Occurrence of Aspergillus chevalieri and A. niger on Herbal Tea and Their Potential to Produce Ochratoxin A (OTA)
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Collection and Fungal Isolation
2.2. Macro- and Microscopic Identification of Fungi
2.3. DNA Extraction, PCR Amplification, Sequencing, and Phylogenetic Analyses
2.4. OTA Extraction and Quantification
3. Results
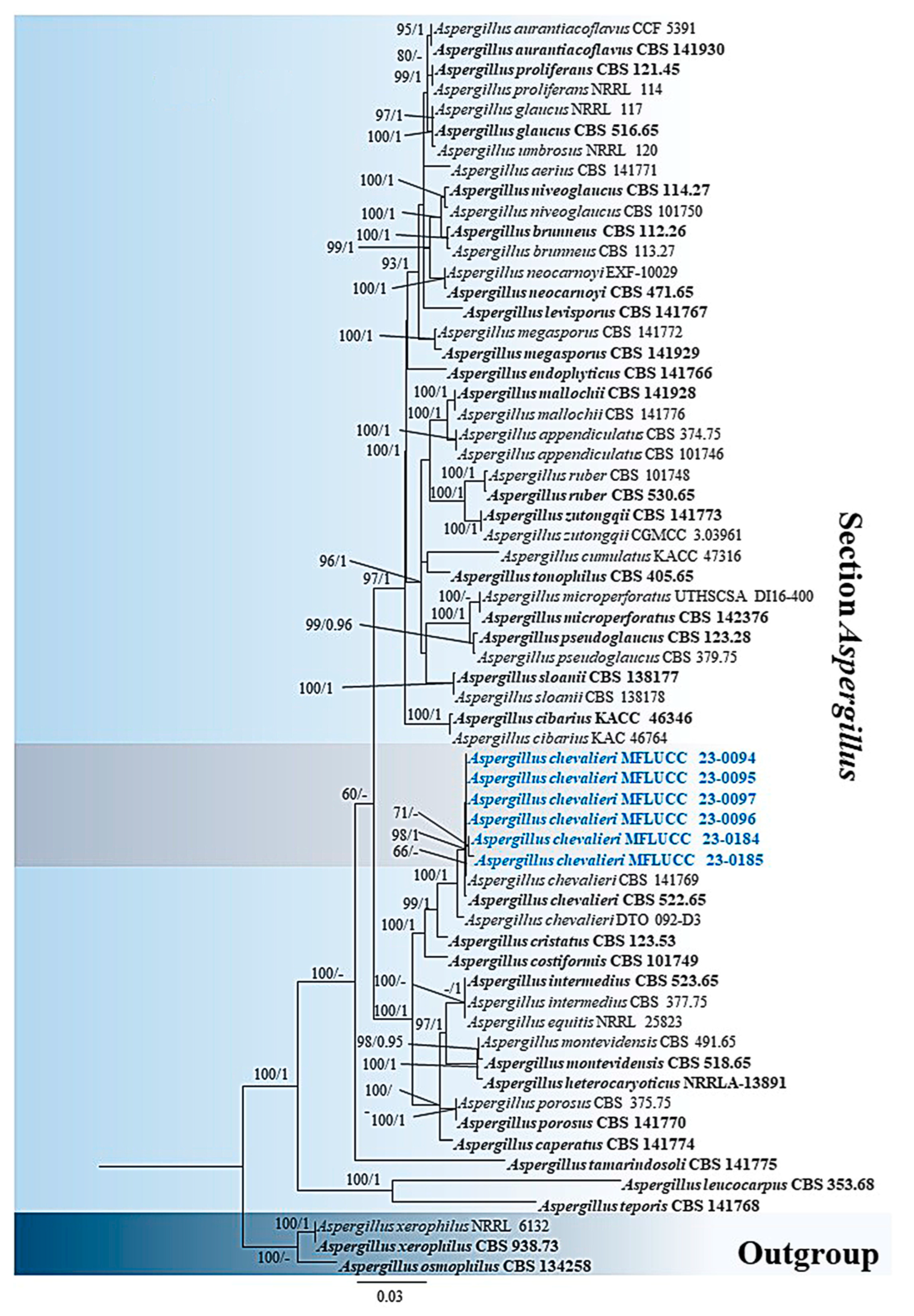
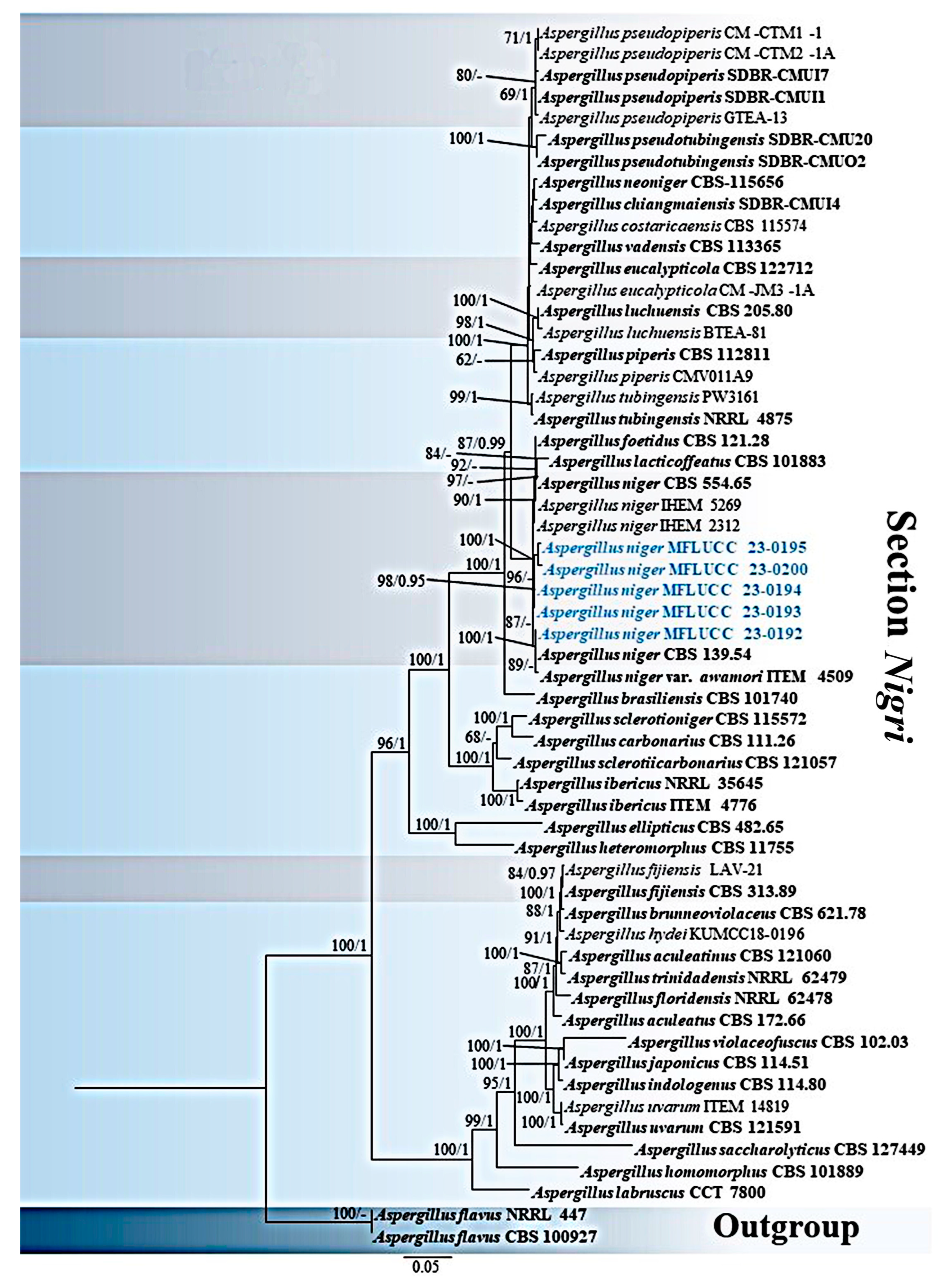
3.1. Phylogenetic Analyses
3.2. Taxonomy
3.2.1. Aspergillus chevalieri (L. Mangin) Thom & Church, The Genus Aspergillus: 111 (1926)

3.2.2. Aspergillus niger Tiegh., Annls Sci. Nat., Bot., sér. 5 8: 240 (1867)

3.3. Mycotoxin Detection
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Long, T.; Hu, R.; Cheng, Z.; Xu, C.; Hu, Q.; Liu, Q.; Gu, R.; Huang, Y.; Long, C. Ethnobotanical study on herbal tea drinks in Guangxi, China. J. Ethnobiol. Ethnomed. 2023, 19, 10. [Google Scholar] [CrossRef] [PubMed]
- Nurmilah, S.; Cahyana, Y.; Utama, G.L. Metagenomics analysis of the polymeric and monomeric phenolic dynamic changes related to the indigenous bacteria of black tea spontaneous fermentation. Biotechnol. Rep. 2022, 36, e00774. [Google Scholar]
- FAO—Food and Agriculture Organization of the United Nations. International Tea Market: Market Situation, Prospects and Emerging Issues. 2022. Available online: https://www.fao.org/documents/card/en?details=cc3017en (accessed on 5 September 2023).
- Chandrasekara, A.; Shahidi, F. Herbal beverages: Bioactive compounds and their role in disease risk reduction—A review. J. Tradit. Complement. Med. 2018, 8, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Shen, S.; Huang, L.; Deng, G.; Wei, Y.; Ning, J.; Wang, Y. Revelation of volatile contributions in green teas with different aroma types by GC–MS and GC–IMS. Food Res. Int. 2023, 169, 112845. [Google Scholar] [CrossRef] [PubMed]
- Tipduangta, P.; Julsrigival, J.; Chaithatwatthana, K.; Pongterdsak, N.; Tipduangta, P.; Chansakaow, S. Antioxidant properties of Thai traditional herbal teas. Beverages 2019, 5, 44. [Google Scholar] [CrossRef]
- Chan, E.W.C.; Eng, S.Y.; Tan, Y.P.; Wong, Z.C.; Lye, P.Y.; Tan, L.N. Antioxidant and sensory properties of Thai herbal teas with emphasis on Thunbergia laurifolia Lindl. Chiang Mai J. Sci. 2012, 39, 599–609. [Google Scholar]
- Ramphinwa, M.L.; Mchau, G.R.A.; Mashau, M.E.; Madala, N.E.; Chimonyo, V.G.P.; Modi, T.A.; Mabhaudhi, T.; Thibane, V.S.; Mudau, F.N. Eco-physiological response of secondary metabolites of teas: Review of quality attributes of herbal tea. Front. Sustain. Food Syst. 2023, 7, 990334. [Google Scholar] [CrossRef]
- Vu, D.C.; Alvarez, S. Phenolic, carotenoid and saccharide compositions of vietnamese Camellia sinensis teas and herbal teas. Molecules 2021, 26, 6496. [Google Scholar] [CrossRef]
- Ashiq, S.; Hussain, M.; Ahmad, B. Natural occurrence of mycotoxins in medicinal plants: A review. Fungal Genet. Biol. 2014, 66, 1–10. [Google Scholar] [CrossRef]
- Chomchalow, N.; Hicks, A. Health potential of Thai traditional beverages. AU J. Technol. 2001, 5, 20–30. [Google Scholar]
- Pinitsoontorn, C.; Suwantrai, S.; Boonsiri, P. Antioxidant activity and oxalate content of selected Thai herbal teas. Asia-Pac. J. Sci. Technol. 2012, 17, 162–168. [Google Scholar]
- Sarwar, S.; Lockwood, B. Herbal teas. Nutrafoods 2010, 9, 7–17. [Google Scholar] [CrossRef]
- Prathumtet, J.; Surasorn, C.; Paopa, T. Total phenolic content and antioxidant activity of three flower infusion tea in Sakon Nakhon Province. J. Health Sci. 2019, 28, 1110–1115. [Google Scholar]
- Ngoitaku, C.; Kwannate, P.; Riangwong, K. Total phenolic content and antioxidant activities of edible flower tea products from Thailand. Int. Food Res. J. 2016, 23, 2286–2290. [Google Scholar]
- Sedova, I.; Kiseleva, M.; Tutelyan, V. Mycotoxins in tea: Occurrence, methods of determination and risk evaluation. Toxins 2018, 10, 444. [Google Scholar] [CrossRef] [PubMed]
- Babu, A.K.; Kumaresan, G.; Raj, V.A.A.; Velraj, R. Review of leaf drying: Mechanism and influencing parameters, drying methods, nutrient preservation, and mathematical models. Renew. Sustain. Energy Rev. 2018, 90, 536–556. [Google Scholar] [CrossRef]
- Chalyy, Z.; Kiseleva, M.; Sedova, I.; Tutelyan, V. Mycotoxins in herbal tea: Transfer into the infusion. World Mycotoxin J. 2021, 14, 539–551. [Google Scholar] [CrossRef]
- Reinholds, I.; Bogdanova, E.; Pugajeva, I.; Bartkevics, V. Mycotoxins in herbal teas marketed in Latvia and dietary exposure assessment. Food Addit. Contam. Part B 2019, 12, 199–208. [Google Scholar] [CrossRef]
- Begum, N.; Qin, C.; Ahanger, M.A.; Raza, S.; Khan, M.I.; Ashraf, M.; Ahmed, N.; Zhang, L. Role of arbuscular mycorrhizal fungi in plant growth regulation: Implications in abiotic stress tolerance. Front. Plant Sci. 2019, 10, 1068. [Google Scholar] [CrossRef]
- Benedict, K.; Chiller, T.M.; Mody, R.K. Invasive fungal infections acquired from contaminated food or nutritional supplements: A review of the literature. Foodborne Pathog. Dis. 2016, 13, 343–349. [Google Scholar] [CrossRef]
- Hyde, K.D.; Xu, J.; Rapior, S.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.S.; Brahamanage, R.S.; Brooks, S.; et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019, 97, 1–136. [Google Scholar] [CrossRef]
- Reinholds, I.; Rusko, J.; Pugajeva, I.; Berzina, Z.; Jansons, M.; Kirilina-Gutmane, O.; Tihomirova, K.; Bartkevics, V. The occurrence and dietary exposure assessment of mycotoxins, biogenic amines, and heavy metals in mould-ripened blue cheeses. Foods 2020, 9, 93. [Google Scholar] [CrossRef] [PubMed]
- Xu, J. Assessing global fungal threats to humans. mLife 2022, 1, 223–240. [Google Scholar] [CrossRef]
- Davis, K.F.; Gephart, J.A.; Emery, K.A.; Leach, A.M.; Galloway, J.N.; D’Odorico, P. Meeting future food demand with current agricultural resources. Glob. Environ. Change 2016, 39, 125–132. [Google Scholar] [CrossRef]
- Antonissen, G.; Martel, A.; Pasmans, F.; Ducatelle, R.; Verbrugghe, E.; Vandenbroucke, V.; Li, S.; Haesebrouck, F.; Van Immerseel, F.; Croubels, S. The impact of Fusarium mycotoxins on human and animal host susceptibility to infectious diseases. Toxins 2014, 6, 430–452. [Google Scholar] [CrossRef]
- Liew, W.P.P.; Mohd-Redzwan, S. Mycotoxin: Its impact on gut health and microbiota. Front. Cell. Infect. Microbiol. 2018, 8, 60. [Google Scholar] [CrossRef]
- Zain, M.E. Impact of mycotoxins on humans and animals. J. Saudi Chem. Soc. 2011, 5, 129–144. [Google Scholar] [CrossRef]
- Fisher, M.C.; Hawkins, N.J.; Sanglard, D.; Gurr, S.J. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 2018, 360, 739–742. [Google Scholar] [CrossRef]
- Kumar, D.; Kalita, P. Reducing postharvest losses during storage of grain crops to strengthen food security in developing countries. Foods 2017, 6, 8. [Google Scholar] [CrossRef]
- Paterson, R.R.M.; Lima, N. Further mycotoxin effects from climate change. Food Res. Int. 2011, 44, 2555–2566. [Google Scholar] [CrossRef]
- Sinphithakkul, P.; Poapolathep, A.; Klangkaew, N.; Imsilp, K.; Logrieco, A.F.; Zhang, Z.; Poapolathep, S. Occurrence of multiple mycotoxins in various types of rice and barley samples in Thailand. J. Food Prot. 2019, 82, 1007–1015. [Google Scholar] [CrossRef]
- Abdulkadar, A.H.W.; Al-Ali, A.A.; Al-Kildi, A.M.; Al-Jedah, J.H. Mycotoxins in food products available in Qatar. Food Control. 2004, 15, 543–548. [Google Scholar] [CrossRef]
- Arroyo-Manzanares, N.; Huertas-Pérez, J.F.; García-Campaña, A.M.; Gámiz-Gracia, L. Mycotoxin analysis: New proposals for sample treatment. Adv. Chem. 2014, 2014, 547506. [Google Scholar] [CrossRef]
- Karaca, A.; Cetin, S.C.; Turgay, O.C.; Kizilkaya, R. Effects of heavy metals on soil enzyme activities. In Soil Heavy Metals; Sherameti, I., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 19, pp. 237–262. [Google Scholar]
- Karbancıoğlu-Güler, F.; Heperkan, D. Natural occurrence of ochratoxin A in dried figs. Anal. Chim. Acta 2008, 617, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Minaeva, L.P.; Aleshkina, A.I.; Markova, Y.M.; Polyanina, A.S.; Pichugina, T.V.; Bykova, I.B.; Stetsenko, V.V.; Efimochkina, N.R.; Sheveleva, S.A. Studying the contamination of tea and herbal infusions with mold fungi as potential mykotoxin producers: The first step to risk assessment (message 1). Health Risk Anal. 2019, 1, 93–102. [Google Scholar] [CrossRef]
- Awuchi, C.G.; Ondari, E.N.; Eseoghene, I.J.; Twinomuhwezi, H.; Amagwula, I.O.; Morya, S. Fungal growth and mycotoxins produc-tion: Types, toxicities, control strategies, and detoxification. In Fungal Reproduction and Growth; IntechOpen: London, UK, 2022. [Google Scholar]
- Dhakal, A.; Hashmi, M.F.; Sbar, E. Aflatoxin Toxicity. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK557781/ (accessed on 10 August 2023).
- Foroud, N.A.; Baines, D.; Gagkaeva, T.Y.; Thakor, N.; Badea, A.; Steiner, B.; Bürstmayr, M.; Bürstmayr, H. Trichothecenes in cereal grains—An update. Toxins 2019, 11, 634. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Frank, J.M.; Houbraken, J.A.M.P.; Kuijpers, A.F.; Samson, R.A. New ochratoxin A producing species of Aspergillus section Circumdati. Stud. Mycol. 2004, 50, 20–43. [Google Scholar]
- Kumar, A.; Pathak, H.; Bhadauria, S.; Sudan, J. Aflatoxin contamination in food crops: Causes, detection, and management: A review. Food Prod. Process. Nutr. 2021, 3, 17. [Google Scholar] [CrossRef]
- Rheeder, J.P.; Marasas, W.F.O.; Vismer, H.F. Production of fumonisin analogs by Fusarium species. Appl. Environ. Microbiol. 2002, 68, 2101–2105. [Google Scholar] [CrossRef]
- Ropejko, K.; Twarużek, M. Zearalenone and its metabolites-general overview, occurrence, and toxicity. Toxins 2021, 13, 35. [Google Scholar] [CrossRef]
- Sánchez-Rangel, D.; Juan-Badillo, A.S.; Plasencia, J. Fumonisin production by Fusarium verticillioides strains isolated from maize in Mexico and development of a polymerase chain reaction to detect potential toxigenic strains in grains. J. Agric. Food Chem. 2005, 53, 8565–8571. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Liu, F.; Wang, Q.; Selvaraj, J.N.; Xing, F.; Zhao, Y.; Liu, Y. Ochratoxin A producing fungi, biosynthetic pathway and regulatory mechanisms. Toxins 2016, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, J.; Chen, Y.; Zhu, J. Zearalenone and its masked forms in cereals and cereal-derived products: A review of the characteristics, incidence, and fate in food processing. J. Fungi 2022, 8, 976. [Google Scholar] [CrossRef] [PubMed]
- Boonmee, S.; Atanasova, V.; Chéreau, S.; Marchegay, G.; Hyde, K.D.; Richard-Forget, F. Efficiency of hydroxycinnamic phenolic acids to inhibit the production of ochratoxin A by Aspergillus westerdijkiae and Penicillium verrucosum. Int. J. Mol. Sci. 2020, 21, 8548. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Wu, F.; Liu, F.; Wang, Q.; Zhang, X.; Selvaraj, J.N.; Zhao, Y.; Xing, F.; Yin, W.B.; et al. A consensus ochratoxin A biosynthetic pathway: Insights from the genome sequence of Aspergillus ochraceus and a comparative genomic analysis. Appl. Environ. Microbiol. 2018, 84, e01009-18. [Google Scholar] [CrossRef]
- Magan, N.; Aldred, D. Post-harvest control strategies: Minimizing mycotoxins in the food chain. Int. J. Food Microbiol. 2007, 119, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Jiang, H.; Li, F. Dynamic ochratoxin A production by strains of Aspergillus niger intended used in food industry of China. Toxins 2019, 11, 122. [Google Scholar] [CrossRef]
- Chen, L.; Guo, W.; Zheng, Y.; Zhou, J.; Liu, T.; Chen, W.; Liang, D.; Zhao, M.; Zhu, Y.; Wu, Q.; et al. Occurrence and characterization of fungi and mycotoxins in contaminated medicinal herbs. Toxins 2020, 12, 30. [Google Scholar] [CrossRef]
- Fraga, M.E.; Curvello, F.; Gatti, M.J.; Cavaglieri, L.R.; Dalcero, A.M.; da Rocha Rosa, C.A. Potential aflatoxin and ochratoxin A production by Aspergillus species in poultry feed processing. Veter Res. Commun. 2007, 31, 343–353. [Google Scholar] [CrossRef]
- Nazar, M.; Ali, M.; Fatima, T.; Gubler, C.J. Toxicity of flavoglaucin from Aspergillus chevalieri in rabbits. Toxicol. Lett. 1984, 23, 233–237. [Google Scholar] [CrossRef]
- Wilkinson, S.; Spilsbury, J.F. Gliotoxin from Aspergillus chevalieri (Mangin) Thom et Church. Nature 1965, 206, 619. [Google Scholar] [CrossRef] [PubMed]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- De Hoog, G.S.; Queiroz-Telles, F.; Haase, G.; Fernandez-Zeppenfeldt, G.; Attili Angelis, D.; Gerrits Van Den Ende, A.H.; Matos, T.; Peltroche-Llacsahuanga, H.; Pizzirani-Kleiner, A.A.; Rainer, J.; et al. Black fungi: Clinical and pathogenic approaches. Med. Mycol. 2000, 38, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Hubka, V.; Ezekiel, C.N.; Hong, S.B.; Novßkovß, A.; Chen, A.J.; Arzanlou, M.; Larsen, T.O.; Sklenßř, F.; Mahakarnchanakul, W.; et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 2019, 93, 1–63. [Google Scholar] [CrossRef] [PubMed]
- Visagie, C.M.; Hirooka, Y.; Tanney, J.B.; Whitfield, E.; Mwange, K.; Meijer, M.; Amend, A.S.; Seifert, K.A.; Samson, R.A. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud. Mycol. 2014, 78, 63–139. [Google Scholar] [CrossRef] [PubMed]
- Kretzer, A.; Li, Y.; Szaro, T.; Bruns, T.D. Internal transcribed spacer sequences from 38 recognized species of Suillus sensu lato: Phylogenetic and taxonomic implications. Mycologia 1996, 88, 776–785. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- O’Donnell, K.; Nirenberg, H.I.; Aoki, T.; Cigelnik, E. A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 2000, 41, 61–78. [Google Scholar] [CrossRef]
- Hong, S.B.; Go, S.J.; Shin, H.D.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 2005, 97, 1316–1329. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Hubka, V.; Lyskova, P.; Frisvad, J.C.; Peterson, S.W.; Skorepova, M.; Kolarik, M. Aspergillus pragensis sp. nov. discovered during molecular re-identification of clinical isolates belonging to Aspergillus section Candidi. Medical. Mycol. 2014, 52, 565–576. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hubka, V.; Nováková, A.; Peterson, S.W.; Frisvad, J.C.; Sklenář, F.; Matsuzawa, T.; Kubátová, A.; Kolařík, M. A reappraisal of Aspergillus section Nidulantes with descriptions of two new sterigmatocystin-producing species. Plant Syst. Evol. 2016, 302, 1267–1299. [Google Scholar] [CrossRef]
- Samson, R.A.; Visagie, C.M.; Houbraken, J.; Hong, S.B.; Hubka, V.; Klaassen, C.H.; Perrone, G.; Seifert, K.A.; Susca, A.; Tanney, J.B.; et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 7, 141–173. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, A.J.; Bhunjun, C.S.; Maharachchikumbura, S.S.N.; Liu, J.K. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 2020, 11, 2652–2676. [Google Scholar] [CrossRef]
- Bragulat, M.R.; Abarca, M.L.; Cabañes, F.J. An easy screening method for fungi producing ochratoxin A in pure culture. Int. J. Food Microbiol. 2001, 71, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C. Physiological criteria and mycotoxin production as AIDS in identification of common asymmetric penicillia. Appl. Environ. Microbiol. 1981, 141, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.W.; Wu, D.L.; Li, W.D.; Hao, Z.H.; Wu, X.L.; Xing, Y.J.; Shi, J.R.; Li, Y.; Dong, F. Occurrence of Fusarium mycotoxins in freshly harvested highland barley (qingke) grains from Tibet, China. Mycotoxin Res. 2023, 39, 193–200. [Google Scholar] [CrossRef]
- Jeong, S.E.; Chung, S.H.; Hong, S.Y. Natural occurrence of aflatoxins and ochratoxin A in meju and soybean paste produced in South Korea. Appl. Biol. Chem. 2019, 62, 65. [Google Scholar] [CrossRef]
- Nakhjavan, B.; Ahmed, N.S.; Khosravifard, M. Development of an improved method of sample extraction and quantitation of multi-mycotoxin in feed by LC-MS/MS. Toxins 2020, 12, 462. [Google Scholar] [CrossRef]
- Thom, C.; Church, M. The Aspergilli; Williams and Wilkins Co.: Baltimore, MD, USA, 1926. [Google Scholar]
- Chen, A.J.; Hubka, V.; Frisvad, J.C.; Visagie, C.M.; Houbraken, J.; Meijer, M.; Varga, J.; Demirel, R.; Jurjević, Ž.; Kubátová, A.; et al. Polyphasic taxonomy of Aspergillus section Aspergillus (formerly Eurotium), and its occurrence in indoor environments and food. Stud. Mycol. 2017, 88, 37–135. [Google Scholar] [CrossRef]
- Hubka, V.; Kolarík, M.; Kubátová, A.; Peterson, S.W. Taxonomic revision of Eurotium and transfer of species to Aspergillus. Mycologia 2013, 105, 912–937. [Google Scholar] [CrossRef] [PubMed]
- Kulik, M.M.; Holaday, C.E. Aflatoxin: A metabolic product of several fungi. Mycopath. Mycol. Appl. 1966, 30, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, H.W.; Kelton, W. Production of sterigmatocystin by some species of the genus Aspergillus and its toxicity to chicken embryos. Appl. Microbiol. 1975, 30, 589–591. [Google Scholar] [CrossRef] [PubMed]
- Stein, R.A.; Bulboacă, A.E. Mycotoxins. In Foodborne Diseases, 3rd ed.; Dodd, C., Aldsworth, T., Stein, R.A., Cliver, D., Riemann, H., Eds.; Elsevier: Amsterdam, The Netherlands; Academic Press: London, UK, 2017; pp. 407–446. [Google Scholar]
- Bian, C.; Kusaya, Y.; Sklenář, F.; D’hooge, E.; Yaguchi, T.; Ban, S.; Visagie, C.M.; Houbraken, J.; Takahashi, H.; Hubka, V. Reducing the number of accepted species in Aspergillus series Nigri. Stud. Mycol. 2022, 102, 95–132. [Google Scholar] [CrossRef]
- Crous, P.W.; Verkley, G.J.M.; Groenewald, J.Z.; Samson, R.A. Fungal Biodiversity; CBS Laboratory Manual Series 1; Westerdijk Fungal Biodiversity Institute: Utrecht, The Netherlands, 2009. [Google Scholar]
- Frisvad, J.C.; Petersen, L.M.; Lyhne, E.K.; Larsen, T.O. Formation of sclerotia and production of indoloterpenes by Aspergillus niger and other species in section Nigri. PLoS ONE 2014, 9, 94857. [Google Scholar] [CrossRef]
- Samson, R.A.; Houbraken, J.; Thrane, U.; Frisvad, J.C.; Andersen, B. Food and Indoor Fungi, 2nd ed.; CBS Laboratory Manual Series No. 2; CBS-KNAW Fungal Biodiversity Centre: Utrecht, The Netherlands, 2010. [Google Scholar]
- Zulkifli, N.A.; Zakaria, L. Morphological and molecular diversity of Aspergillus from corn grain used as livestock feed. HAYATI J. Biosci. 2017, 24, 26–34. [Google Scholar] [CrossRef]
- Wang, Q.; Gong, J.; Chisti, Y.; Sirisansaneeyakul, S. Fungal isolates from a pu-erh type tea fermentation and their ability to convert tea polyphenols to theabrownins. J. Food Sci. 2015, 80, M809–M817. [Google Scholar] [CrossRef]
- Pakshir, K.; Mirshekari, Z.; Nouraei, H.; Zareshahrabadi, Z.; Zomorodian, K.; Khodadadi, H.; Hadaegh, A. Mycotoxins detection and fungal contamination in black and green tea by HPLC-based method. J. Appl. Toxicol. 2020, 2020, 2456210. [Google Scholar] [CrossRef]
- Mannani, N.; Tabarani, A.; Abdennebi, E.H.; Zinedine, A. Assessment of aflatoxin levels in herbal green tea available on the Moroccan market. Food Control. 2020, 108, 106882. [Google Scholar] [CrossRef]
- Mannaa, M.; Kim, K.D. Influence of temperature and water activity on deleterious fungi and mycotoxin production during grain storage. Mycobiology 2017, 45, 240–254. [Google Scholar] [CrossRef]
- Medina, A.; Rodriguez, A.; Magan, N. Climate change and mycotoxigenic fungi: Impacts on mycotoxin production. Curr. Opin. Food Sci. 2015, 5, 99–104. [Google Scholar] [CrossRef]
- Molnár, K.; Rácz, C.; Dövényi-Nagy, T.; Bakó, K.; Pusztahelyi, T.; Kovács, S.; Adácsi, C.; Pócsi, I.; Dobos, A. The effect of environmental factors on mould counts and AFB1 toxin production by Aspergillus flavus in maize. Toxins 2023, 15, 227. [Google Scholar] [CrossRef] [PubMed]
- Kovalsky-Paris, M.P.; Liu, Y.J.; Nahrer, K.; Binder, E.M. Climate change impacts on mycotoxin production. Clim. Chang. Mycotoxins 2015, 25, 133–152. [Google Scholar]
- Singh, B.K.; Delgado-Baquerizo, M.; Egidi, E.; Guirado, E.; Leach, J.E.; Liu, H.; Trivedi, P. Climate change impacts on plant pathogens, food security and paths forward. Nat. Rev. Microbiol. 2023, 21, 640–656. [Google Scholar] [CrossRef]
- Wang, X.; Jarmusch, S.A.; Frisvad, J.C.; Larsen, T.O. Current status of secondary metabolite pathways linked to their related biosynthetic gene clusters in Aspergillus section Nigri. Nat. Prod. Rep. 2023, 40, 237–274. [Google Scholar] [CrossRef]
- European, Union. Maximum Levels of Ochratoxin A in Certain Foodstuffs. Commission Regulation (EU) No 2022/1370. Off. J. Eur. Union 2022. Available online: http://data.europa.eu/eli/reg/2006/1881/2014-07-01 (accessed on 5 September 2023).
- Greco, M.V.; Pardo, A.G.; Ludemann, V.; Martino, P.E.; Pose, G.N. Mycoflora and natural incidence of selected mycotoxins in rabbit and chinchilla feeds. Sci. World. J. 2012, 2012, 956056. [Google Scholar] [CrossRef]

| No. | Herbal Tea | Scientific Name and Family | Herbal Tea Substrate | Number of Samples (Package) | Local Markets |
|---|---|---|---|---|---|
| 1 | Black Tea | Camellia sinensis, Theaceae | leaves | 6 | Doi Mae Salong Fah Thai Mae Sai |
| 2 | Bael Fruit | Aegle marmelos, Rutaceae | fruits | 4 | Doi Mae Salong Fah Thai Mae Sai |
| 3 | Goji Berry | Lycium sp., Solanaceae | fruits | 2 | Doi Mae Salong Fah Thai |
| 4 | Green Tea | Camellia sinensis, Theaceae | leaves | 6 | Doi Mae Salong Mae Sai Lan Muang |
| 5 | Jasmine | Jasminum sp., Oleaceae | flowers | 2 | Doi Mae Salong |
| 6 | Lavender | Lavandula sp., Lamiaceae | flowers | 4 | Lan Muang Mae Sai |
| 7 | Rose | Rosa sp., Rosaceae | flowers | 2 | Doi Mae Salong |
| Gene | Primers (Forward/Reverse) | PCR Condition | References |
|---|---|---|---|
| β-tubulin (BenA) | Bt2a/Bt2b | 1 cycle at 94 °C for 3 min; 35 cycles of 94 °C for 30 s; 55 °C for 50 s; 72 °C for 1 min; and a final extension at 72 °C for 10 min | Frisvad et al. [58], Hubka et al. [66,67], Samson et al. [68] |
| Calmodulin (CaM) | CMD5/CMD6 CL1/CL2A | 1 cycle at 95 °C for 5 min; 35 cycles of 94 °C for 45 s; 55 °C for 45 s; 72 °C for 1 min; and a final extension at 72 °C for 10 min | Frisvad et al. [58], Hubka et al. [66,67], Samson et al. [68] |
| Internal transcribed spacer (ITS) | ITS5/ITS4 | 1 cycle at 94 °C for 3 min; 35 cycles of 94 °C for 30 s; 55 °C for 50 s; 72 °C for 1 min; and a final extension at 72 °C for 10 min | Frisvad et al. [58], Hubka et al. [66,67], Samson et al. [68] |
| RNA polymerase II second largest subunit(RPB2) | RPB2f-5f/RPB2f-7cr | 1 cycle at 95 °C for 5 min; 40 cycles of 95 °C for 1 min; 57 °C for 1 min; 72 °C for 1 min; and a final extension at 72 °C for 10 min | Frisvad et al. [58], Samson et al. [68] |
| Taxa | Culture Collection No. | GenBank Accession No. | |||
|---|---|---|---|---|---|
| ITS | BenA | CAM | RPB2 | ||
| A. aerius | CBS 141771 | LT670916 | LT670990 | LT670991 | LT670992 |
| A. appendiculatus | CBS 101746 | HE615133 | HE801334 | HE801319 | HE801308 |
| A. appendiculatus | CBS 374.75 T | HE615132 | HE801333 | HE801318 | HE801307 |
| A. aurantiacoflavus | CBS 141930 T | LT670917 | LT670993 | LT670994 | LT670995 |
| A. aurantiacoflavus | CCF 5391 | LT670918 | LT670996 | LT670997 | LT670998 |
| A. brunneus | CBS 113.27 | EF652056 | EF651904 | EF651997 | EF651938 |
| A. brunneus | CBS 112.26 T | EF652060 | EF651907 | EF651998 | EF651939 |
| A. caperatus | CBS 141774 T | LT670922 | LT671008 | LT671009 | LT671010 |
| A. chevalieri | DTO 092-D3 | LT670929 | LT671029 | LT671030 | LT671031 |
| A. chevalieri | CBS 141769 | LT670927 | LT671023 | LT671024 | LT671025 |
| A. chevalieri | CBS 522.65 T | EF652068 | EF651911 | EF652002 | EF651954 |
| A. chevalieri | MFLUCC 23-0094 | OR478693 | OR508967 | N/A | N/A |
| A. chevalieri | MFLUCC 23-0095 | OR478694 | OR508966 | OR508963 | N/A |
| A. chevalieri | MFLUCC 23-0096 | OR478695 | OR508968 | OR508964 | N/A |
| A. chevalieri | MFLUCC 23-0097 | OR478696 | OR508969 | OR508965 | N/A |
| A. chevalieri | MFLUCC 23-0184 | OR502380 | OR573934 | OR604627 | OR604629 |
| A. chevalieri | MFLUCC 23-0185 | OR501403 | OR573933 | N/A | OR604628 |
| A. cibarius | KACC 46346 T | JQ918177 | JQ918180 | JQ918183 | JQ918186 |
| A. cibarius | KAC 46764 | JQ918178 | JQ918184 | JQ918181 | JQ918187 |
| A. costiformis | CBS 101749 T | HE615136 | HE801338 | HE801320 | HE801309 |
| A. cristatus | CBS 123.53 T | EF652078 | EF651914 | EF652001 | EF651957 |
| A. cumulatus | KACC 47316 | KF928303 | KF928297 | KF928300 | KF928294. |
| A. endophyticus | CBS 141766 T | LT670941 | LT671067 | LT671068 | LT671069 |
| A. equitis | NRRL 25823 | EF652073 | EF651895 | EF652015 | EF651961 |
| A. glaucus | CBS 516.65 T | EF652052 | EF651989 | EF651887 | EF651934 |
| A. glaucus | NRRL 117 | EF652053 | EF651990 | EF651888 | EF651935 |
| A. heterocaryoticus | NRRLA-13891 T | EU021619 | EU021670 | EU021687 | EU021659 |
| A. intermedius | CBS 377.75 | HE974459 | HE974432 | HE974437 | HE974425 |
| A. intermedius | CBS 523.65 T | EF652074 | EF651892 | EF652012 | EF651958 |
| A. leucocarpus | CBS 353.68 T | EF652087 | EF651925 | EF652023 | EF651972 |
| A. levisporus | CBS 141767 T | LT670950 | LT671094 | LT671095 | LT671096 |
| A. mallochii | CBS 141928 T | KX450907 | KX450889 | KX450902 | KX450894 |
| A. mallochii | CBS 141776 | KX450908 | KX450890 | KX450903 | KX450895 |
| A. megasporus | CBS 141772 | KX450911 | KX450893 | KX450906 | KX450898 |
| A. megasporus | CBS 141929 T | KX450910 | KX450892 | KX450905 | KX450897 |
| A. microperforatus | CBS 142376 T | LT627271 | LT627296 | LT627321 | LT627346 |
| A. microperforatus | UTHSCSADI16-400 | LT627270 | LT627295 | LT627320 | LT627345 |
| A. montevidensis | CBS 518.65 | EF652076 | EF651897 | EF652017 | EF651963 |
| A. montevidensis | CBS 491.65 T | EF652077 | EF651898 | EF652020 | EF651964 |
| A. neocarnoyi | EXF-10029 | LT670955 | LT671109 | LT671110 | LT671111 |
| A. neocarnoyi | CBS 471.65 T | EF652057 | EF651903 | EF651985 | EF651942 |
| A. niveoglaucus | CBS 101750 | HE615135 | HE801331 | HE801323 | HE801312 |
| A. niveoglaucus | CBS 114.27 T | EF652058 | EF651905 | EF651993 | EF651943 |
| A. osmophilus | CBS 134258 T | KC473921 | KC473924 | KC473918 | KX512310 |
| A. porosus | CBS 375.75 | LT670963 | LT671136 | LT671137 | LT671138 |
| A. porosus | CBS 141770 T | LT670961 | LT671130 | LT671131 | LT671132 |
| A. proliferans | CBS 121.45 T | EF652064 | EF651891 | EF651988 | EF651941 |
| A. proliferans | NRRL 114 | EF652051 | EF651886 | EF651987 | EF651933 |
| A. pseudoglaucus | CBS 379.75 | HE615131 | HE801336 | HE801322 | HE801311 |
| A. pseudoglaucus | CBS 123.28 T | EF652050 | EF651917 | EF652007 | EF651952 |
| A. ruber | CBS 530.65 T | EF652066 | EF651920 | EF652009 | EF651947 |
| A. ruber | CBS 101748 | HE615134 | HE801337 | HE801325 | HE801315 |
| A. sloanii | CBS 138178 | KJ775542 | KJ775076 | KJ775313 | KX450900 |
| A. sloanii | CBS 138177 T | KJ775540 | KJ775074 | KJ775309 | KX463365 |
| A. tamarindosoli | CBS 141775 T | LT670981 | LT671191 | LT671192 | LT671193 |
| A. teporis | CBS 141768 T | LT670982 | LT671194 | LT671195 | LT671196 |
| A. tonophilus | CBS 405.65 T | EF652081 | EF651919 | EF652000 | EF651969 |
| A. umbrosus | NRRL120 | EF652054 | EF651889 | EF651991 | EF651936 |
| A. xerophilus | CBS 938.73 T | EF652085 | EF651923 | EF651983 | EF651970 |
| A. xerophilus | NRRL 6132 | EF652086 | EF651924 | EF651984 | EF651971 |
| A. zutongqii | CGMCC 3.06103 | LT670989 | LT671215 | LT671216 | LT671217 |
| A. zutongqii | CBS 141773 T | LT670986 | LT671206 | LT671207 | LT671208 |
| Taxa | Culture Collection No. | Gene Bank Accession No. | |||
|---|---|---|---|---|---|
| ITS | BenA | CAM | RPB2 | ||
| A. aculeatinus | CBS 121060 T | EU159211 | EU159220 | EU159241 | HF559233 |
| A. aculeatus | CBS 172.66 T | EF661221 | HE577806 | EF661148 | EF661046 |
| A. awamori | ITEM 4509 T | AM087614 | AY820001 | AJ964874 | HE984360 |
| A. brasiliensis | CBS 101740 T | FJ629321 | FJ629272 | FN594543 | KY006765 |
| A. brunneoviolaceus | CBS 621.78 T | AJ280003 | EF661105 | EF661147 | EF661045 |
| A. carbonarius | CBS 111.26 T | EF661204 | EF661099 | EF661167 | EF661068 |
| A. chiangmaiensis | SDBR-CMUI4 | MW588209 | MW602898 | MK457199 | MW602899 |
| A. costaricaensis | CBS 115574 T | DQ900602 | FJ629277 | FN594545 | HE984361 |
| A. ellipticus | CBS 482.65 T | EF661194 | EF661122 | EF661170 | EF661051 |
| A. eucalypticola | CBS 122712 T | EU482439 | EU482435 | EU482433 | MN969070 |
| A. flavus | CBS 100927 T | AF027863 | EF661485 | EF661508 | EF661440 |
| A. flavus | NRRL 447 T | EF661560 | EF661483 | EF661506 | EF661438 |
| A. fijiensis | CBS 313.89 T | FJ491680 | FJ491688 | FJ491695 | N/A |
| A. floridensis | NRRL 62478 T | N/A | HE984412 | HE984429 | HE984376 |
| A. foetidus | CBS 121 28 T | FJ491683 | FJ491690 | FJ491694 | N/A |
| A. homomorphus | CBS 101899 T | EF166063 | AY820015 | FN594549 | N/A |
| A. heteromorphus | CBS 11755 T | EU821305 | EF661103 | EF661169 | EF661050 |
| A. hydei | KUMCC 18-0196 | MT152332 | MT161679 | MT178247 | MT384370 |
| A. ibericus | ITEM 4776 T | NR 119514 | AM419748 | AJ971805 | N/A |
| A. ibericus | NRRL 35645 T | EF661201 | EF661101 | EF661164 | N/A |
| A. indologenus | CBS 114.80 T | AJ280005 | AY585539 | AM419750 | HE984366 |
| A. japonicus | CBS 114.51 T | AJ279985 | HE577804 | FN594551 | N/A |
| A. labruscus | CCT 7800 T | KU708544 | KT986014 | KT986008 | N/A |
| A. lacticoffeatus | CBS 101883 T | FJ629336 | AY819998 | EU163270 | HE984367 |
| A. luchuensis | CBS 205.80 T | JX500081 | JX500062 | JX500071 | LC179910 |
| A. niger | CBS 554 65 T | EF661186 | EF661089 | EF661154 | EF661058 |
| A. niger | IHEM 2312 | MH613218 | MH614521 | MH645010 | OP082127 |
| A. niger | IHEM 5296 | MH613217 | MH614519 | MH645009 | OP082167 |
| A. niger | MFLUCC 23-0192 | OR502379 | OR594234 | OR502379 | OR604630 |
| A. niger | MFLUCC 23-0193 | OR501408 | OR594235 | OR501408 | OR604631 |
| A. niger | MFLUCC 23-0194 | OR501405 | OR594237 | OR501405 | OR604634 |
| A. niger | MFLUCC 23-0195 | OR500483 | OR594236 | OR500483 | OR604632 |
| A. niger | MFLUCC 23-0200 | OR501402 | OR573928 | OR501402 | OR604633 |
| A. neoniger | CBS 115656 T | FJ491682 | FJ491691 | FJ491700 | KC796429 |
| A. piperis | CBS 112811 T | EU821316 | FJ629303 | EU163267 | KC796427 |
| A. piperis | CMV011A9 | N/A | MK451187 | MK451493 | MK450798 |
| A. pseudopiperis | SDBR-CMUI7 T | MK457204 | MK457206 | MK457205 | MK457208 |
| A. pseudopiperis | SDBR-CMUI1 T | MW588212 | MW602913 | MW602912 | MW602914 |
| A. pseudotubingensis | SDBR CMUO2 T | MK457204 | MK457206 | MK457205 | MK457208 |
| A. pseudotubingensis | SDBR CMU20 T | MW588212 | MW602913 | MW602912 | MW602914 |
| A. sclerotiicarbonarius | CBS 121057 T | EU159216 | EU159229 | EU159235 | N/A |
| A. saccharolyticus | CBS 127449 T | HM853552 | HM853553 | HM853554 | HF559235 |
| A. sclerotioniger | CBS 115572 T | DQ900606 | AY819996 | FN594557 | HE984369 |
| A. trinidadensis | NRRL 62479 T | N/A | HE984420 | HE984434 | HE984379 |
| A. tubingensis | PW3161 | AB987902 | LC000547 | LC000560 | LC000573 |
| A. tubingensis | NRRL 4875 T | EF661193 | EF661086 | EF661151 | EF661055 |
| A. uvarum | CBS 121591 T | AM745757 | AM745751 | AM745755 | HE984370 |
| A. uvarum | ITEM 14819 | N/A | HE984421 | HE984435 | HE984380 |
| A. vadensis | CBS 113365 T | AY585549 | AY585531 | FN594560 | HE984371 |
| A. violaceofuscus | CBS 102.03 T | FJ491677 | FJ491686 | FJ491697 | HF559234 |
| A. welwitschiae | CBS 139.54 T | MH857271 | FJ629291 | KC480196 | MN969100 |
| No. | Isolate No. | Taxa | Herbal Tea/Market |
|---|---|---|---|
| 1 | MFLUCC 23-0094 | A. chevalieri | Jasmine/Doi Mae Salong |
| 2 | MFLUCC 23-0095 | A. chevalieri | Jasmine/Doi Mae Salong |
| 3 | MFLUCC 23-0096 | A. chevalieri | Jasmine/Doi Mae Salong |
| 4 | MFLUCC 23-0097 | A. chevalieri | Rose/Doi Mae Salong |
| 5 | MFLUCC 23-0184 | A. chevalieri | Bael fruit/Doi Mae Salong |
| 6 | MFLUCC 23-0185 | A. chevalieri | Black tea/Doi Mae Salong |
| 7 | MFLUCC 23-0192 | A. niger | Goji berry/Doi Mae Salong |
| 8 | MFLUCC 23-0193 | A. niger | Bael fruit/Mae Sai |
| 9 | MFLUCC 23-0194 | A. niger | Lavender/Mae Sai |
| 10 | MFLUCC 23-0195 | A. niger | Green tea/Fah Thai |
| 11 | MFLUCC 23-0200 | A. niger | Green tea/Fah Thai |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noorabadi, M.T.; Gomes de Farias, A.R.; Mapook, A.; Hyde, K.D.; Boonmee, S. Occurrence of Aspergillus chevalieri and A. niger on Herbal Tea and Their Potential to Produce Ochratoxin A (OTA). Diversity 2023, 15, 1183. https://doi.org/10.3390/d15121183
Noorabadi MT, Gomes de Farias AR, Mapook A, Hyde KD, Boonmee S. Occurrence of Aspergillus chevalieri and A. niger on Herbal Tea and Their Potential to Produce Ochratoxin A (OTA). Diversity. 2023; 15(12):1183. https://doi.org/10.3390/d15121183
Chicago/Turabian StyleNoorabadi, Maryam T., Antonio Roberto Gomes de Farias, Ausana Mapook, Kevin D. Hyde, and Saranyaphat Boonmee. 2023. "Occurrence of Aspergillus chevalieri and A. niger on Herbal Tea and Their Potential to Produce Ochratoxin A (OTA)" Diversity 15, no. 12: 1183. https://doi.org/10.3390/d15121183
APA StyleNoorabadi, M. T., Gomes de Farias, A. R., Mapook, A., Hyde, K. D., & Boonmee, S. (2023). Occurrence of Aspergillus chevalieri and A. niger on Herbal Tea and Their Potential to Produce Ochratoxin A (OTA). Diversity, 15(12), 1183. https://doi.org/10.3390/d15121183






