Aldehyde Dehydrogenase Diversity in Azospirillum Genomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Selection
2.2. Aldehyde Dehydrogenases Sequence Searching
2.3. Comparative Genome Analyses
2.4. Multiple Sequence Alignment and Phylogenetic Analyses
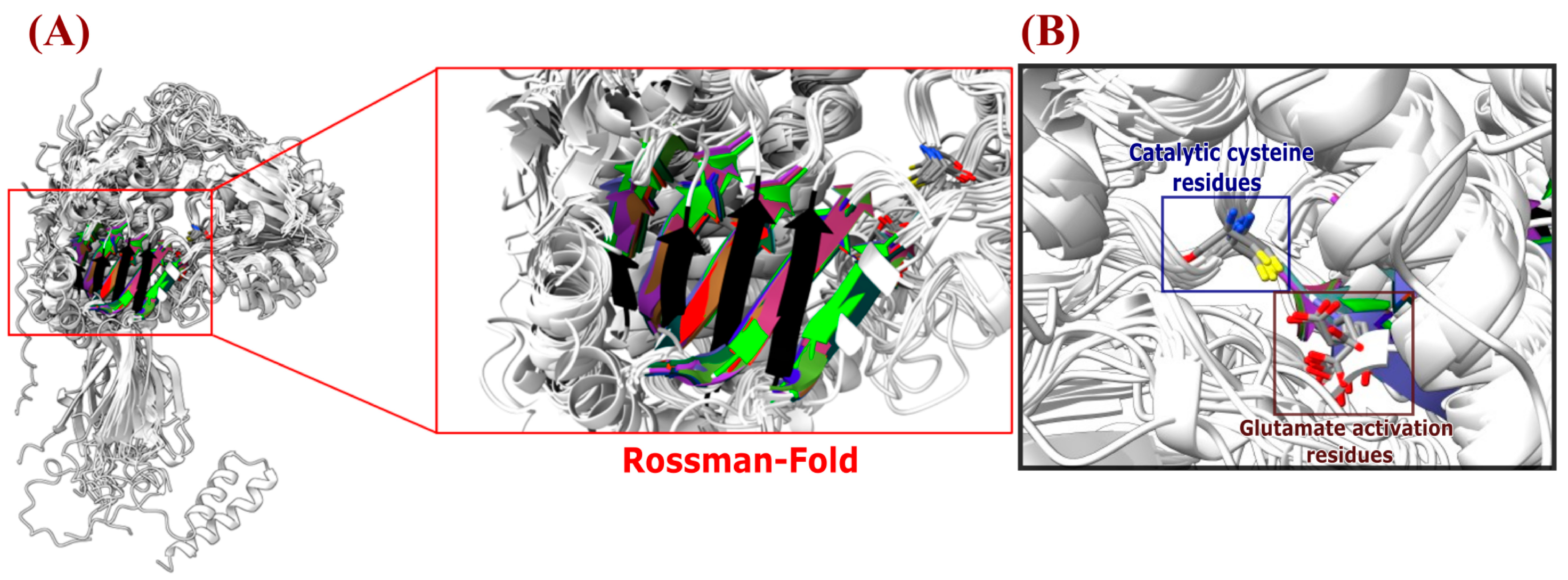
2.5. Protein Modelling, Molecular Conservation, and Structural Analysis
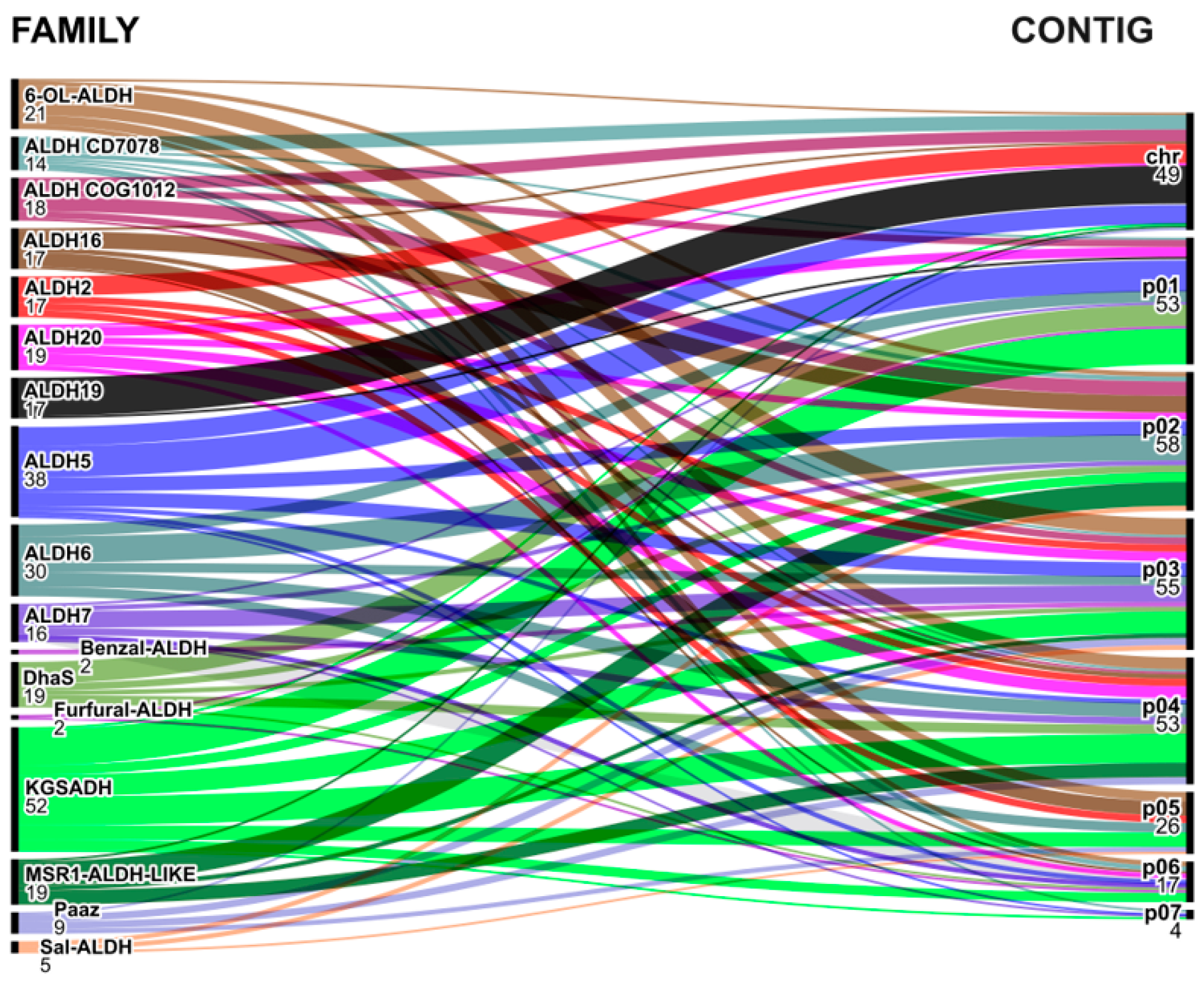
2.6. Aldehyde Dehydrogenase Gene Localization in Azospirillum Genomes
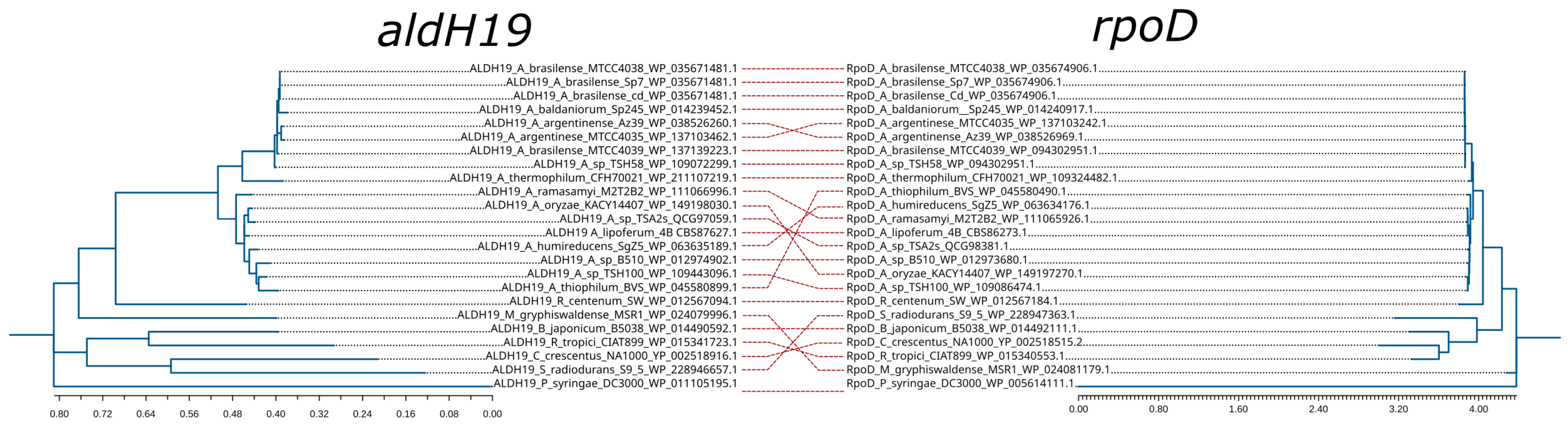
2.7. Aldehyde Dehydrogenase Gene Identification as Potential Phylogenetic Markers
3. Results
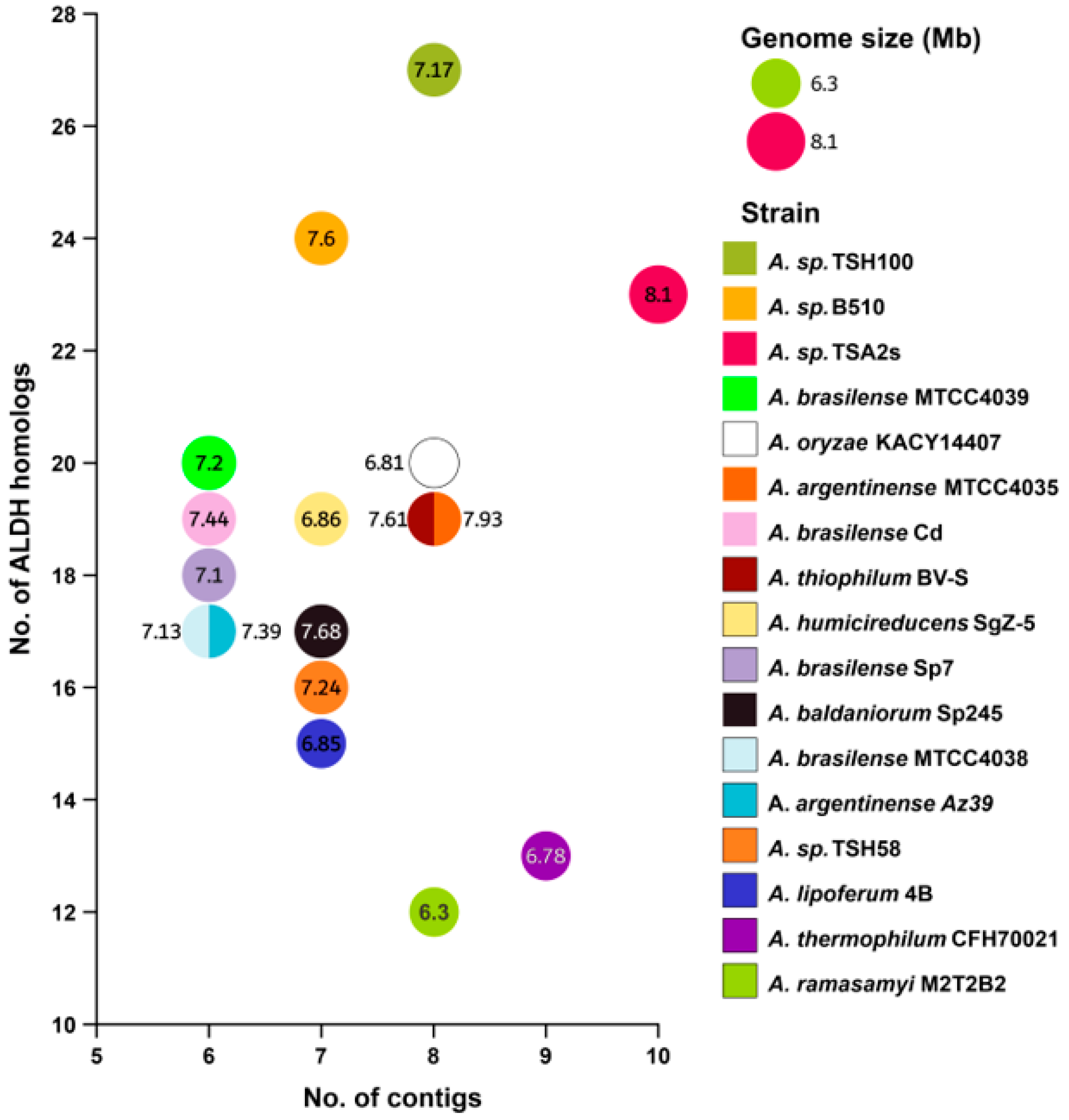
3.1. Selection of Azospirillum Genomes
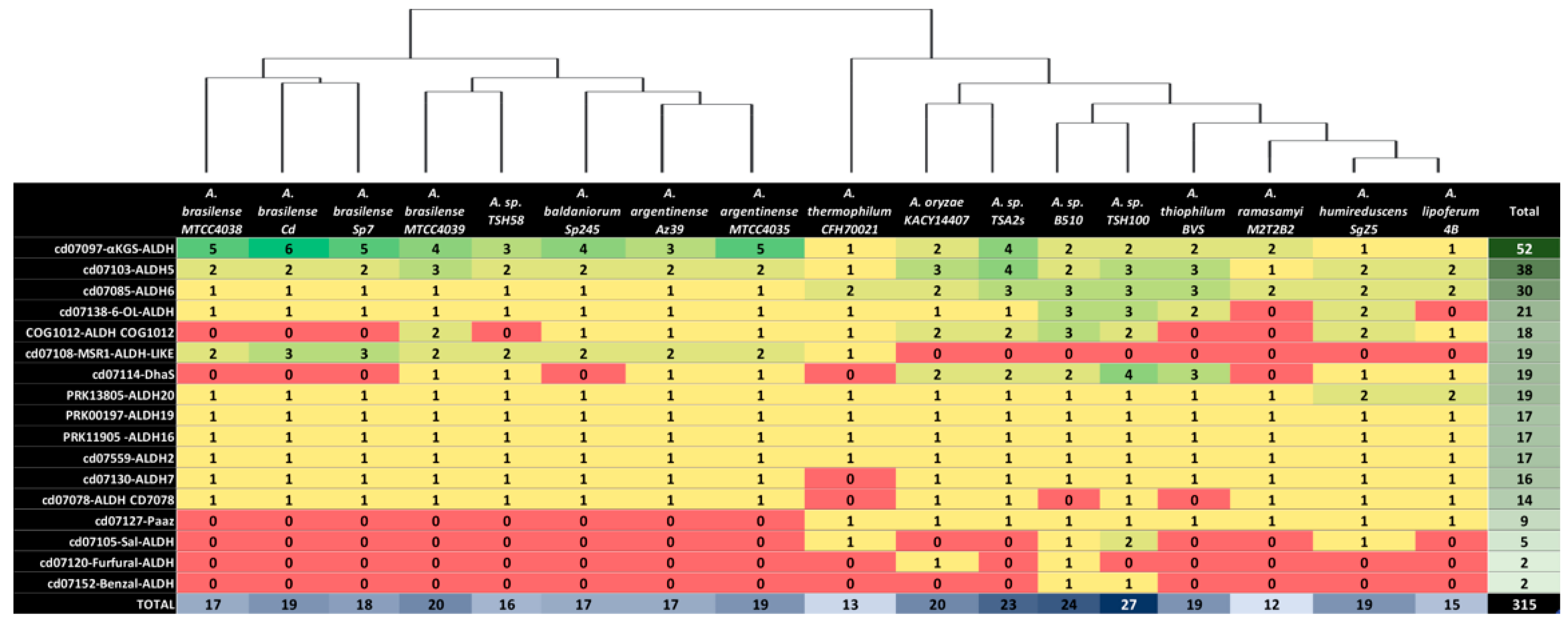
3.2. Number of Aldehyde Dehydrogenases Identified for Each Azospirillum Strain
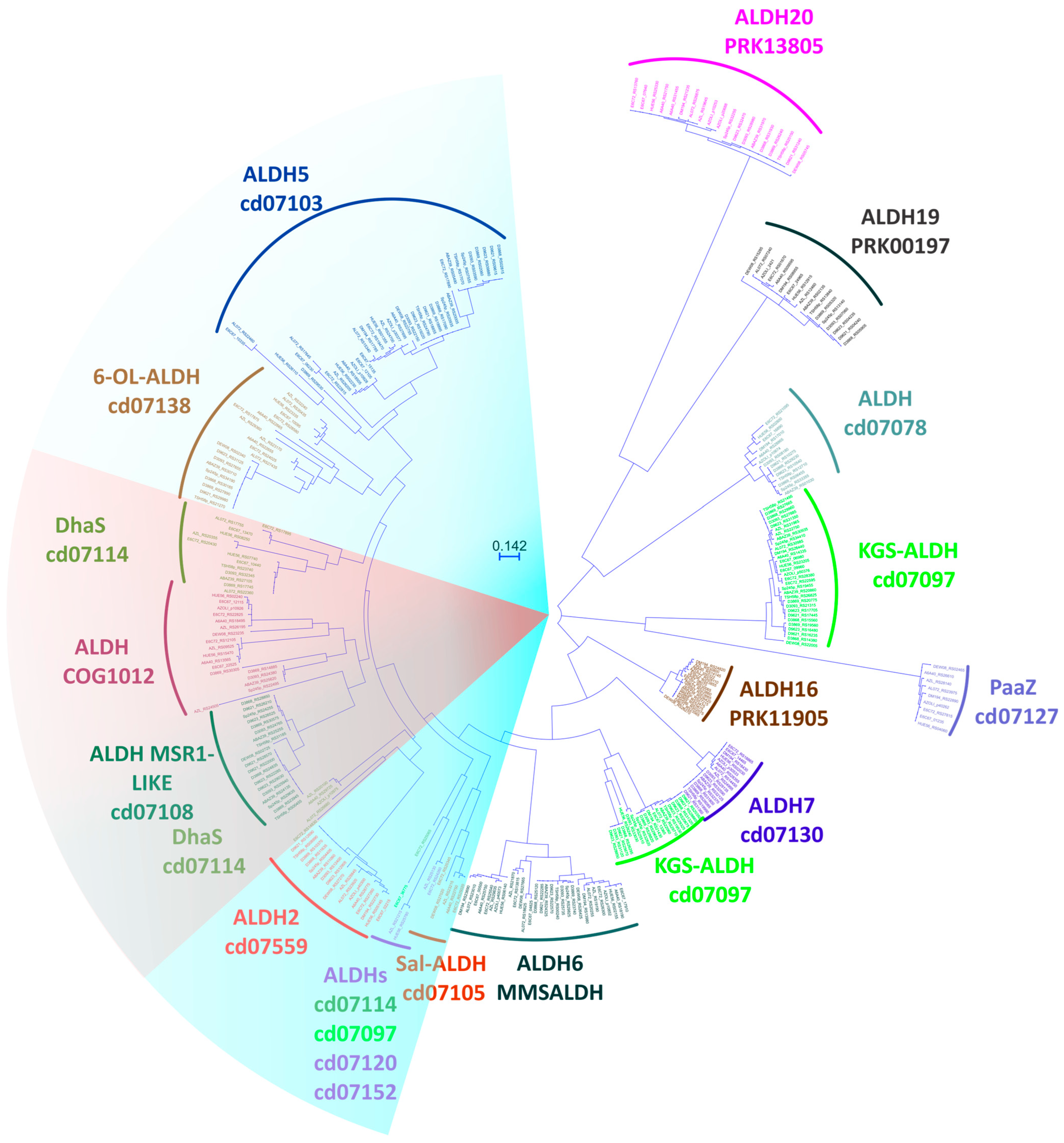
3.3. Sequence Alignment and Clustering of Aldehyde Dehydrogenases
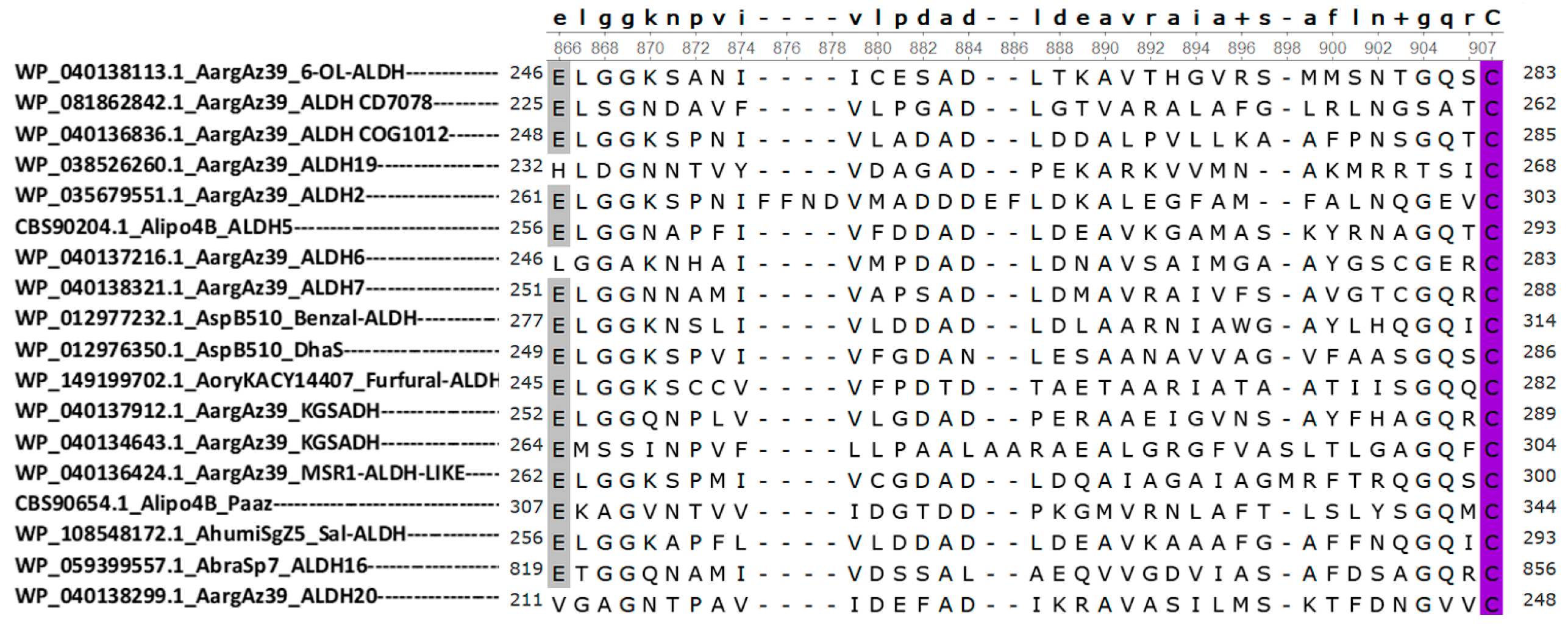
3.4. Alignment and Phylogenetic Analysis
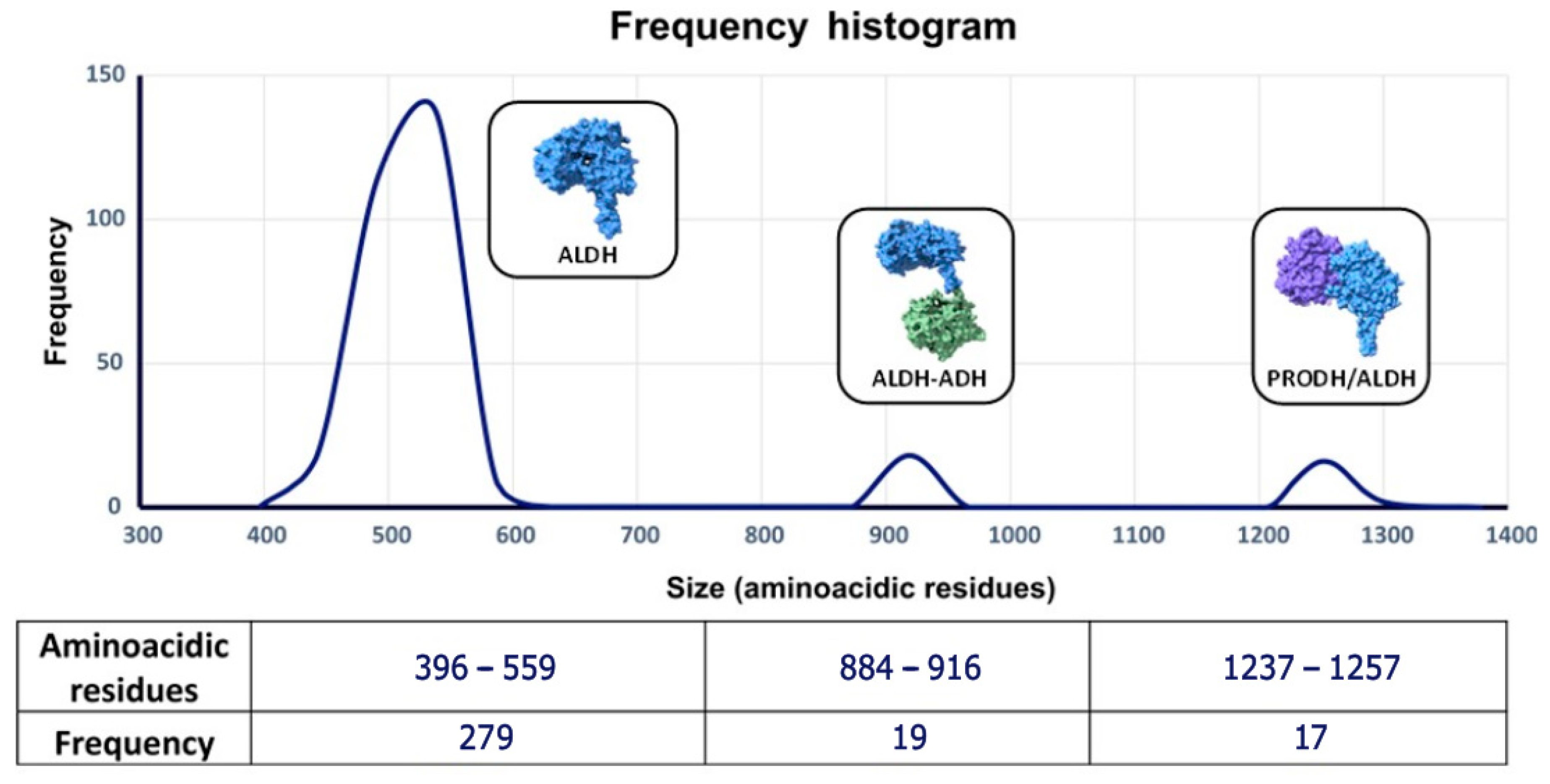
3.5. Comparison of Structural Models of Aldehyde Dehydrogenase Families
3.6. Analyzing Aldehyde Dehydrogenase Families Found in Azospirillum Genomes
- ALDH5
- 6-OL-ALDH
- MSR1, DhaS, and COG1012 group
- MSR1-ALDH-LIKE
- DhaS (cd7114)
- ALDH COG1012
- KGSADH
- ALDH6
- ALDH20
- ALDH2
- ALDH16
- ALDH19
- ALDH7
- ALDH CD7078
- Paaz
- ALDH cd07105
- ALDH cd07120
- ALDH cd07152
3.7. Aldehyde Dehydrogenases as Phylogenetic Marker
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kuykendall, J.R.; Kuykendall, N.S. 15.19—Aldehydes. In Comprehensive Toxicology, 3rd ed.; McQueen, C.A., Ed.; Elsevier: Oxford, UK, 2018; pp. 352–388. ISBN 978-0-08-100601-6. [Google Scholar]
- Deza-Ponzio, R.; Herrera, M.L.; Bellini, M.J.; Virgolini, M.B.; Hereñú, C.B. Aldehyde Dehydrogenase 2 in the Spotlight: The Link between Mitochondria and Neurodegeneration. Neurotoxicology 2018, 68, 19–24. [Google Scholar] [CrossRef]
- Muzio, G.; Maggiora, M.; Paiuzzi, E.; Oraldi, M.; Canuto, R.A. Aldehyde Dehydrogenases and Cell Proliferation. Free. Radic. Biol. Med. 2012, 52, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Shortall, K.; Djeghader, A.; Magner, E.; Soulimane, T. Insights into Aldehyde Dehydrogenase Enzymes: A Structural Perspective. Front. Mol. Biosci. 2021, 8, 659550. [Google Scholar] [CrossRef]
- Nosova, T.; Jokelainen, K.; Kaihovaara, P.; Jousimies-Somer, H.; Siitonen, A.; Heine, R.; Salaspuro, M. Aldehyde Dehydrogenase Activity and Acetate Production by Aerobic Bacteria Representing the Normal Flora of Human Large Intestine. Alcohol Alcohol 1996, 31, 555–564. [Google Scholar] [CrossRef]
- Tola, A.J.; Jaballi, A.; Germain, H.; Missihoun, T.D. Recent Development on Plant Aldehyde Dehydrogenase Enzymes and Their Functions in Plant Development and Stress Signaling. Genes 2021, 12, 51. [Google Scholar] [CrossRef]
- Vasiliou, V.; Nebert, D.W. Analysis and Update of the Human Aldehyde Dehydrogenase (ALDH) Gene Family. Hum Genom. 2005, 2, 138–143. [Google Scholar] [CrossRef]
- Riveros-Rosas, H.; Julián-Sánchez, A.; Moreno-Hagelsieb, G.; Muñoz-Clares, R.A. Aldehyde Dehydrogenase Diversity in Bacteria of the Pseudomonas Genus. Chem.-Biol. Interact. 2019, 304, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-H.; Chen, F.-F.; Wang, C.-E.; Fu, H.-H.; Fang, X.-Q.; Ye, J.-R.; Shi, J.-Y. Indole-3-Acetic Acid in Burkholderia Pyrrocinia JK-SH007: Enzymatic Identification of the Indole-3-Acetamide Synthesis Pathway. Front. Microbiol. 2019, 10, 2559. [Google Scholar] [CrossRef]
- Kim, Y.-G.; Lee, S.; Kwon, O.-S.; Park, S.-Y.; Lee, S.-J.; Park, B.-J.; Kim, K.-J. Redox-Switch Modulation of Human SSADH by Dynamic Catalytic Loop. EMBO J. 2009, 28, 959–968. [Google Scholar] [CrossRef] [PubMed]
- Ferrández, A.; García, J.L.; Díaz, E. Transcriptional Regulation of the Divergent paaCatabolic Operons for Phenylacetic Acid Degradation in Escherichia coli. J. Biol. Chem. 2000, 275, 12214–12222. [Google Scholar] [CrossRef]
- Koppaka, V.; Thompson, D.C.; Chen, Y.; Ellermann, M.; Nicolaou, K.C.; Juvonen, R.O.; Petersen, D.; Deitrich, R.A.; Hurley, T.D.; Vasiliou, V. Aldehyde Dehydrogenase Inhibitors: A Comprehensive Review of the Pharmacology, Mechanism of Action, Substrate Specificity, and Clinical Application. Pharmacol. Rev. 2012, 64, 520–539. [Google Scholar] [CrossRef] [PubMed]
- Hempel, J.; Nicholas, H.; Lindahl, R. Aldehyde Dehydrogenases: Widespread Structural and Functional Diversity within a Shared Framework. Protein Sci. 1993, 2, 1890–1900. [Google Scholar] [CrossRef]
- Brocker, C.; Vasiliou, M.; Carpenter, S.; Carpenter, C.; Zhang, Y.; Wang, X.; Kotchoni, S.O.; Wood, A.J.; Kirch, H.-H.; Kopečný, D.; et al. Aldehyde Dehydrogenase (ALDH) Superfamily in Plants: Gene Nomenclature and Comparative Genomics. Planta 2013, 237, 189–210. [Google Scholar] [CrossRef] [PubMed]
- Nebert, D.W.; Wain, H.M. Update on Human Genome Completion and Annotations: Gene Nomenclature. Hum. Genom. 2003, 1, 66–71. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wisniewski-Dyé, F.; Borziak, K.; Khalsa-Moyers, G.; Alexandre, G.; Sukharnikov, L.O.; Wuichet, K.; Hurst, G.B.; McDonald, W.H.; Robertson, J.S.; Barbe, V.; et al. Azospirillum Genomes Reveal Transition of Bacteria from Aquatic to Terrestrial Environments. PLoS Genet. 2011, 7, e1002430. [Google Scholar] [CrossRef]
- Pedraza, R.O.; Filippone, M.P.; Fontana, C.; Salazar, S.M.; Ramírez-Mata, A.; Sierra-Cacho, D.; Baca, B.E. Chapter 6—Azospirillum. In Beneficial Microbes in Agro-Ecology; Amaresan, N., Senthil Kumar, M., Annapurna, K., Kumar, K., Sankaranarayanan, A., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 73–105. ISBN 978-0-12-823414-3. [Google Scholar]
- Reis, V.M.; Baldani, V.L.D.; Baldani, J.I. Isolation, Identification and Biochemical Characterization of Azospirillum Spp. and Other Nitrogen-Fixing Bacteria. In Handbook for Azospirillum: Technical Issues and Protocols; Cassán, F.D., Okon, Y., Creus, C.M., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 3–26. ISBN 978-3-319-06542-7. [Google Scholar]
- Cassán, F.; López, G.; Nievas, S.; Coniglio, A.; Torres, D.; Donadio, F.; Molina, R.; Mora, V. What Do We Know About the Publications Related with Azospirillum? A Metadata Analysis. Microb. Ecol. 2021, 81, 278–281. [Google Scholar] [CrossRef]
- Datasets—NCBI—NIH. Available online: https://www.ncbi.nlm.nih.gov/datasets/genomes/?taxon=191&utm_source=data-hub (accessed on 3 February 2022).
- McGinnis, S.; Madden, T.L. BLAST: At the Core of a Powerful and Diverse Set of Sequence Analysis Tools. Nucleic Acids Res 2004, 32, W20–W25. [Google Scholar] [CrossRef]
- Singh, V.S.; Dubey, B.K.; Pandey, P.; Rai, S.; Tripathi, A.K. Co-Metabolism of Ethanol in Azospirillum brasilense Sp7 Is Mediated by Fructose and Glycerol and Regulated Negatively by an Alternative Sigma Factor RpoH2. J. Bacteriol. 2021, 203, JB0026921. [Google Scholar] [CrossRef]
- Kerfeld, C.A.; Scott, K.M. Using BLAST to Teach “E-Value-Tionary” Concepts. PLoS Biol. 2011, 9, e1001014. [Google Scholar] [CrossRef]
- Eren, A.M.; Esen, Ö.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An Advanced Analysis and Visualization Platform for ’omics Data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s Conserved Domain Database. Nucleic Acids Res. 2015, 43, D222-226. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the Functional Annotation of Proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. jModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- UniProt: The Universal Protein Knowledgebase—PMC. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5210571/ (accessed on 30 August 2023).
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Mauri, M.; Elli, T.; Caviglia, G.; Uboldi, G.; Azzi, M. RAWGraphs: A Visualisation Platform to Create Open Outputs. In Proceedings of the Proceedings of the 12th Biannual Conference on Italian SIGCHI Chapter, Cagliari, Italy, 18–20 September 2017; Association for Computing Machinery: New York, NY, USA, 2017; pp. 1–5. [Google Scholar]
- Maroniche, G.A.; García, J.E.; Salcedo, F.; Creus, C.M. Molecular Identification of Azospirillum Spp.: Limitations of 16S rRNA and Qualities of rpoD as Genetic Markers. Microbiol. Res. 2017, 195, 1–10. [Google Scholar] [CrossRef]
- Tripathi, A.K.; Singh, C. Azospirillum brasilense Genome Assembly ASM131501v1. Available online: https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_001315015.1/ (accessed on 21 October 2022).
- Singh, C.; Pandey, P.; Singh, D.N.; Pandey, R.; Shasany, A.K.; Tripathi, A.K. Whole-Genome Sequences of Four Indian Isolates of Azospirillum brasilense. Microbiol. Resour. Announc. 2019, 8, e00633-19. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.; Sakai, Y.; Senoo, K.; Ishii, S. Potentially Mobile Denitrification Genes Identified in Azospirillum Sp. Strain TSH58. Appl. Environ. Microbiol. 2019, 85, e02474-18. [Google Scholar] [CrossRef] [PubMed]
- Rivera, D.; Revale, S.; Molina, R.; Gualpa, J.; Puente, M.; Maroniche, G.; Paris, G.; Baker, D.; Clavijo, B.; McLay, K.; et al. Complete Genome Sequence of the Model Rhizosphere Strain Azospirillum brasilense Az39, Successfully Applied in Agriculture. Genome Announc. 2014, 2, e00683-14. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.K.; Singh, C. ASM827496v1—Genome—Assembly—NCBI. Available online: https://www.ncbi.nlm.nih.gov/assembly/GCF_008274965.1/?shouldredirect=false (accessed on 21 July 2022).
- Zhou, S.; Han, L.; Wang, Y.; Yang, G.; Zhuang, L.; Hu, P. Azospirillum humicireducens Sp. Nov., a Nitrogen-Fixing Bacterium Isolated from a Microbial Fuel Cell. Int. J. Syst. Evol. Microbiol. 2013, 63, 2618–2624. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Park, Y.-J.; Shin, J.-H. ASM1334728v1—Genome—Assembly—NCBI. Available online: https://www.ncbi.nlm.nih.gov/assembly/GCF_013347285.1/ (accessed on 21 November 2023).
- Anandham, R.; Heo, J.; Krishnamoorthy, R.; SenthilKumar, M.; Gopal, N.O.; Kim, S.-J.; Kwon, S.-W. Azospirillum ramasamyi Sp. Nov., a Novel Diazotrophic Bacterium Isolated from Fermented Bovine Products. Int. J. Syst. Evol. Microbiol. 2019, 69, 1369–1375. [Google Scholar] [CrossRef]
- Kaneko, T.; Minamisawa, K.; Isawa, T.; Nakatsukasa, H.; Mitsui, H.; Kawaharada, Y.; Nakamura, Y.; Watanabe, A.; Kawashima, K.; Ono, A.; et al. Complete Genomic Structure of the Cultivated Rice Endophyte Azospirillum sp B510. DNA Res. 2010, 17, 37–50. [Google Scholar] [CrossRef]
- Gao, N.; Shen, W.; Nishizawa, T.; Isobe, K.; Guo, Y.; Ying, H.; Senoo, K. Genome Sequences of Two Azospirillum sp. Strains, TSA2S and TSH100, Plant Growth-Promoting Rhizobacteria with N2O Mitigation Abilities. Microbiol. Resour. Announc. 2019, 8, e00459-19. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Ming, H.; Ding, C.-L.; Ji, W.-L.; Cheng, L.-J.; Niu, M.; Zhang, Y.; Zhang, L.-Y.; Meng, X.-L.; Nie, G.-X. Azospirillum thermophilum sp. Nov., Isolated from a Hot Spring. Int. J. Syst. Evol. Microbiol. 2020, 70, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Fomenkov, A.; Vincze, T.; Grabovich, M.; Anton, B.P.; Dubinina, G.; Orlova, M.; Belousova, E.; Roberts, R.J. Complete Genome Sequence of a Strain of Azospirillum thiophilum Isolated from a Sulfide Spring. Genome Announc. 2016, 4, e01521-15. [Google Scholar] [CrossRef]
- Pony, P.; Rapisarda, C.; Terradot, L.; Marza, E.; Fronzes, R. Filamentation of the Bacterial Bi-Functional Alcohol/Aldehyde Dehydrogenase AdhE Is Essential for Substrate Channeling and Enzymatic Regulation. Nat. Commun. 2020, 11, 1426. [Google Scholar] [CrossRef]
- Tretter, L.; Adam-Vizi, V. Alpha-Ketoglutarate Dehydrogenase: A Target and Generator of Oxidative Stress. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 2335–2345. [Google Scholar] [CrossRef]
- Watanabe, S.; Yamada, M.; Ohtsu, I.; Makino, K. α-Ketoglutaric Semialdehyde Dehydrogenase Isozymes Involved in Metabolic Pathways of D-Glucarate, D-Galactarate, and Hydroxy-L-Proline: Molecular and Metabolic Convergent Evolution. J. Biol. Chem. 2007, 282, 6685–6695. [Google Scholar] [CrossRef] [PubMed]
- Malaspina, P.; Picklo, M.J.; Jakobs, C.; Snead, O.C.; Gibson, K.M. Comparative Genomics of Aldehyde Dehydrogenase 5a1 (Succinate Semialdehyde Dehydrogenase) and Accumulation of Gamma-Hydroxybutyrate Associated with Its Deficiency. Hum. Genom. 2009, 3, 106–120. [Google Scholar] [CrossRef]
- Talfournier, F.; Stines-Chaumeil, C.; Branlant, G. Methylmalonate-Semialdehyde Dehydrogenase from Bacillus subtilis: Substrate Specificity and Coenzyme A Binding. J. Biol. Chem. 2011, 286, 21971–21981. [Google Scholar] [CrossRef]
- Steele, M.I.; Lorenz, D.; Hatter, K.; Park, A.; Sokatch, J.R. Characterization of the mmsAB Operon of Pseudomonas aeruginosa PAO Encoding Methylmalonate-Semialdehyde Dehydrogenase and 3-Hydroxyisobutyrate Dehydrogenase. J. Biol. Chem. 1992, 267, 13585–13592. [Google Scholar] [CrossRef] [PubMed]
- Kostichka, K.; Thomas, S.M.; Gibson, K.J.; Nagarajan, V.; Cheng, Q. Cloning and Characterization of a Gene Cluster for Cyclododecanone Oxidation in Rhodococcus ruber SC1. J. Bacteriol. 2001, 183, 6478–6486. [Google Scholar] [CrossRef]
- Membrillo-Hernández, J.; Echave, P.; Cabiscol, E.; Tamarit, J.; Ros, J.; Lin, E.C.C. Evolution of the adhE Gene Product of Escherichia coli from a Functional Reductase to a Dehydrogenase: GENETIC AND BIOCHEMICAL STUDIES OF THE MUTANT PROTEINS. J. Biol. Chem. 2000, 275, 33869–33875. [Google Scholar] [CrossRef]
- Richter, M.; Kube, M.; Bazylinski, D.A.; Lombardot, T.; Glöckner, F.O.; Reinhardt, R.; Schüler, D. Comparative Genome Analysis of Four Magnetotactic Bacteria Reveals a Complex Set of Group-Specific Genes Implicated in Magnetosome Biomineralization and Function. J. Bacteriol. 2007, 189, 4899–4910. [Google Scholar] [CrossRef]
- McClerklin, S.A.; Lee, S.G.; Harper, C.P.; Nwumeh, R.; Jez, J.M.; Kunkel, B.N. Indole-3-Acetaldehyde Dehydrogenase-Dependent Auxin Synthesis Contributes to Virulence of Pseudomonas syringae Strain DC3000. PLoS Pathog. 2018, 14, e1006811. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Li, S.; Zhang, N.; Cui, X.; Zhou, X.; Zhang, G.; Shen, Q.; Zhang, R. Analysis and Cloning of the Synthetic Pathway of the Phytohormone Indole-3-Acetic Acid in the Plant-Beneficial Bacillus amyloliquefaciens SQR9. Microb. Cell Fact. 2015, 14, 130. [Google Scholar] [CrossRef]
- Xie, B.; Xu, K.; Zhao, H.X.; Chen, S.F. Isolation of Transposon Mutants from Azospirillum brasilense Yu62 and Characterization of Genes Involved in Indole-3-Acetic Acid Biosynthesis. FEMS Microbiol. Lett. 2005, 248, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Malinich, E.A.; Bauer, C.E. Transcriptome Analysis of Azospirillum brasilense Vegetative and Cyst States Reveals Large-Scale Alterations in Metabolic and Replicative Gene Expression. Microb. Genom. 2018, 4, e000200. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; White, C.E.; diCenzo, G.C.; Zhang, Y.; Stogios, P.J.; Savchenko, A.; Finan, T.M. L-Hydroxyproline and d-Proline Catabolism in Sinorhizobium meliloti. J. Bacteriol. 2016, 198, 1171–1181. [Google Scholar] [CrossRef]
- Korasick, D.A.; Končitíková, R.; Kopečná, M.; Hájková, E.; Vigouroux, A.; Moréra, S.; Becker, D.F.; Šebela, M.; Tanner, J.J.; Kopečný, D. Structural and Biochemical Characterization of Aldehyde Dehydrogenase 12, the Last Enzyme of Proline Catabolism in Plants. J. Mol. Biol. 2019, 431, 576–592. [Google Scholar] [CrossRef]
- Vasiliou, V.; Bairoch, A.; Tipton, K.F.; Nebert, D.W. Eukaryotic Aldehyde Dehydrogenase (ALDH) Genes: Human Polymorphisms, and Recommended Nomenclature Based on Divergent Evolution and Chromosomal Mapping. Pharmacogenetics 1999, 9, 421–434. [Google Scholar]
- Končitíková, R.; Vigouroux, A.; Kopečná, M.; Andree, T.; Bartoš, J.; Šebela, M.; Moréra, S.; Kopečný, D. Role and Structural Characterization of Plant Aldehyde Dehydrogenases from Family 2 and Family 7. Biochem. J. 2015, 468, 109–123. [Google Scholar] [CrossRef]
- Sathyanarayanan, N.; Cannone, G.; Gakhar, L.; Katagihallimath, N.; Sowdhamini, R.; Ramaswamy, S.; Vinothkumar, K.R. Molecular Basis for Metabolite Channeling in a Ring Opening Enzyme of the Phenylacetate Degradation Pathway. Nat. Commun. 2019, 10, 4127. [Google Scholar] [CrossRef] [PubMed]
- Denome, S.A.; Stanley, D.C.; Olson, E.S.; Young, K.D. Metabolism of Dibenzothiophene and Naphthalene in Pseudomonas Strains: Complete DNA Sequence of an Upper Naphthalene Catabolic Pathway. J. Bacteriol. 1993, 175, 6890–6901. [Google Scholar] [CrossRef] [PubMed]
- Nichols, N.N.; Mertens, J.A. Identification and Transcriptional Profiling of Pseudomonas putida Genes Involved in Furoic Acid Metabolism. FEMS Microbiol. Lett. 2008, 284, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Gillooly, D.J.; Robertson, A.G.; Fewson, C.A. Molecular Characterization of Benzyl Alcohol Dehydrogenase and Benzaldehyde Dehydrogenase II of Acinetobacter calcoaceticus. Biochem. J. 1998, 330 Pt 3, 1375–1381. [Google Scholar] [CrossRef]
- Krishna, R.V.; Beilstein, P.; Leisinger, T. Biosynthesis of Proline in Pseudomonas aeruginosa. Properties of Gamma-Glutamyl Phosphate Reductase and 1-Pyrroline-5-Carboxylate Reductase. Biochem. J. 1979, 181, 223–230. [Google Scholar] [CrossRef]
- Cherney, L.T.; Cherney, M.M.; Garen, C.R.; Niu, C.; Moradian, F.; James, M.N.G. Crystal Structure of N-Acetyl-Gamma-Glutamyl-Phosphate Reductase from Mycobacterium tuberculosis in Complex with NADP(+). J. Mol. Biol. 2007, 367, 1357–1369. [Google Scholar] [CrossRef]
- Brocker, C.; Lassen, N.; Estey, T.; Pappa, A.; Cantore, M.; Orlova, V.V.; Chavakis, T.; Kavanagh, K.L.; Oppermann, U.; Vasiliou, V. Aldehyde Dehydrogenase 7A1 (ALDH7A1) Is a Novel Enzyme Involved in Cellular Defense against Hyperosmotic Stress. J. Biol. Chem. 2010, 285, 18452–18463. [Google Scholar] [CrossRef]
- Liu, L.-K.; Becker, D.F.; Tanner, J.J. Structure, Function, and Mechanism of Proline Utilization A (PutA). Arch. Biochem. Biophys. 2017, 632, 142–157. [Google Scholar] [CrossRef]
- Hou, Q.; Bartels, D. Comparative Study of the Aldehyde Dehydrogenase (ALDH) Gene Superfamily in the Glycophyte Arabidopsis thaliana and Eutrema halophytes. Ann. Bot. 2015, 115, 465–479. [Google Scholar] [CrossRef] [PubMed]
- van Lis, R.; Popek, M.; Couté, Y.; Kosta, A.; Drapier, D.; Nitschke, W.; Atteia, A. Concerted Up-Regulation of Aldehyde/Alcohol Dehydrogenase (ADHE) and Starch in Chlamydomonas reinhardtii Increases Survival under Dark Anoxia. J. Biol. Chem. 2017, 292, 2395–2410. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Yang, J.; Jang, J.; Choi, J.-S.; Roe, A.J.; Byron, O.; Seok, C.; Song, J.-J. Aldehyde-Alcohol Dehydrogenase Undergoes Structural Transition to Form Extended Spirosomes for Substrate Channeling. Commun. Biol. 2020, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Page, R.; Nelson, M.S.; von Delft, F.; Elsliger, M.-A.; Canaves, J.M.; Brinen, L.S.; Dai, X.; Deacon, A.M.; Floyd, R.; Godzik, A.; et al. Crystal Structure of Gamma-Glutamyl Phosphate Reductase (TM0293) from Thermotoga maritima at 2.0 A Resolution. Proteins 2004, 54, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Baugh, L.; Gallagher, L.A.; Patrapuvich, R.; Clifton, M.C.; Gardberg, A.S.; Edwards, T.E.; Armour, B.; Begley, D.W.; Dieterich, S.H.; Dranow, D.M.; et al. Combining Functional and Structural Genomics to Sample the Essential Burkholderia structome. PLoS ONE 2013, 8, e53851. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh Behbahani, B.; Jooyandeh, H.; Falah, F.; Vasiee, A. Gamma-aminobutyric Acid Production by Lactobacillus brevis A3: Optimization of Production, Antioxidant Potential, Cell Toxicity, and Antimicrobial Activity. Food Sci. Nutr. 2020, 8, 5330–5339. [Google Scholar] [CrossRef]
- Chen, C.-H.; Joshi, A.U.; Mochly-Rosen, D. The Role of Mitochondrial Aldehyde Dehydrogenase 2 (ALDH2) in Neuropathology and Neurodegeneration. Acta Neurol. Taiwan 2016, 25, 111–123. [Google Scholar]
- Harrison, P.W.; Lower, R.P.J.; Kim, N.K.D.; Young, J.P.W. Introducing the Bacterial ‘Chromid’: Not a Chromosome, Not a Plasmid. Trends Microbiol. 2010, 18, 141–148. [Google Scholar] [CrossRef]
- Fritz, K.S.; Petersen, D.R. An Overview of the Chemistry and Biology of Reactive Aldehydes. Free Radic. Biol. Med. 2013, 59, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Ghosh, A. Evolution, Family Expansion, and Functional Diversification of Plant Aldehyde Dehydrogenases. Gene 2022, 829, 146522. [Google Scholar] [CrossRef]
- Schultz, J.; Copley, R.R.; Doerks, T.; Ponting, C.P.; Bork, P. SMART: A Web-Based Tool for the Study of Genetically Mobile Domains. Nucleic Acids Res. 2000, 28, 231–234. [Google Scholar] [CrossRef]
- Langendorf, C.G.; Key, T.L.G.; Fenalti, G.; Kan, W.-T.; Buckle, A.M.; Caradoc-Davies, T.; Tuck, K.L.; Law, R.H.P.; Whisstock, J.C. The X-Ray Crystal Structure of Escherichia coli Succinic Semialdehyde Dehydrogenase; Structural Insights into NADP+/Enzyme Interactions. PLoS ONE 2010, 5, e9280. [Google Scholar] [CrossRef]
- Lee, S.G.; Harline, K.; Abar, O.; Akadri, S.O.; Bastian, A.G.; Chen, H.-Y.S.; Duan, M.; Focht, C.M.; Groziak, A.R.; Kao, J.; et al. The Plant Pathogen Enzyme AldC Is a Long-Chain Aliphatic Aldehyde Dehydrogenase. J. Biol. Chem. 2020, 295, 13914–13926. [Google Scholar] [CrossRef]
- Baburam, C.; Feto, N.A. Mining of Two Novel Aldehyde Dehydrogenases (DHY-SC-VUT5 and DHY-G-VUT7) from Metagenome of Hydrocarbon Contaminated Soils. BMC Biotechnol. 2021, 21, 18. [Google Scholar] [CrossRef]
- Cook, S.D.; Nichols, D.S.; Smith, J.; Chourey, P.S.; McAdam, E.L.; Quittenden, L.; Ross, J.J. Auxin Biosynthesis: Are the Indole-3-Acetic Acid and Phenylacetic Acid Biosynthesis Pathways Mirror Images? Plant Physiol. 2016, 171, 1230–1241. [Google Scholar] [CrossRef]
- Cook, S.D. An Historical Review of Phenylacetic Acid. Plant Cell Physiol. 2019, 60, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Baudoin, E.; Lerner, A.; Mirza, M.S.; El Zemrany, H.; Prigent-Combaret, C.; Jurkevich, E.; Spaepen, S.; Vanderleyden, J.; Nazaret, S.; Okon, Y.; et al. Effects of Azospirillum brasilense with Genetically Modified Auxin Biosynthesis Gene ipdC upon the Diversity of the Indigenous Microbiota of the Wheat Rhizosphere. Res. Microbiol. 2010, 161, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Jijón-Moreno, S.; Marcos-Jiménez, C.; Pedraza, R.O.; Ramírez-Mata, A.; de Salamone, I.G.; Fernández-Scavino, A.; Vásquez-Hernández, C.A.; Soto-Urzúa, L.; Baca, B.E. The ipdC, hisC1 and hisC2 Genes Involved in Indole-3-Acetic Production Used as Alternative Phylogenetic Markers in Azospirillum brasilense. Antonie van Leeuwenhoek 2015, 107, 1501–1517. [Google Scholar] [CrossRef]
- Carreño-Lopez, R.; Campos-Reales, N.; Elmerich, C.; Baca, B.E. Physiological Evidence for Differently Regulated Tryptophan-Dependent Pathways for Indole-3-Acetic Acid Synthesis in Azospirillum brasilense. Mol. Gen. Genet. 2000, 264, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, M.; Srivastava, S. Organization of the ipdC Region Regulates IAA Levels in Different Azospirillum brasilense Strains: Molecular and Functional Analysis of ipdC in Strain SM. Environ. Microbiol. 2008, 10, 1365–1373. [Google Scholar] [CrossRef] [PubMed]
- Riveros-Rosas, H.; González-Segura, L.; Julián-Sánchez, A.; Díaz-Sánchez, Á.G.; Muñoz-Clares, R.A. Structural Determinants of Substrate Specificity in Aldehyde Dehydrogenases. Chem.-Biol. Interact. 2013, 202, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Kodaki, T.; Makino, K. A Novel Alpha-Ketoglutaric Semialdehyde Dehydrogenase: Evolutionary Insight into an Alternative Pathway of Bacterial L-Arabinose Metabolism. J. Biol. Chem. 2006, 281, 28876–28888. [Google Scholar] [CrossRef]
- Hosoya, S.; Yamane, K.; Takeuchi, M.; Sato, T. Identification and Characterization of the Bacillus subtilis d-Glucarate/Galactarate Utilization Operon ycbCDEFGHJ. FEMS Microbiol. Lett. 2002, 210, 193–199. [Google Scholar] [CrossRef]
- Dobbelaere, S.; Croonenborghs, A.; Thys, A.; Vande Broek, A.; Vanderleyden, J. Phytostimulatory Effect of Azospirillum brasilense Wild Type and Mutant Strains Altered in IAA Production on Wheat. Plant Soil 1999, 212, 155–164. [Google Scholar] [CrossRef]
- Xie, C.-H.; Yokota, A. Azospirillum Oryzae sp. Nov., a Nitrogen-Fixing Bacterium Isolated from the Roots of the Rice Plant Oryza Sativa. Int. J. Syst. Evol. Microbiol. 2005, 55, 1435–1438. [Google Scholar] [CrossRef]
- Luu, R.A.; Schneider, B.J.; Ho, C.C.; Nesteryuk, V.; Ngwesse, S.E.; Liu, X.; Parales, J.V.; Ditty, J.L.; Parales, R.E. Taxis of Pseudomonas putida F1 toward Phenylacetic Acid Is Mediated by the Energy Taxis Receptor Aer2. Appl. Environ. Microbiol. 2013, 79, 2416–2423. [Google Scholar] [CrossRef]

| Genus | Specie | Strain | Country | Isolated from | Last Annotation Updated | Reference |
|---|---|---|---|---|---|---|
| Azospirillum | brasilense | Sp7 | Brazil | Agricultural field | 2021 | [37] |
| Azospirillum | argentinense | MTCC4035 | India | Agricultural field | 2021 | [38] |
| Azospirillum | brasilense | MTCC4038 | India | Agricultural field | 2021 | [38] |
| Azospirillum | brasilense | MTCC4039 | India | Agricultural field | 2021 | [38] |
| Azospirillum | baldaniorum | Sp245 | Brazil | Wheat root | 2021 | [39] |
| Azospirillum | argentinense | Az39 | Argentina | Wheat rhizosphere | 2012 | [40] |
| Azospirillum | lipoferum | 4B | Brazil | Wheat | 2012 | [40] |
| Azospirillum | brasilense | cd | USA | Grass | 2021 | [41] |
| Azospirillum | humicireducens | SgZ-5 | China | Humus from microbial fuel cell | 2022 | [42] |
| Azospirillum | oryzae | KACC 14407 | Republic of Korea | Soil | 2021 | [43] |
| Azospirillum | ramasamyi | M2T2B2 | Republic of Korea | Cow dung | 2021 | [44] |
| Azospirillum | sp | B510 | Japan | Rice soil | 2021 | [45] |
| Azospirillum | sp | TSA2s | Japan | Rice soil | 2019 | [46] |
| Azospirillum | sp | TSH100 | Japan | Rice soil | 2019 | [46] |
| Azospirillum | sp | TSH 58 | Japan | Rice soil | 2021 | [39] |
| Azospirillum | termophilum | CFH70021 | China | Hot spring | 2021 | [47] |
| Azospirillum | thiophilum | BV-S | Russia | Bacterial mat from sulfide spring | 2021 | [48] |
| # | CDD Cluster | ALDH’s Per-CDD Cluster | Anvi’o COGs | ALDH Family | ALDH Family Putative Function | References |
|---|---|---|---|---|---|---|
| 1 | cd07097 | 52 | 3 | αKGS-ALDH | Alpha ketoglutarate semialdehyde dehydrogenase | [50,51] |
| 2 | cd07103 | 38 | 3 | ALDH5 | Succinate-5-semialdehyde dehydrogenase | [52] |
| 3 | cd07085 | 30 | 1 | ALDH6 | Methyl malonate semialdehyde dehydrogenase | [53,54] |
| 4 | cd07138 | 21 | 2 | 6-OL-ALDH | 6-oxolauric aldehyde dehydrogenase | [55] |
| 5 | PRK13805 | 19 | 1 | ALDH20 | Alcohol dehydrogenase/aldehyde dehydrogenase | [49,56] |
| 6 | cd07108 | 19 | 1 | MSR1-ALDH-LIKE | No function described yet | [57] |
| 7 | cd07114 | 19 | 5 | DhaS | IAC aldehyde dehydrogenase | [58,59,60] |
| 8 | COG1012 | 18 | 6 | ALDH COG1012 | No especific function described yet | -- |
| 9 | cd07559 | 17 | 1 | ALDH2 | AC/IAC aldehyde dehydrogenase | [2,60,61] |
| 10 | PRK11905 | 17 | 1 | ALDH16 | Proline dehydrogenase/pyrroline-5-carboxylate dehydrogenase | [62] |
| 11 | PRK00197 | 17 | 1 | ALDH19 | Gamma-glutamyl phosphate reductase | [63] |
| 12 | cd07130 | 16 | 1 | ALDH7 | α-aminoadipic semialdehyde dehydrogenase | [64,65] |
| 13 | cd07078 | 14 | 1 | ALDH CD7078 | No function described yet | --- |
| 14 | cd07127 | 9 | 1 | Paaz | Aldehyde dehydrogenase part of oxepin alphaproteobacteria system | [66] |
| 15 | cd07105 | 5 | 1 | Sal-ALDH | Salicylaldehyde dehydrogenase | [67] |
| 16 | cd07120 | 2 | 1 | Furfural-ALDH | Aldehyde dehydrogenase participating in Furfural convertion | [68] |
| 17 | cd07152 | 2 | 1 | Benzal-ALDH | Benzaldehyde dehydrogenase | [69] |
| Total | 315 | 31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuatlayotl-Olarte, R.; Xiqui-Vázquez, M.L.; Reyes-Carmona, S.R.; Mancilla-Simbro, C.; Baca, B.E.; Ramírez-Mata, A. Aldehyde Dehydrogenase Diversity in Azospirillum Genomes. Diversity 2023, 15, 1178. https://doi.org/10.3390/d15121178
Cuatlayotl-Olarte R, Xiqui-Vázquez ML, Reyes-Carmona SR, Mancilla-Simbro C, Baca BE, Ramírez-Mata A. Aldehyde Dehydrogenase Diversity in Azospirillum Genomes. Diversity. 2023; 15(12):1178. https://doi.org/10.3390/d15121178
Chicago/Turabian StyleCuatlayotl-Olarte, Ricardo, María Luisa Xiqui-Vázquez, Sandra Raquel Reyes-Carmona, Claudia Mancilla-Simbro, Beatriz Eugenia Baca, and Alberto Ramírez-Mata. 2023. "Aldehyde Dehydrogenase Diversity in Azospirillum Genomes" Diversity 15, no. 12: 1178. https://doi.org/10.3390/d15121178
APA StyleCuatlayotl-Olarte, R., Xiqui-Vázquez, M. L., Reyes-Carmona, S. R., Mancilla-Simbro, C., Baca, B. E., & Ramírez-Mata, A. (2023). Aldehyde Dehydrogenase Diversity in Azospirillum Genomes. Diversity, 15(12), 1178. https://doi.org/10.3390/d15121178






