC-DNA 930, OR800671, AJRPC-DNA 931, OR800672, AJRPC-DNA 932, OR800673, AJRPC-DNA 933, OR800674, AJRPC-DNA 1736, OR800688, AJRPC-DNA 1737, OR800689, AJRPC-DNA 1738, OR800690, AJRPC-DNA 1779, OR800706, AJRPC-DNA 1780, OR800707, AJRPC-DNA 1781, OR800708, AJRPC-DNA 1782, OR800709, AJRPC-DNA 1783, OR800710, AJRPC-DNA 1784, OR800711, AJRPC-DNA 1785, OR800712, AJRPC-DNA 1786, OR800713, Iran: Kohgiluyeh and Boyer-Ahmad prov., Beshar River, near Allah Abad village, Karun drainage, 30.45029, 51.75346.
Turcinoemacheilus bahaii.—AJRPC-DNA 203, OR800659, AJRPC-DNA 1822, OR800723, AJRPC-DNA 1823, OR800724, AJRPC-DNA 1831, OR800729, AJRPC-DNA 1832, OR800725, AJRPC-DNA 1833, OR800726, AJRPC-DNA 1834, OR800727, AJRPC-DNA 1835, OR800728, Iran: Esfahan prov.: Khorbeh stream at Khorbeh, 32.51877, 50.22835.
Turcinoemacheilus christofferi.—AJRPC-DNA 1836, OR800662, AJRPC-DNA 1713, OR800698, Iran: Lorestan prov., Gholiyan River, tributary of Bakhtiyari River, Karun drainage, 33.07499, 49.64562.
Turcinoemacheilus hafezi.—AJRPC-DNA 163, OR800657, AJRPC-DNA 1624, OR800676, AJRPC-DNA 1625, OR800677, AJRPC-DNA 1626, OR800678, AJRPC-DNA 1627, OR800679, AJRPC-DNA 1628, OR800680, AJRPC-DNA 1629, OR800681, AJRPC-DNA 1630, OR800682, Iran: Chaharmahal and Bakhtiari prov.: Filabad stream at Filabad, 32.29427, 50.50578.
Turcinoemacheilus moghbeli.—AJRPC-DNA 19A, OR800649, Iran: Kermanshah prov., Goleyn River at Goleyn Village, 34.25603, 45.96339.—AJRPC-DNA 1635, OR800687, AJRPC-DNA 1636, OR800688, AJRPC-DNA 40A, OR800650, AJRPC-DNA 40B, OR800651, AJRPC-DNA 435B, OR800666, AJRPC-DNA 435C, OR800667, Iran: Kermanshah prov., Leyleh River at Sepidbarg, tributary of Sirvan drainage, 34.87334, 46.35020.—AJRPC-DNA 52A, OR800652, AJRPC-DNA 52B, OR800653, AJRPC-DNA 52C, OR800654, AJRPC-DNA 1739, OR800702, AJRPC-DNA 1744, OR800704, Iran: Kermanshah prov., Zemkan River near Siyah Taher Village, 34.73312 46.20455.
Turcinoemacheilus saadii.—AJRPC-DNA 12, OR800646, AJRPC-DNA 13, OR800647, AJRPC-DNA 14, OR800648, AJRPC-DNA 1631, OR800683, AJRPC-DNA 1632, OR800684, AJRPC-DNA 1633, OR800685, AJRPC-DNA 1634, OR800686, AJRPC-DNA 1642, OR800693, AJRPC-DNA 1643, OR800694, AJRPC-DNA 1644, OR800695, AJRPC-DNA 1645, OR800696, AJRPC-DNA 1646, OR800697, Iran: Kermanshah prov., Kangavar stream at Kangavar Kohne, 34.34849, 47.98972.—AJRPC-DNA 1640, OR800691, AJRPC-DNA 1641, OR800692, AJRPC-DNA 419A, OR800663, AJRPC-DNA 419B, OR800664, AJRPC-DNA 419C, OR800665, Iran: Ilam prov.: Seymareh River at Tang-e Ghir, 33.75044, 46.6837.—AJRPC-DNA 1801, OR800722, AJRPC-DNA 1796, OR800721, AJRPC-DNA 1795, OR800720, AJRPC-DNA 1794, OR800719, AJRPC-DNA 1793, OR800718, AJRPC-DNA 1792, OR800717, AJRPC-DNA 1791, OR800716, AJRPC-DNA 1787, OR800714, AJRPC-DNA 1788, OR800715, Iran: Lorestan prov.: Sezar River at chamchit, 33.37798, 48.96580.—AJRPC-DNA 161, OR800656, Iran: Chaharmahal and Bakhtiari prov.: Bazoft near Terki, 32.30322, 49.94893.—AJRPC-DNA 1740, OR800703, Iran: Kohgiluyeh and Boyer-Ahmad prov., Beshar River, near Allah Abad village, Karun drainage, 30.45029, 51.75346.—AJRPC-DNA 201, OR800658, AJRPC-DNA 1639, OR800690, Iran: Chaharmahal and Bakhtiari prov.: Khersan River at Atishgah, 31.24365 50.99066.
Turcinoemacheilus sp.—AJRPC-DNA 321, OR800661, AJRPC-DNA 1758, OR800705, Iran: Kurdistan prov., Lesser Zab at Armardeh, 35.91506, 45.72798.—AJRPC-DNA 153, OR800655, Iran: Kurdistan prov., Choman at Cherush, a tributary of Lesser Zab, 35.95400, 45.69216.