Limnonema gen. nov. (Aerosakkonemataceae, Cyanobacteria): Two Novel Species from Republic of Korea Characterized by Morphological and Molecular Analyses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Culture Conditions
2.2. Morphological and and Ultrastructure Analyses
2.3. Molecular Methods
2.4. Phylogenetic and Molecular Distance Analyses
3. Results
3.1. Taxonomic Treatment and Morphological Characterization
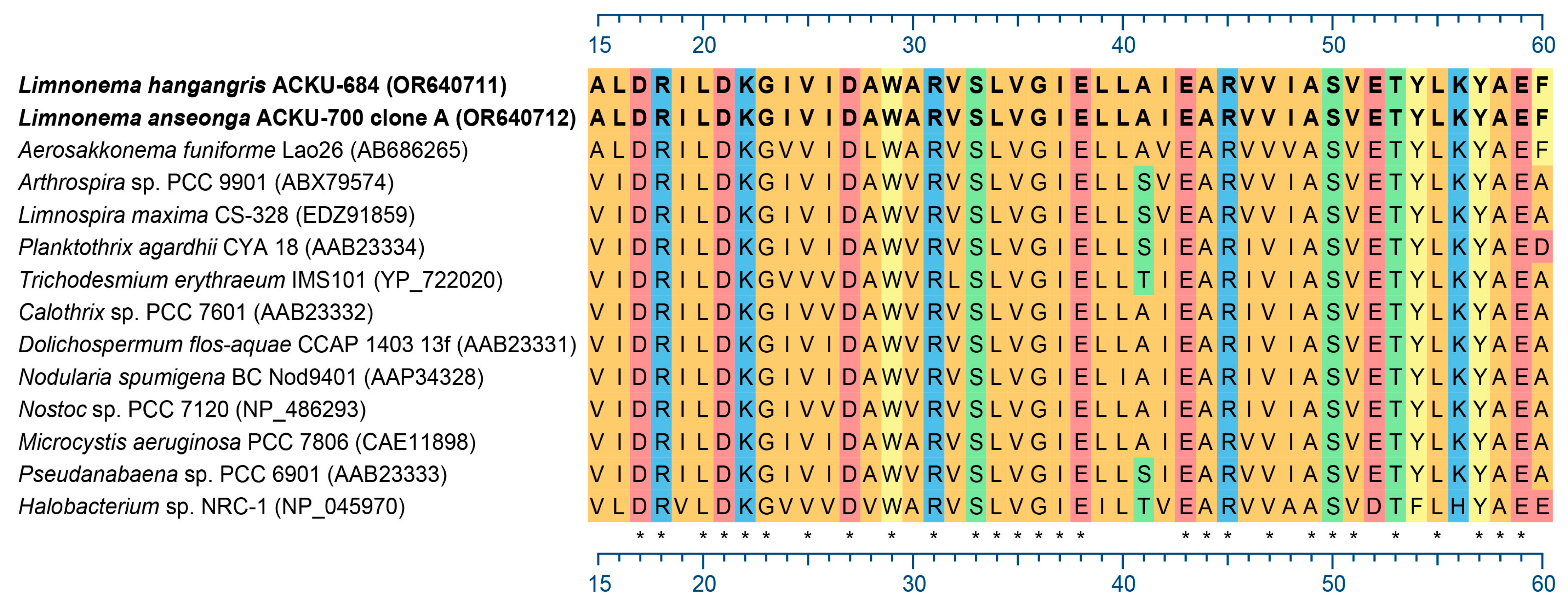
3.2. Detection of Gas Vesicle Protein (gvp)
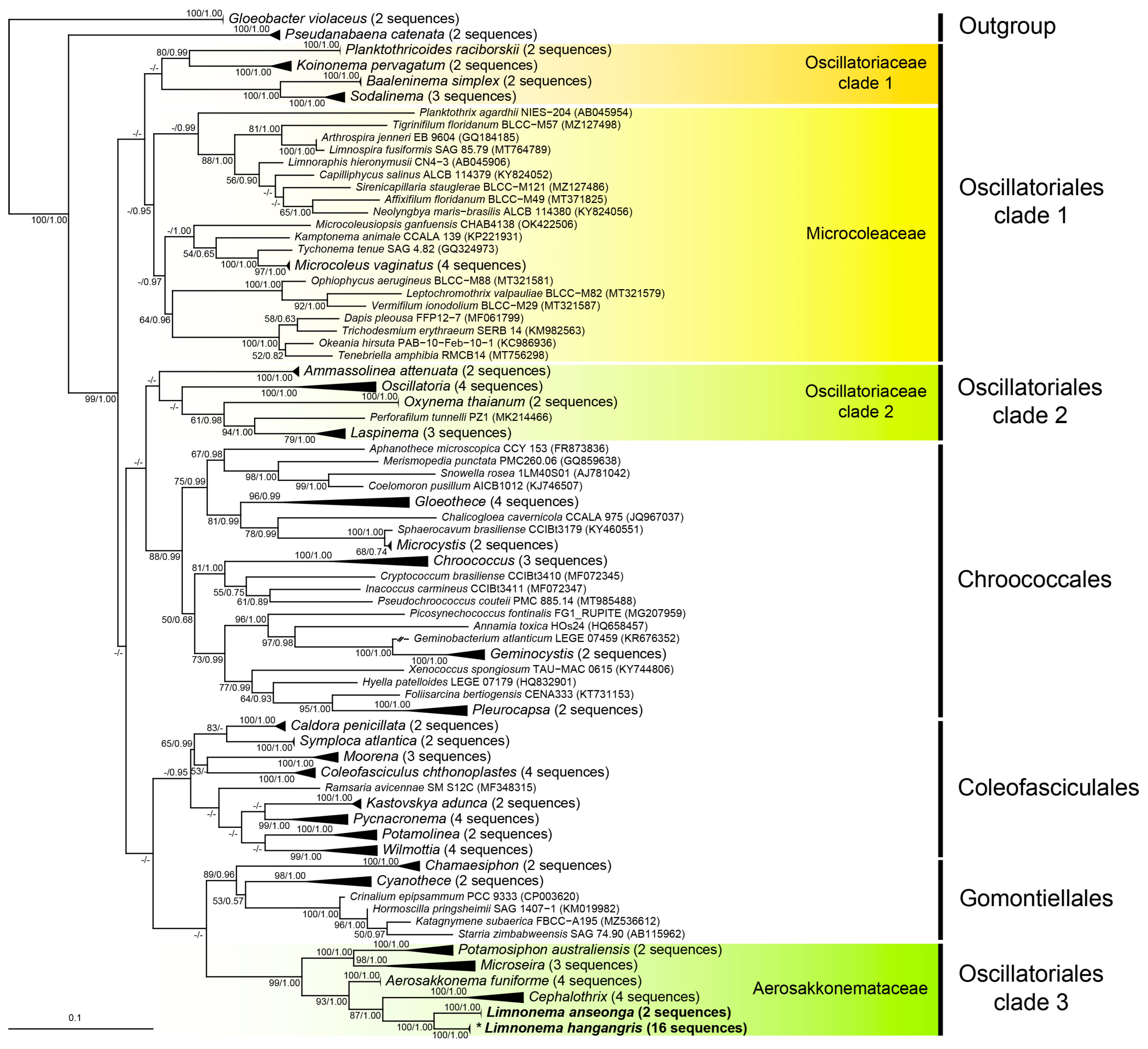
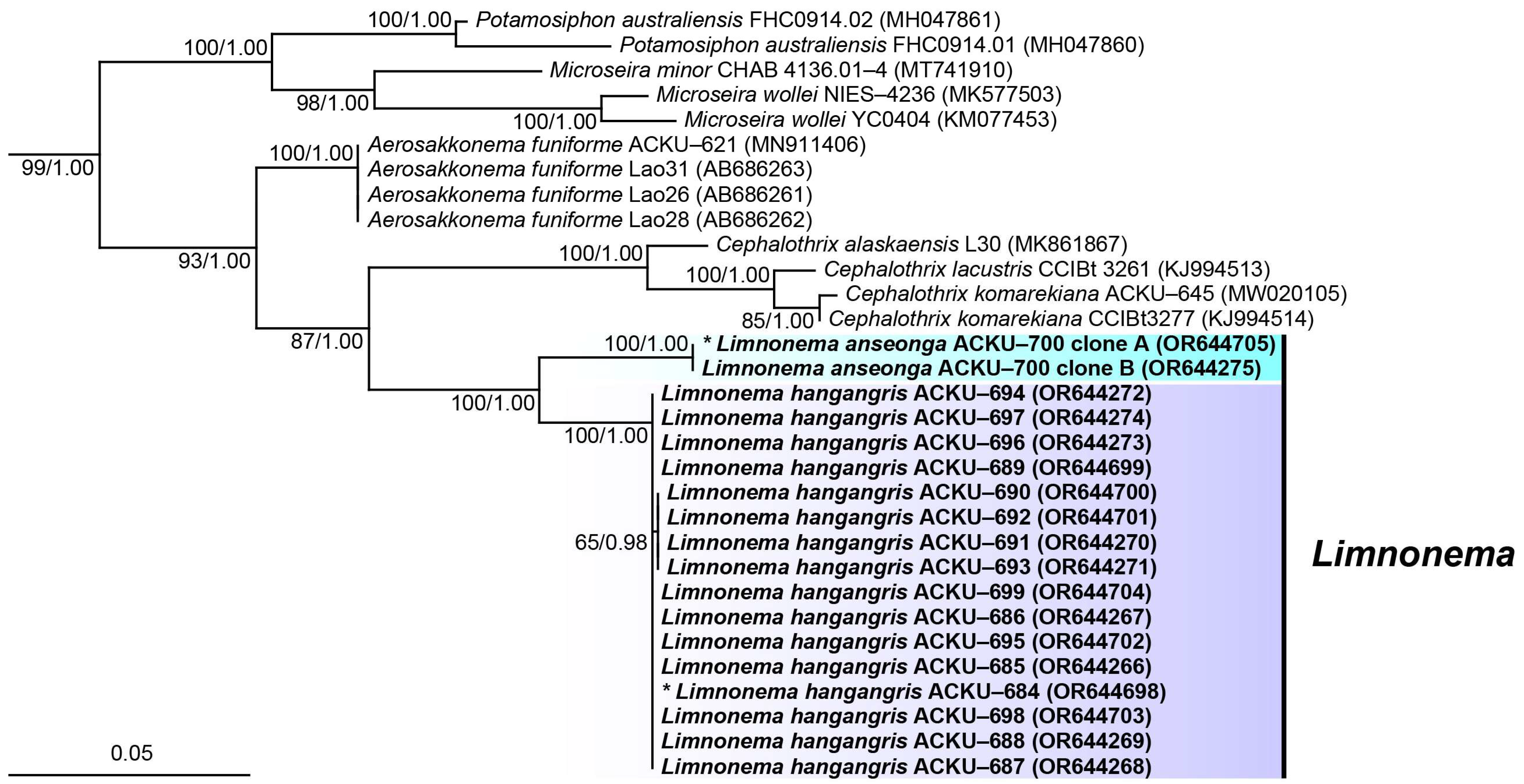
3.3. 16S rRNA Gene Sequences Phylogenetic Tree and Molecular Distance
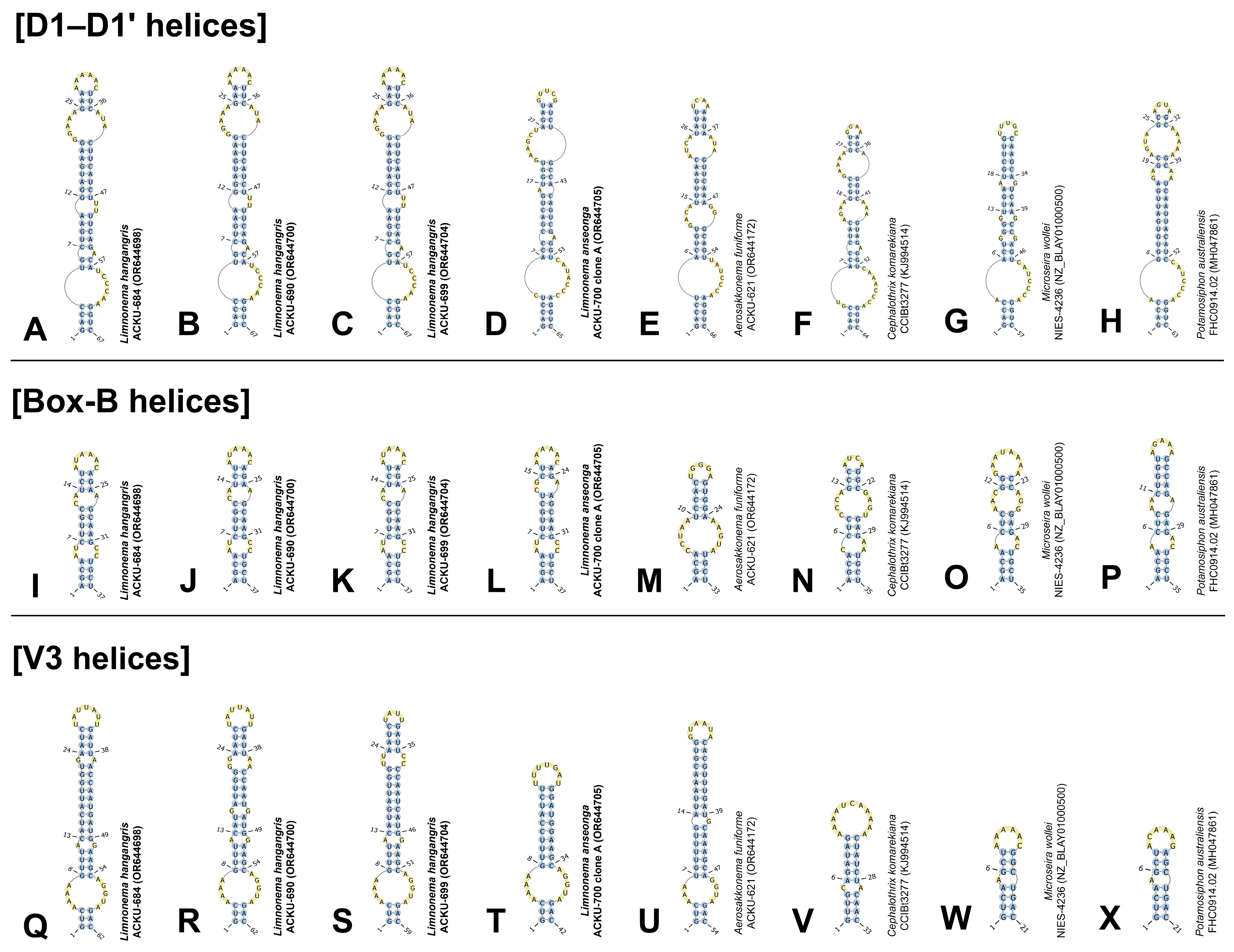
3.4. Secondary Structure of 16S–23S ITS Region
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Casamatta, D.A.; Vis, M.L.; Sheath, R.G. Cryptic species in cyanobacterial systematics: A case study of Phormidium retzii (Oscillatoriales) using RAPD molecular markers and 16S rDNA sequence data. Aquat. Bot. 2003, 77, 295–309. [Google Scholar] [CrossRef]
- Dvořák, P.; Jahodářová, E.; Hašler, P.; Gusev, E.; Poulíčková, A. A new tropical cyanobacterium Pinocchia polymorpha gen. et sp. nov. derived from the genus Pseudanabaena. Fottea 2015, 15, 113–120. [Google Scholar] [CrossRef]
- Castenholz, R.W. Species usage, concept, and evolution in the cyanobacteria (blue-green algae). J. Phycol. 1992, 28, 737–745. [Google Scholar] [CrossRef]
- Komárek, J. The modern classification of cyanoprokaryotes (Cyanobacteria). Oceanol. Hidrobiol. Stud. 2005, 34, 5–17. [Google Scholar]
- Komárek, J.; Anagnostidis, K. Cyanoprokaryota Teil 2: Oscillatoriales. In Süsswasserflora von Mitteleuropa Band 19/2; Büdel, B., Krienitz, L., Gärtner, G., Schagerl, M., Eds.; Elsevier: Heidelberg, Germany, 2005; pp. 1–759. [Google Scholar]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Komárek, J. A polyphasic approach for the taxonomy of cyanobacteria: Principles and applications. Eur. J. Phycol. 2016, 51, 346–353. [Google Scholar] [CrossRef]
- Strunecky, O.; Ivanova, A.P.; Mares, J. An updated classification of cyanobacterial orders and families based on phylogenomic and polyphasic analysis. J. Phycol. 2023, 59, 12–51. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. AlgaeBase. World-Wide Electronic Publication, National University of Ireland, Galway. 2023. Available online: www.algaebase.org (accessed on 14 October 2023).
- Thu, N.K.; Tanabe, Y.; Yoshida, M.; Matsuura, H.; Watanabe, M.M. Aerosakkonema funiformegen. et sp. nov. (Oscillatoriales), a new gas-vacuolated oscillatorioid cyanobacterium isolated from a mesotrophic reservoir. Phycologia 2012, 51, 672–683. [Google Scholar] [CrossRef]
- da Silva Malone, C.F.; Rigonato, J.; Laughinghouse, H.D.; Schmidt, E.C.; Bouzon, Z.L.; Wilmotte, A.; Fiore, M.F.; Sant’Anna, C.L. Cephalothrix gen. nov. (Cyanobacteria): Towards an intraspecific phylogenetic evaluation by multilocus analyses. Int. J. Syst. Evol. Microbiol. 2015, 65, 2993–3007. [Google Scholar] [CrossRef]
- Strunecky, O.; Raabova, L.; Bernardova, A.; Ivanova, A.P.; Semanova, A.; Crossley, J.; Kaftan, D. Diversity of cyanobacteria at the Alaska North Slope with description of two new genera: Gibliniella and Shackletoniella. FEMS Microbiol. Ecol. 2020, 96, 1–60. [Google Scholar] [CrossRef] [PubMed]
- McGregor, G.B.; Sendall, B.C. Phylogeny and toxicology of Lyngbya wollei (Cyanobacteria, Oscillatoriales) from north-eastern Australia, with a description of Microseira gen. nov. J. Phycol. 2015, 51, 109–119. [Google Scholar] [CrossRef]
- Geng, R.; Li, W.; Chao, A.; Guo, X.; Li, H.; Yu, G.; Li, R. Establishment of a New Filamentous Cyanobacterial Genus, Microcoleusiopsis gen. nov. (Microcoleaceae, Cyanobacteria), from Benthic Mats in Open Channel, Jiangxi Province, China. Diversity 2021, 13, 178–190. [Google Scholar] [CrossRef]
- McGregor, G.B.; Sendall, B.C. Potamosiphon australiensis gen. nov., sp nov. (Oscillatoriales), a new filamentous cyanobacterium from subtropical north-eastern Australia. Phytotaxa 2019, 387, 77–93. [Google Scholar] [CrossRef]
- Sciuto, K.; Moro, I. Cyanobacteria: The bright and dark sides of a charming group. Biodivers. Conserv. 2015, 24, 711–738. [Google Scholar] [CrossRef]
- Dadheech, P.K.; Abed, R.M.M.; Mahmoud, H.; Mohan, M.K.; Krienitz, L. Polyphasic characterization of cyanobacteria isolated from desert crusts, and the description of Desertifilum tharensegen. et sp. nov. (Oscillatoriales). Phycologia 2012, 51, 260–270. [Google Scholar] [CrossRef]
- Sournia, A. Phytoplankton Manual; UNESCO: France, Paris, 1978; pp. 1–337. [Google Scholar]
- Kiel, G.; Gaylarde, C.C. Bacterial diversity in biofilms on external surfaces of historic buildings in Porto Alegre. World J. Microbiol. Biotechnol. 2006, 22, 293–297. [Google Scholar] [CrossRef]
- Stanier, R.Y.; Kunisawa, R.; Mandel, M.C.B.G.; Cohen-Bazire, G. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 1971, 35, 171–205. [Google Scholar] [CrossRef]
- Lee, N.-J.; Seo, Y.; Ki, J.-S.; Lee, O.-M. A study of newly recorded genus and species for aerial cyanobacteria Wilmottia murrayi (Oscillatoriales, Cyanobacteria) in Korea. Korean J. Environ. Biol. 2019, 37, 260–267. [Google Scholar] [CrossRef]
- Kim, J.-H.; Jeong, M.S.; Kim, D.-Y.; Her, S.; Wie, M.-B. Zinc oxide nanoparticles induce lipoxygenase-mediated apoptosis and necrosis in human neuroblastoma SH-SY5Y cells. Neurochem. Int. 2015, 90, 204–214. [Google Scholar] [CrossRef]
- Neilan, B.A.; Jacobs, D.; Blackall, L.L.; Hawkins, P.R.; Cox, P.T.; Goodman, A.E. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int. J. Syst. Evol. Microbiol. 1997, 47, 693–697. [Google Scholar] [CrossRef] [PubMed]
- Taton, A.; Grubisic, S.; Brambilla, E.; De Wit, R.; Wilmotte, A. Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): A morphological and molecular approach. Appl. Environ. Microbiol. 2003, 69, 5157–5169. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Page, R.D.M. Tree View: An application to display phylogenetic trees on personal computers. Bioinformatics 1996, 12, 357–358. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Byun, Y.; Han, K. PseudoViewer3: Generating planar drawings of large-scale RNA structures with pseudoknots. Bioinformatics 2009, 25, 1435–1437. [Google Scholar] [CrossRef]
- Turland, N.J.; Wiersema, J.H.; Barrie, F.R.; Greuter, W.; Hawksworth, D.L.; Herendeen, P.S.; Knapp, S.; Kusber, W.-H.; Li, D.-Z.; Marhold, K.; et al. International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code) Adopted by the Nineteenth International Botanical Congress Shenzhen, China, 17 July 2017; Regnum Vegetabile 159; Koeltz Botanical Books: Glashütten, Germany, 2018; pp. 1–254. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
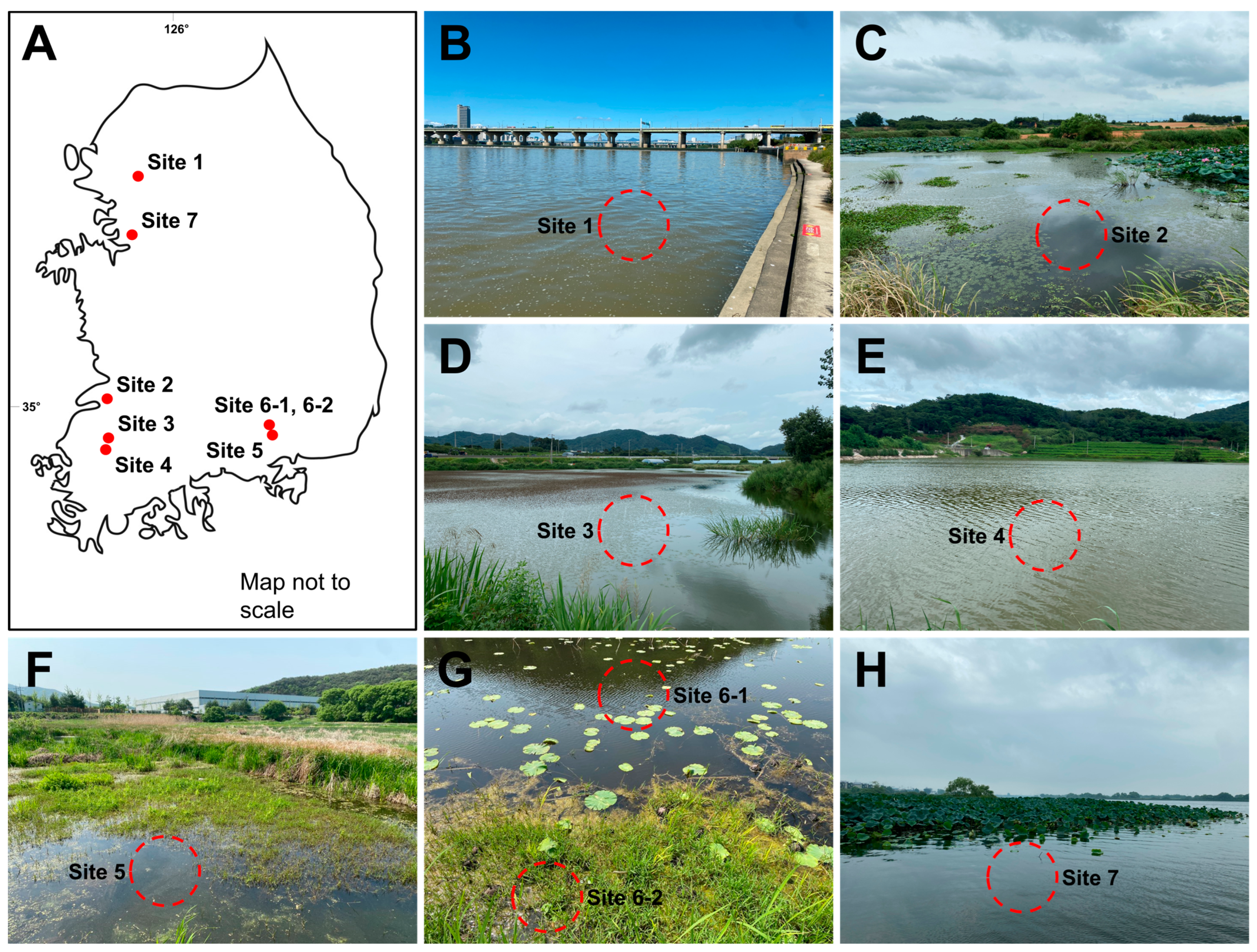
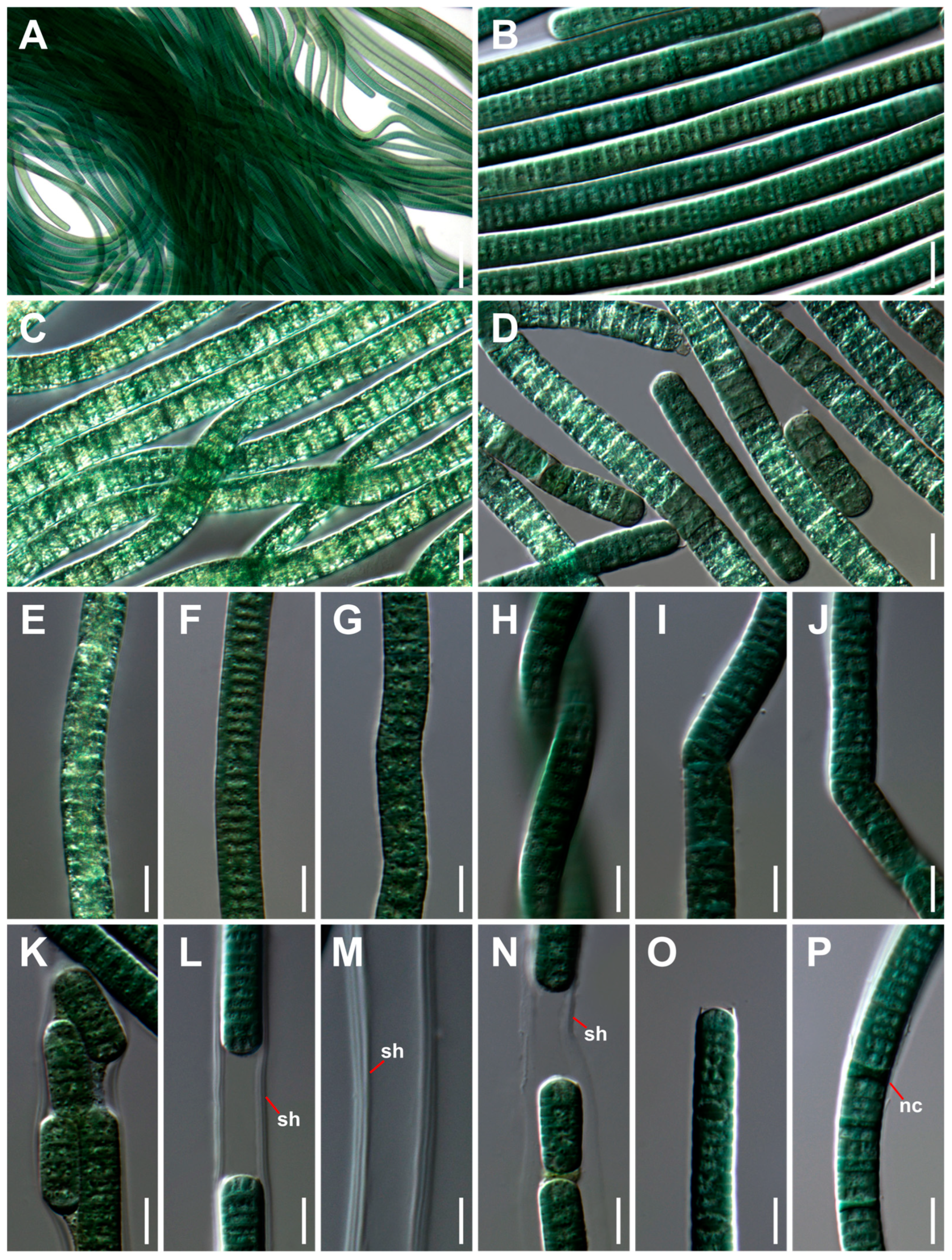
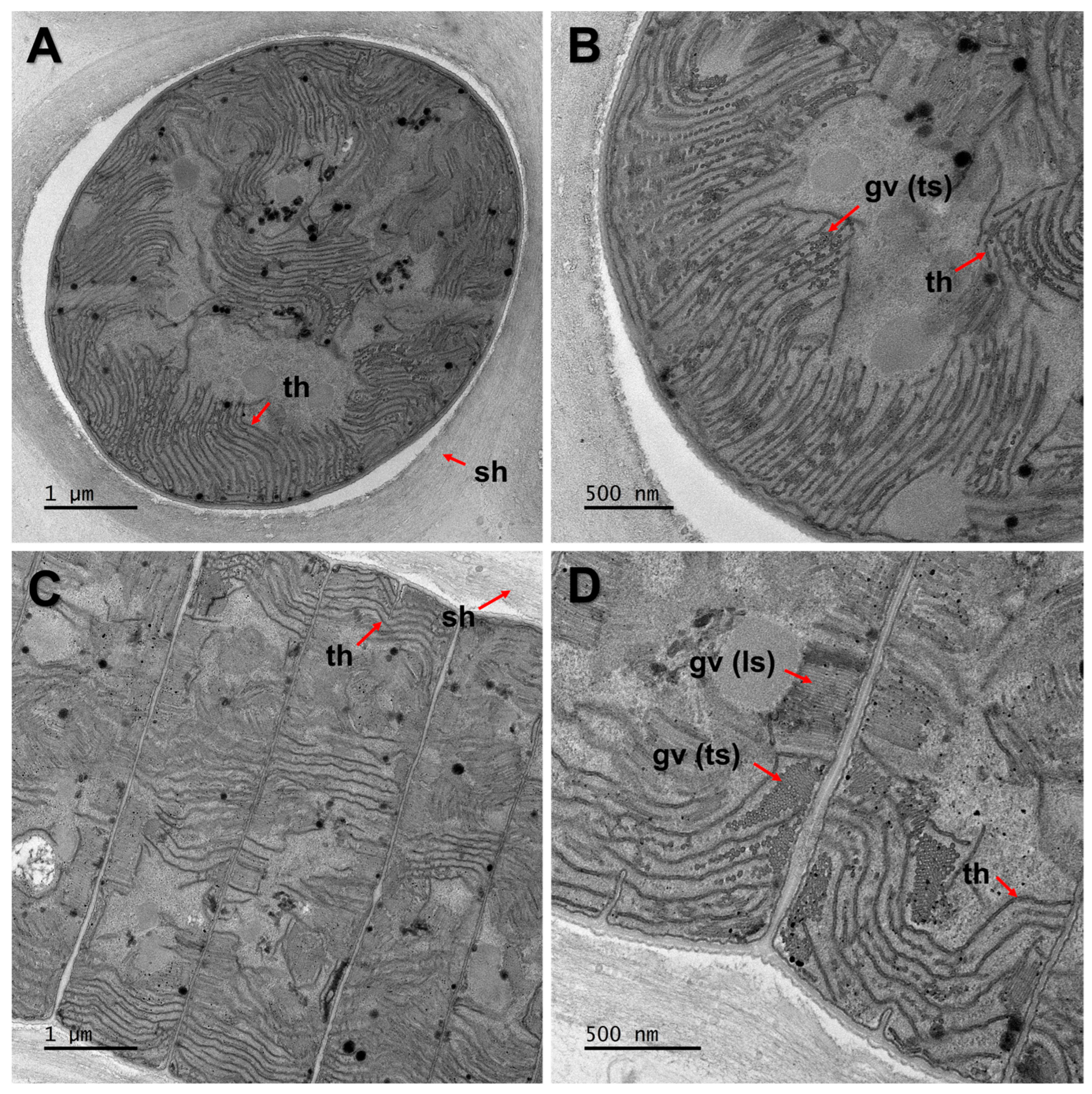
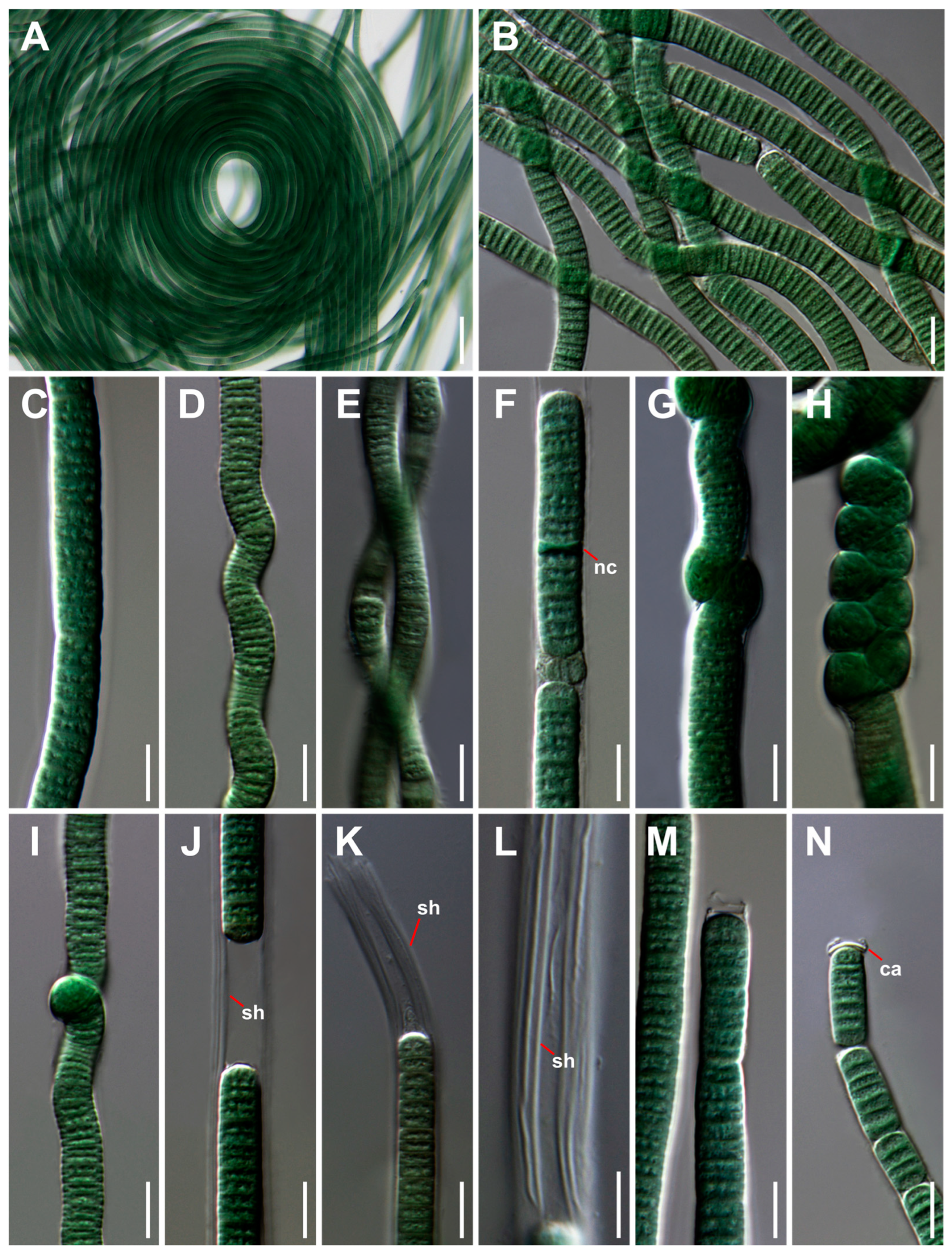
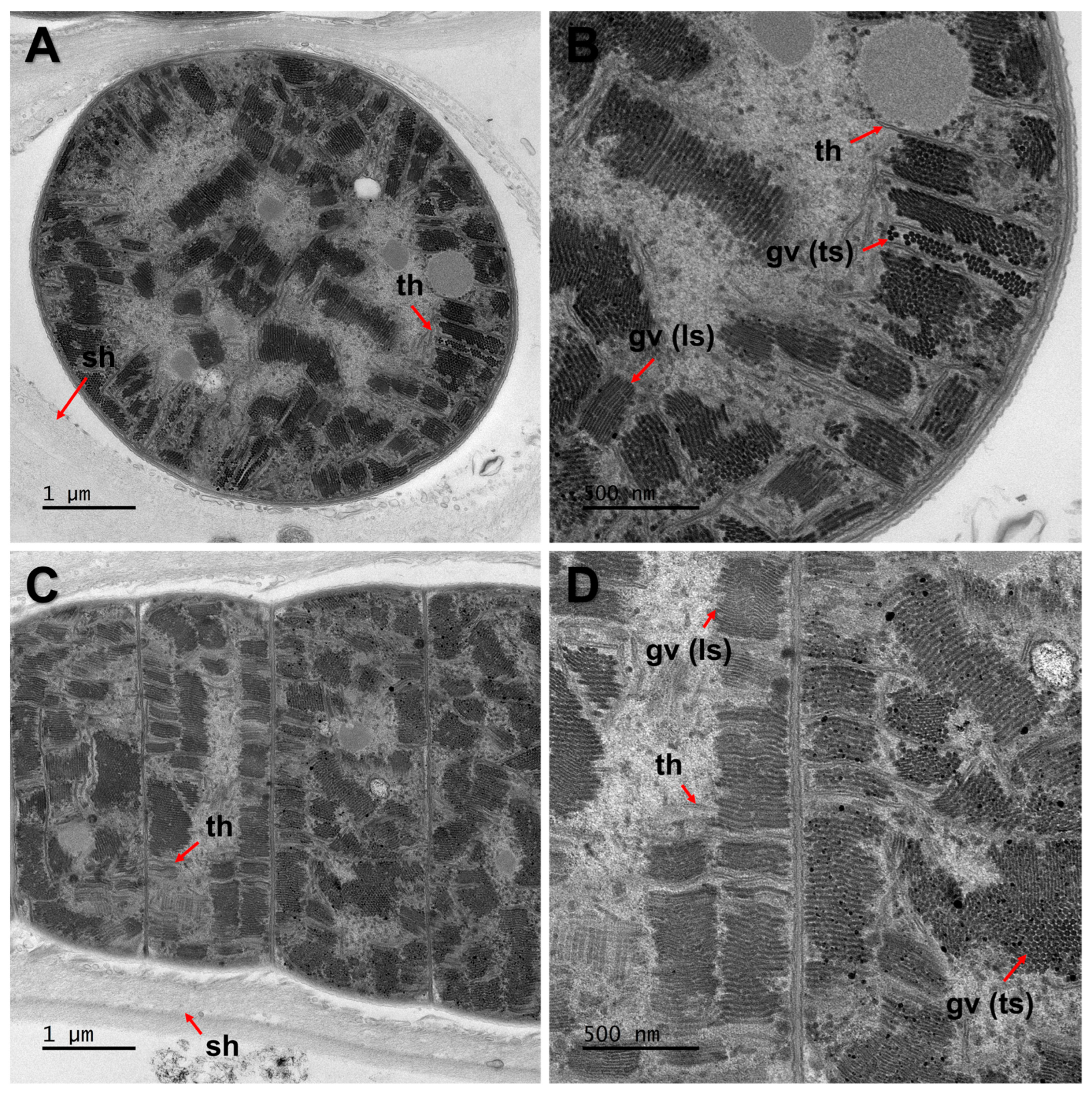
| Strain | Site | Location | GPS | Occurrence | Temperature | pH |
|---|---|---|---|---|---|---|
| Limnonema hangangris | ||||||
| ACKU-684 | Site 1 | Han River, Seoul, Republic of Korea | 37°31′10.2″ N 127°05′27.2″ E | Plankton on the surface of river | 26.3 °C | 7.6 |
| ACKU-685 | ||||||
| ACKU-686 | ||||||
| ACKU-687 | ||||||
| ACKU-688 | ||||||
| ACKU-689 | Site 2 | Yeonjoong Reservoir, Daedong-ri, Julpo-myeon, Buan, Jeollabuk-do, Republic of Korea | 35°36′25.8″ N 126°41′25.5″ E | Plankton on the surface of reservoir | 29.2 °C | 7.0 |
| ACKU-694 | ||||||
| ACKU-690 | Site 3 | Pungjeong Reservoir, Nokjin-ri, Nam-myeon, Jangseong, Jeollanam-do, Republic of Korea | 35°14′54.7″ N 126°47′47.9″ E | Plankton on the surface of reservoir | 29.1 °C | 7.1 |
| ACKU-691 | ||||||
| ACKU-693 | ||||||
| ACKU-692 | Site 4 | Sanmak Reservoir, Deungim-dong, Gwangsan-gu, Gwangju, Jeollanam-do, Republic of Korea | 35°11′34.6″ N 126°46′03.7″ E | Plankton on the surface of reservoir | 32.4 °C | 8.5 |
| ACKU-699 | Site 5 | Beopsu Jilnal Swamp, Ugeo-ri, Beopsu-myeon, Haman, Gyeongsangnam-do, Republic of Korea | 35°19′11.7″ N 128°21′00.2″ E | Plankton on the surface of reservoir in the swamp | 20.1 °C | 6.9 |
| ACKU-698 | Site 6-1 | Daepyeong Reservoir, Daesong-ri, Beopsu-myeon, Haman, Gyeongsangnam-do, Republic of Korea | 35°20′25.0″ N 128°20′04.7″ E | Plankton on the surface of reservoir | 21.6 °C | 7.0 |
| ACKU-695 | Site 6-2 | Epilithic cyanobacteria of gravel at the bottom of reservoir | ||||
| ACKU-696 | ||||||
| ACKU-697 | ||||||
| Limnonema anseonga | ||||||
| ACKU-700 clone A | Site 7 | Anseong Stream, Dodu-ri, Paengseong-eup, Pyeongtaek, Gyeonggi-do, Republic of Korea | 36°58′36.6″ N 126°58′41.4″ E | Plankton on the surface of stream | 25.9 °C | 8.2 |
| ACKU-700 clone B | ||||||
| Apical Cell | Calyptra | Sheath | Gas Vacuoles | Thylakoids | Cell Size (μm) | Habitat/Distribution | ||
|---|---|---|---|---|---|---|---|---|
| Width | Length | |||||||
| Limnonema hangangris ACKU-684 | Rounded | Not observed | Thick, longitudinally lamellated | Observed | Irregular | 6.0–7.9 | 2.2–2.5 | Surface or gravel of freshwater/Korea |
| L. anseonga ACKU-700 clone A | Rounded | Rarely observed | Thick, longitudinally lamellated | Observed | Irregular | 6.4–9.5 | 2.1–2.5 | Surface of freshwater/Korea |
| Aerosakkonema funiforme Lao26 | Rounded or flattened-rounded | Not observed | Not observed | Observed | Irregular | 11.7–16.6 | 2.6–4.1 | Reservoir/Laos |
| Cephalothrix alaskaensis L30 | Rounded | Not observed | Hyaline, firm, sometimes loosening at the end of filaments | Not observed | Parietal | 5.0–6.5 | 1.8–2.0 | Water soil shoreline/Alaska |
| C. komarekiana CCIBt3277 | Strongly capitate | Not observed | Attached to trichome or wide | Observed | Radial | 4.8–6.6 | 2.0–3.4 | Alkaline lake/Brazilian |
| C. lacustris CCIBt3261 | Strongly capitate | Observed (conical) | Attached to trichome | Observed | ND | 5.6–6.7 | 2.2–3.1 | Freshwater pond/Brazilian |
| Microseira wollei YC0404 | Rounded | Not observed | Colorless, hyaline, firm, lamellated | Not observed | ND | 30–48 | 4.0–9.0 | Catchments/Australia |
| M. minor CHAB 4136 | Rounded | Not observed | Colorless, hyaline, firm, lamellated | Not observed | Irregular | 9.0–13.3 | 3.1–7.7 | Rock surface in freshwater river/China |
| Potamosiphon australiensis FHC0914 | Rounded | Not observed | Colourless, sometimes lamellated | Not observed | Irregular | 15.7–17.8 | 1.7–2.5 | Benthic in freshwater coastal streams/Australia |
| No. | Species and Strain | <1> | <2> | <3> | <4> | <5> | <6> | <7> | <8> | <9> | <10> | <11> | <12> | <13> | <14> | <15> | <16> | <17> |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA similarity (%) | ||||||||||||||||||
| <1> | Limnonema hangangris ACKU-684 | 100 | 100 | 100 | 100 | 99.9 | 99.9 | 97.7 | 95.5 | 95.5 | 95.5 | 94.4 | 94.4 | 94.3 | 91.8 | 91.9 | 92.9 | |
| <2> | L. hangangris ACKU-689 | 0.00 | 100 | 100 | 100 | 99.9 | 99.9 | 97.7 | 95.5 | 95.5 | 95.5 | 94.4 | 94.4 | 94.3 | 91.8 | 91.9 | 92.9 | |
| <3> | L. hangangris ACKU-690 | 0.00 | 0.00 | 100 | 100 | 99.9 | 99.9 | 97.7 | 95.5 | 95.5 | 95.5 | 94.4 | 94.4 | 94.3 | 91.8 | 91.9 | 92.9 | |
| <4> | L. hangangris ACKU-692 | 0.00 | 0.00 | 0.00 | 100 | 99.9 | 99.9 | 97.7 | 95.5 | 95.5 | 95.5 | 94.4 | 94.4 | 94.3 | 91.8 | 91.9 | 92.9 | |
| <5> | L. hangangris ACKU-695 | 0.00 | 0.00 | 0.00 | 0.00 | 99.9 | 99.9 | 97.7 | 95.5 | 95.5 | 95.5 | 94.4 | 94.4 | 94.3 | 91.8 | 91.9 | 92.9 | |
| <6> | L. hangangris ACKU-698 | 0.09 | 0.09 | 0.09 | 0.09 | 0.09 | 100 | 97.6 | 95.4 | 95.4 | 95.4 | 94.3 | 94.4 | 94.3 | 91.8 | 91.9 | 92.9 | |
| <7> | L. hangangris ACKU-699 | 0.09 | 0.09 | 0.09 | 0.09 | 0.09 | 0.00 | 97.6 | 95.4 | 95.4 | 95.4 | 94.3 | 94.4 | 94.3 | 91.8 | 91.9 | 92.9 | |
| <8> | L. anseonga ACKU-700 clone A | 2.25 | 2.25 | 2.25 | 2.25 | 2.25 | 2.34 | 2.34 | 95.9 | 95.9 | 95.9 | 94.0 | 93.6 | 93.6 | 91.6 | 91.9 | 93.7 | |
| <9> | Aerosakkonema funiforme Lao26 | 4.46 | 4.46 | 4.46 | 4.46 | 4.46 | 4.56 | 4.56 | 4.08 | 100 | 100 | 95.8 | 95.0 | 95.1 | 92.4 | 92.9 | 94.2 | |
| <10> | A. funiforme Lao28 | 4.46 | 4.46 | 4.46 | 4.46 | 4.46 | 4.56 | 4.56 | 4.08 | 0.00 | 100 | 95.8 | 95.0 | 95.1 | 92.4 | 92.9 | 94.2 | |
| <11> | A. funiforme ACKU-621 | 4.46 | 4.46 | 4.46 | 4.46 | 4.46 | 4.56 | 4.56 | 4.08 | 0.00 | 0.00 | 95.8 | 95.0 | 95.1 | 92.4 | 92.9 | 94.2 | |
| <12> | Cephalothrix alaskaensis L30 | 5.66 | 5.66 | 5.66 | 5.66 | 5.66 | 5.76 | 5.76 | 6.16 | 4.18 | 4.18 | 4.18 | 97.4 | 97.6 | 91.7 | 92.0 | 93.1 | |
| <13> | C. komarekiana CCIBt3277 | 5.76 | 5.76 | 5.76 | 5.76 | 5.76 | 5.76 | 5.76 | 6.66 | 5.06 | 5.06 | 5.06 | 2.53 | 98.9 | 92.1 | 91.9 | 92.8 | |
| <14> | C. lacustris CCIBt3261 | 5.86 | 5.86 | 5.86 | 5.86 | 5.86 | 5.86 | 5.86 | 6.67 | 4.96 | 4.96 | 4.96 | 2.25 | 1.02 | 92.2 | 91.7 | 92.6 | |
| <15> | Microseira wollei YC0404 | 8.61 | 8.61 | 8.61 | 8.61 | 8.61 | 8.61 | 8.61 | 8.91 | 7.88 | 7.88 | 7.88 | 8.71 | 8.29 | 8.19 | 94.7 | 94.2 | |
| <16> | M. minor CHAB 4136.01-4 | 8.39 | 8.39 | 8.39 | 8.39 | 8.39 | 8.29 | 8.29 | 8.29 | 7.16 | 7.16 | 7.16 | 8.21 | 8.42 | 8.63 | 5.25 | 95.0 | |
| <17> | Potamosiphon australiensis FHC0914.02 | 7.47 | 7.47 | 7.47 | 7.47 | 7.47 | 7.47 | 7.47 | 6.55 | 5.85 | 5.85 | 5.85 | 7.06 | 7.57 | 7.78 | 5.95 | 4.96 | |
| p-distance (%) | ||||||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.-H.; Kim, S.-W.; Lee, N.-J.; Kim, D.-H.; Wang, H.-R.; Lee, O.-M. Limnonema gen. nov. (Aerosakkonemataceae, Cyanobacteria): Two Novel Species from Republic of Korea Characterized by Morphological and Molecular Analyses. Diversity 2023, 15, 1174. https://doi.org/10.3390/d15121174
Song J-H, Kim S-W, Lee N-J, Kim D-H, Wang H-R, Lee O-M. Limnonema gen. nov. (Aerosakkonemataceae, Cyanobacteria): Two Novel Species from Republic of Korea Characterized by Morphological and Molecular Analyses. Diversity. 2023; 15(12):1174. https://doi.org/10.3390/d15121174
Chicago/Turabian StyleSong, Ji-Ho, So-Won Kim, Nam-Ju Lee, Do-Hyun Kim, Hye-Ryeung Wang, and Ok-Min Lee. 2023. "Limnonema gen. nov. (Aerosakkonemataceae, Cyanobacteria): Two Novel Species from Republic of Korea Characterized by Morphological and Molecular Analyses" Diversity 15, no. 12: 1174. https://doi.org/10.3390/d15121174
APA StyleSong, J.-H., Kim, S.-W., Lee, N.-J., Kim, D.-H., Wang, H.-R., & Lee, O.-M. (2023). Limnonema gen. nov. (Aerosakkonemataceae, Cyanobacteria): Two Novel Species from Republic of Korea Characterized by Morphological and Molecular Analyses. Diversity, 15(12), 1174. https://doi.org/10.3390/d15121174






