Genetic Diversity of Yuca (Manihot esculenta esculenta; Cassava, Manioc), an Indigenous Crop in the Peruvian Amazon
Abstract
:1. Introduction
2. Materials and Methods
3. Results
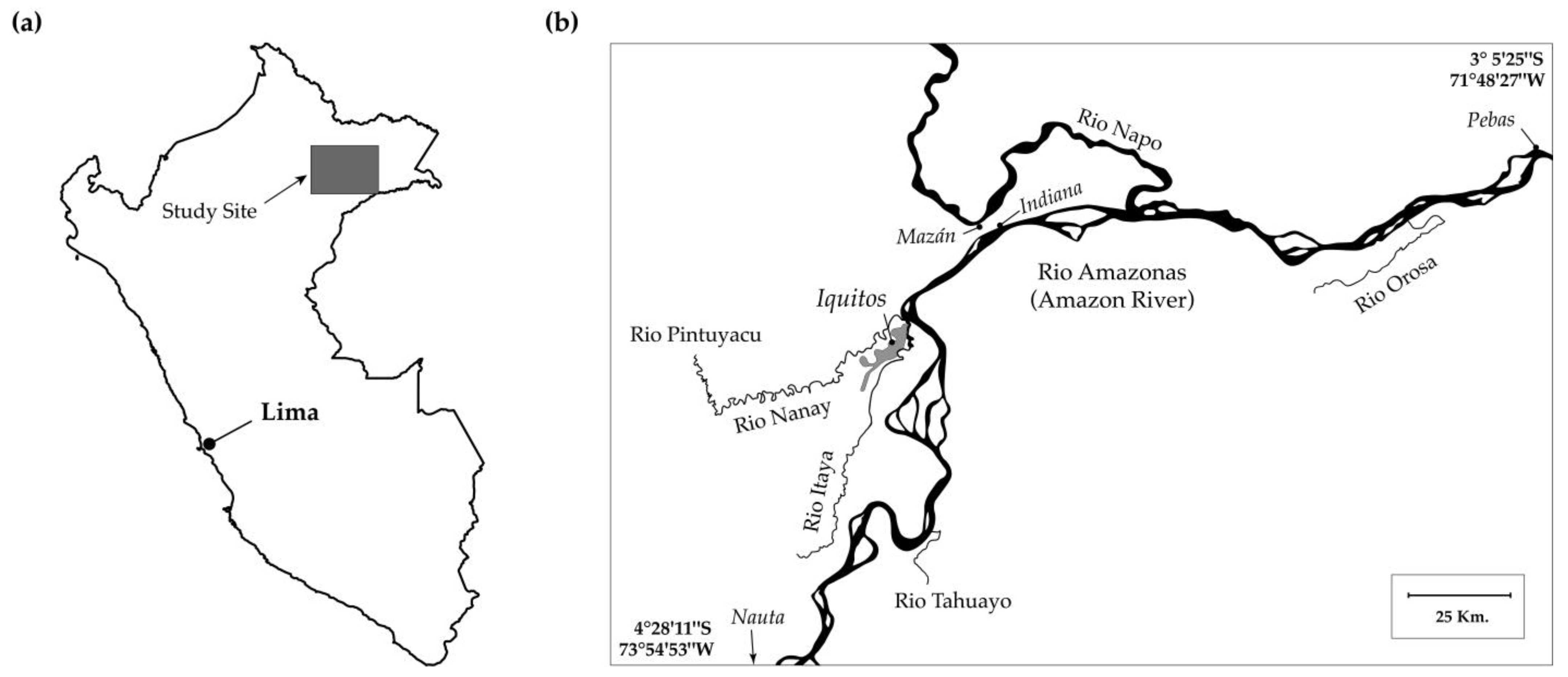
3.1. River Distances and Direct Distances
3.2. Diversity in STRs
3.3. NJ Tree
3.4. Population Structure
3.5. Landrace Names and Relatedness
4. Discussion
4.1. Genetic Diversity
4.2. Relationships between Landraces
4.3. Geography and Population Structure
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Allem, A.C. The Origins and Taxonomy of Cassava. In Cassava: Biology, Production, and Utilization; Hillocks, R.J., Thresh, J.M., Bellotti, A.C., Eds.; CAB International: Wallingford, UK, 2002; pp. 1–16. [Google Scholar]
- Balagopalan, C. Cassava Utilization in Food, Feed, and Industry. In Cassava: Biology, Production, and Utilization; Hillocks, R.J., Thresh, J.M., Bellotti, A.C., Eds.; CAB International: Wallingford, UK, 2002; pp. 301–318. [Google Scholar]
- Olsen, K.; Schaal, B. Microsatellite Variation in Cassava (Manihot esculenta, Euphorbiaceae) and Its Wild Relatives: Further Evidence for a Southern Amazonian Origin of Domestication. Am. J. Bot. 2001, 88, 131–142. [Google Scholar] [CrossRef]
- Dickau, R.; Ranere, A.J.; Cooke, R.G. Starch Grain Evidence for the Preceramic Dispersals of Maize and Root Crops into Tropical Dry and Humid Forests of Panama. Proc. Natl. Acad. Sci. USA 2007, 104, 3651–3656. [Google Scholar] [CrossRef]
- Isendahl, C. The Domestication and Early Spread of Manioc (Manihot esculenta Crantz): A Brief Synthesis. Lat. Am. Antiq. 2011, 22, 452–468. [Google Scholar] [CrossRef]
- Jones, W.O. Manioc in Africa; Stanford University Press: Stanford, CT, USA, 1959. [Google Scholar]
- Hillocks, R.J. Cassava in Africa. In Cassava: Biology, Production, and Utilization; Hillocks, R.J., Thresh, J.M., Bellotti, A.C., Eds.; CAB International: Wallingford, UK, 2002; pp. 41–54. [Google Scholar]
- Alves, A.A.C. Cassava Botany and Physiology. In Cassava: Biology, Production, and Utilization; Hillocks, R.J., Thresh, J.M., Bellotti, A.C., Eds.; CAB International: Wallingford, UK, 2002; pp. 67–90. [Google Scholar]
- McMahon, J.M.; White, W.L.B.; Sayre, R.T. Cyanogenesis in Cassava. J. Exp. Biol. 1999, 46, 731–741. [Google Scholar]
- Arias, J.C.; Ramos, L.A.; Acosta, L.E.; Camacho, H.A.; Marín, G. Diversidad De Yucas Entre Los Ticuna: Riqueza Cultural Y GenéTica De Un Producto Tradicional; Instituto Amazónico de Investigaciones Científicas, Sinchi: Bogotá, Columbia, 2004. [Google Scholar]
- Boster, J.S. Exchange of Varieties and Information between Aguaruna Manioc Cultivators. Am. Anthropol. 1986, 88, 428–436. [Google Scholar] [CrossRef]
- Fraser, J.A. The Diversity of Bitter Manioc (Manihot esculenta Crantz) Cultivation in a Whitewater Amazonian Landscape. Diversity 2010, 2, 586–609. [Google Scholar] [CrossRef]
- Salick, J.; Cellinese, N.; Knapp, S. Indigenous Diversity of Cassava: Generation Maintenance, Use and Loss among the Amuesha, Peruvian Upper Amazon. Econ. Bot. 1997, 51, 6–19. [Google Scholar] [CrossRef]
- Wilson, W.M.; DuFour, D.L. Why “Bitter” Cassava? Productivity of “Bitter” and “Sweet” Cassava in a Tukanoan Indian Settlement in the Northwest Amazon. Econ. Bot. 2002, 56, 49–57. [Google Scholar] [CrossRef]
- Sánchez, H.I.; López, P. Diversidad De Yuca (Manihot esculenta Crantz) En Jenaro Herrera, Loreto, Perú; Documento Técnico No. 28; IIAP: Iquitos, Peru, 2001. [Google Scholar]
- Chiwona-Karltun, L.; Brimer, L.; Saka, J.D.K.; Mhone, A.R.; Mkumbira, J.; Johansson, L.; Bokanga, M.; Mahungu, N.M.; Rosling, H. Bitter Taste in Cassava Roots Correlates with Cyanogenic Glucoside Levels. J. Sci. Food Agric. 2004, 84, 581–590. [Google Scholar] [CrossRef]
- Wooding, S.P.; Payahua, C.N. Ethnobotanical Diversity of Cassava (Manihot esculenta Crantz) in the Peruvian Amazon. Diversity 2022, 14, 252. [Google Scholar] [CrossRef]
- Duputié, A.; Massol, F.; David, P.; Haxaire, C.; McKey, D. Traditional Amerindian Cultivators Combine Directional and Ideotypic Selection for Sustainable Management of Cassava Genetic Diversity. J. Evol. Biol. 2009, 22, 1317–1325. [Google Scholar] [CrossRef]
- Boster, J.S. Classification, Cultivation and Selection of Aguaruna Varieties of Manihot esculenta (Euphorbiaceae). Adv. Econ. Bot. 1984, 1, 34–47. [Google Scholar]
- Boster, J.S. Selection for Perceptual Distinctiveness: Evidence from Aguaruna Jívaro Varieties of Manihot esculenta. Econ. Bot. 1984, 39, 310–325. [Google Scholar] [CrossRef]
- Rogers, D.J.; Fleming, H.S. A Monograph of Manihot esculenta with an Explanation of the Taximetrics Methods Used. Econ. Bot. 1973, 27, 1–113. [Google Scholar] [CrossRef]
- Wang, W.; Feng, B.; Xiao, J.; Xia, Z.; Zhou, X.; Li, P.; Zhang, W.; Wang, Y.; Moller, B.L.; Zhang, P.; et al. Cassava Genome from a Wild Ancestor to Cultivated Varieties. Nat. Comm. 2014, 5, 5110. [Google Scholar] [CrossRef]
- Bredeson, J.V.; Lyons, J.B.; Prochnik, S.E.; Wu, G.A.; Ha, C.M.; Edsinger-Gonzales, E.; Grimwood, J.; Schmutz, J.; Rabbi, I.Y.; Egesi, C.; et al. Sequencing Wild and Cultivated Cassava and Related Species Reveals Extensive Interspecific Hybridization and Genetic Diversity. Nat. Biotechnol. 2016, 34, 562–570. [Google Scholar] [CrossRef]
- Ceballos, N.; Morante, N.; Calle, F.; Lenis, J.; Salazar, S. Developing New Cassava Varieties: Tools, Techniques, and Strategies. In Achieving Sustainable Cultivation of Cassava. Volume 2: Genetics, Breeding, Pests and Diseases; Hershey, C., Ed.; Burleigh Dodds Science Publishing: London, UK, 2017; pp. 49–90. [Google Scholar]
- Machado, C.L.R.; Crespo-Lopez, M.E.; Augusto-Oliveira, M.; Arrifano, G.P.; Macchi, B.M.; Lopes-Araujo, A.; Santos-Sacramento, L.; Souza-Monteiro, J.R.; Alvarez-Leite, J.I.; Souza, C.B.A. Eating in the Amazon: Nutritional Status of the Riverine Populations and Possible Nudge Interventions. Foods 2021, 10, 1015. [Google Scholar] [CrossRef]
- Piperata, B.A.; Spence, J.E.; Da-Gloria, P.; Hubbe, M. The Nutrition Transition in Amazonia: Rapid Economic Change and Its Impact on Growth and Development in Ribeirinhos. Am. J. Phys. Anthropol. 2011, 146, 1–13. [Google Scholar] [CrossRef]
- Piperata, B.A. Nutritional Status of Ribeirinhos in Brazil and the Nutrition Transition. Am. J. Phys. Anthropol. 2007, 133, 868–878. [Google Scholar] [CrossRef]
- Olsen, K.M.; Schaal, B.A. Evidence on the Origin of Cassava: Phylogeography of Manihot esculenta. Proc. Natl. Acad. Sci. USA 1999, 96, 5586–5591. [Google Scholar] [CrossRef]
- Acosta Muñoz, L.E.; Valderrama, A.M.M. Enterramientos De Masas De Yuca Del Pueblo Ticuna: Tecnología Tradicional En La Várzea Del Amazonas Colombiano; Instituto Amazónico de Investigaciones Científicas/SINCHI: Leticia, Colombia, 2004. [Google Scholar]
- Chavarriaga-Aguirre, P.; Maya, M.M.; Bonierbale, M.W.; Kresovich, S.; Fregene, M.A.; Tohme, J.; Kochert, G. Microsatellites in Cassava (Manihot Esculenta Crantz): Discovery, Inheritance and Variability. Theor. Appl. Genet. 1998, 97, 493–501. [Google Scholar] [CrossRef]
- Mba, R.E.C.; Stephenson, P.; Edwards, K.; Melzer, S.; Nkumbira, J.; Gullberg, U.; Apel, K.; Gale, M.; Tohme, H.; Fregene, M. Simple Sequence Repeat (Ssr) Markers Survey of the Cassava (Manihot esculenta Crantz) Genome: Towards an Ssr-Based Molecular Genetic Map of Cassava. Theor. Appl. Genet. 2001, 102, 21–31. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing, Version 4.0.3; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- Hijmans, R.J.; Williams, E.; Vennes, C. Geosphere: Spherical Trigonometry, Version 1.5-18. R Package; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Amestoy, P.R.; Azzalini, A.; Badics, T.; Benison, G.; Bowman, A.; Bahm, W.; Briggs, K.; Bruggeman, J.; Buchmueller, J.; Butts, C.T.; et al. Igraph: Network Analysis and Visualization, Version 1.4.1. R Package; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Dray, S.; Dufour, A.-B.; Thiolouse, J. Ade4: Analysis of Ecological Data: Exploratory and Euclidean Methods in Environmental Sciences, Version 1.7-22. R Package; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Kamvar, Z.N.; Tabima, J.F.; Everhart, S.E.; Brooks, J.C.; Krueger-Hadfield, S.A.; Sotka, E.; Knaus, B.J.; Meirmans, P.G.; Chevalier, F.D.; Folarin, D.; et al. Poppr: Genetic Analysis of Populations with Mixed Reproduction, Version 2.9.3. R Package; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Paradis, E. Pegas: An R Package for Population Genetics with an Integrated-Modular Approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef]
- Paradis, E.; Blomberg, S.P.; Bolker, B.; Brown, J.; Claramunt, S.; Claude, J.; Cuong, H.S.; Desper, R.; Didier, G.; Durand, B.; et al. Ape: Analyses of Phylogenetics and Evolution, Version 5.7. R Package; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Coulon, A. Genhet: An Easy-to-Use R Function to Estimate Individual Heterozygosity. Mol. Ecol. Res. 2009, 10, 167–169. [Google Scholar] [CrossRef]
- Nei, M. Analysis of Gene Diversity in Subdivided Populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W. Allozyme Diversity in Plant Species. In Plant Population Genetics, Breeding, and Genetic Resources; Brown, H.D., Clegg, M.T., Kahler, A.L., Weir, B.S., Eds.; Sinauer Associates: Sunderland, MA, USA, 1989; pp. 43–63. [Google Scholar]
- Jost, L. Gst and Its Relatives Do Not Measure Differentiation. Mol. Ecol. 2008, 17, 4015–4026. [Google Scholar] [CrossRef]
- Alves-Pereira, A.; Clement, C.R.; Picanco-Rodrigues, D.; Veasey, E.A.; Dequigiovanni, G.; Ramos, S.L.F.; Pinheiro, J.B.; Zucchi, M.I. Patterns of Nuclear and Chloroplast Genetic Diversity and Structure of Manioc Along Major Brazilian Amazonian Rivers. Ann. Bot. 2018, 121, 625–639. [Google Scholar] [CrossRef]
- Bradbury, E.J.; Duputié, A.; Delêtre, M.; Roullier, C.; Narváez-Trujillo, A.; Manu-Aduening, J.A.; Emshwiller, E.; McKey, D. Geographic Differences in Patterns of Genetic Differentiation among Bitter and Sweet Manioc (Manihot esculenta Subsp. Esculenta; Euphorbiaceae). Am. J. Bot. 2013, 100, 857–866. [Google Scholar] [CrossRef]
- Hurtado, P.; Olsen, K.M.; Buitrago, C.; Ospina, C.; Marin, J.; Duque, M.; de Vicente, C.; Wongtiem, P.; Wenzel, P.; Killian, A.; et al. Comparison of Simple Sequence Repeat (Ssr) and Diversity Array Technology (Dart) Markers for Assessing Genetic Diversity in Cassava (Manihot esculenta Crantz). Plant Gen. Res. Char. Util. 2008, 6, 208–214. [Google Scholar] [CrossRef]
- Kizito, E.B.; Chiwona-Karltun, L.; Egwang, T.; Fregene, M.; Westerbergh, A. Genetic Diversity and Variety Composition of Cassava on Small-Scale Farms in Uganda: An Interdisciplinary Study Using Genetic Markers and Farmer Interviews. Genetica 2007, 130, 301–318. [Google Scholar] [CrossRef]
- Peroni, N.; Kageyama, P.Y.; Begossi, A. Molecular Differentiation, Diversity, and Folk Classification of “Sweet” and “Bitter” Cassava (Manihot esculenta) in Caiçara and Caboclo Management Systems (Brazil). Genet. Resour. Crop Evol. 2007, 54, 1333–1349. [Google Scholar] [CrossRef]
- Hedrick, P.W. What Is the Evidence for Heterozygote Advantage Selection? Trends Ecol. Evol. 2012, 27, 698–704. [Google Scholar] [CrossRef]
- Gemmell, N.J.; Slate, J. Heterozygote Advantage for Fecundity. PLoS ONE 2006, 1, e125. [Google Scholar] [CrossRef]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic Species as a Window on Diversity and Conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef]
- Avise, J.C. Phylogeography: Retrospect and Prospect. J. Biogeogr. 2009, 36, 3–15. [Google Scholar] [CrossRef]
- Boster, J.S. Inferring Decision Making from Preferences and Behavior: An Analysis of Auranuna Jívaro Manioc Selection. Hum. Ecol. 1984, 12, 343–358. [Google Scholar] [CrossRef]







| Marker | Forward Primer | Reverse Primer | |
|---|---|---|---|
| GA12 | GATTCCTCTAGCAGTTAAGC | CGATGATGCTCTTCGGAGGG | |
| GA13 | TTCCCTCGCTAGAACTTGTC | CTATTTGACCGTCTTCGCCG | |
| GA21 | GGCTTCATCATGGAAAAACC | CAATGCTTTACGGAAGAGCC | |
| GA126 | AGTGGAAATAAGCCATGTGATG | CCCATAATTGATGCCAGGTT | Chavarriaga-Aquirre et al. [30] |
| GA127 | CTCTAGCTATGGATTAGATCT | GTAGCTTCGAGTCGTGGGAGA | |
| GA131 | TTCCAGAAAGACTTCCGTTCA | CTCAACTACTGCACTGCACTC | |
| GA136 | CGTTGATAAAGTGGAAAGAGCA | ACTCCACTCCCGATGCTCGC | |
| GA161 | TGTTCTTGATCTTCTGCTGCA | TGATTGTGGACGTGGGTAGA | |
| SSRY32 | CAAATTTGCAACAATAGAGAACA | TCCACAAAGTCGTCCATTACA | |
| SSRY46 | TCAGGAACAATACTCCATCGAA | CGCTAAAGAAGCTGTCGAGC | Mba et al. [31] |
| SSRY70 | CGCTATTAGAATTGCCAGCAC | CGCTTGTTGTATCCATTGGC | |
| SSRY83 | TGGCTAGATGGTGATTATTGCTT | TGCTTACTCTTTGATTCCACG | |
| SSRY169 | ACAGCTCTAAAAACTGCAGCC | AACGTAGGCCCTAACTAACCC |
| Marker | Alleles | Length Range | Obs. Hz. | Exp. Hz. | HWE p-Value |
|---|---|---|---|---|---|
| GA12 | 15 | 123–153 | 0.45 | 0.75 | <0.001 |
| GA13 | 4 | 133–139 | 0.00 | 0.14 | <0.001 |
| GA21 | 7 | 104–118 | 0.37 | 0.74 | <0.001 |
| GA126 | 10 | 180–214 | 0.66 | 0.79 | <0.001 |
| GA127 | 10 | 205–237 | 0.40 | 0.82 | <0.001 |
| GA131 | 8 | 99–115 | 0.50 | 0.83 | <0.001 |
| GA136 | 8 | 135–159 | 0.26 | 0.76 | <0.001 |
| GA161 | 10 | 90–128 | 0.84 | 0.80 | <0.001 |
| SSRY32 | 3 | 296–300 | 0.02 | 0.35 | <0.001 |
| SSRY46 | 3 | 264–268 | 0.00 | 0.18 | <0.001 |
| SSRY70 | 5 | 245–253 | 0.15 | 0.73 | <0.001 |
| SSRY83 | 2 | 239–241 | 0.00 | 0.04 | 0.013 |
| SSRY169 | 2 | 100–102 | 0.00 | 0.17 | <0.001 |
| Mean | 6.69 | 0.28 | 0.55 | <0.001 |
| Landrace | PHt | Obs. Hs | Exp. Hs | IR | HL |
|---|---|---|---|---|---|
| amarilla | 0.33 | 1.39 | 0.66 | 0.34 | 0.48 |
| andioca | 0.31 | 1.10 | 0.56 | 0.37 | 0.57 |
| añera | 0.31 | 1.10 | 0.56 | 0.47 | 0.56 |
| arpón | 0.30 | 1.09 | 0.63 | 0.28 | 0.50 |
| blanca | 0.23 | 0.82 | 0.42 | 0.55 | 0.66 |
| blanca (dark stem) | 0.54 | 1.92 | 0.98 | -0.06 | 0.23 |
| blanca (light stem) | 0.31 | 1.10 | 0.56 | 0.43 | 0.57 |
| brava (cultivated) | 0.31 | 1.10 | 0.56 | 0.53 | 0.57 |
| brava (feral) | 0.23 | 0.82 | 0.42 | 0.57 | 0.67 |
| bufeo | 0.42 | 1.43 | 0.78 | 0.21 | 0.39 |
| cerveza | 0.31 | 1.10 | 0.56 | 0.39 | 0.56 |
| cogollo colorado | 0.00 | 0.00 | 0.00 | 1.00 | 1.00 |
| cogollo morado | 0.15 | 0.55 | 0.28 | 0.68 | 0.77 |
| colorada | 0.23 | 0.82 | 0.42 | 0.55 | 0.66 |
| gallinazo | 0.00 | 0.00 | 0.00 | 1.00 | 1.00 |
| iguano | 0.38 | 1.37 | 0.70 | 0.30 | 0.45 |
| indianino | 0.31 | 1.10 | 0.56 | 0.41 | 0.55 |
| inviernino | 0.00 | 0.00 | 0.00 | 1.00 | 1.00 |
| lobera | 0.31 | 1.10 | 0.56 | 0.41 | 0.56 |
| lobera colorada | 0.46 | 1.64 | 0.84 | 0.12 | 0.33 |
| lobera negra | 0.42 | 1.37 | 0.72 | 0.23 | 0.44 |
| lupuna | 0.00 | 0.00 | 0.00 | 1.00 | 1.00 |
| lupunillo | 0.33 | 1.10 | 0.58 | 0.38 | 0.54 |
| mano de tunche | 0.15 | 0.55 | 0.28 | 0.77 | 0.78 |
| morada | 0.31 | 1.10 | 0.56 | 0.42 | 0.62 |
| morada amarilla | 0.50 | 1.83 | 0.94 | 0.00 | 0.26 |
| motelillo | 0.38 | 1.37 | 0.70 | 0.24 | 0.45 |
| napino | 0.31 | 1.10 | 0.56 | 0.39 | 0.55 |
| palmera | 0.23 | 0.82 | 0.42 | 0.56 | 0.66 |
| palo blanco | 0.31 | 1.10 | 0.56 | 0.42 | 0.57 |
| palo negro | 0.33 | 1.18 | 0.63 | 0.29 | 0.50 |
| palo negro Antonio (narrow leaf) | 0.36 | 1.18 | 0.64 | 0.31 | 0.49 |
| palo negro Antonio (wide leaf) | 0.36 | 1.14 | 0.64 | 0.42 | 0.49 |
| palo negro Arimuya | 0.17 | 0.59 | 0.31 | 0.62 | 0.75 |
| piririca | 0.38 | 1.37 | 0.70 | 0.23 | 0.44 |
| posheco enano | 0.31 | 1.10 | 0.56 | 0.42 | 0.57 |
| posheco gigante | 0.31 | 1.10 | 0.56 | 0.38 | 0.54 |
| señorita | 0.25 | 0.88 | 0.47 | 0.48 | 0.62 |
| umishina | 0.38 | 1.37 | 0.70 | 0.23 | 0.44 |
| ungurahui | 0.46 | 1.64 | 0.84 | 0.06 | 0.35 |
| vidrio | 0.23 | 0.82 | 0.42 | 0.55 | 0.66 |
| viejillo | 0.15 | 0.55 | 0.28 | 0.72 | 0.78 |
| virginia | 0.23 | 0.82 | 0.42 | 0.60 | 0.68 |
| Minimum | 0.00 | 0.00 | 0.00 | −0.06 | 0.23 |
| Maximum | 0.54 | 1.92 | 0.98 | 1.00 | 1.00 |
| Mean | 0.29 | 1.01 | 0.52 | 0.45 | 0.59 |
| Variance Source | d.f. | Sum of Squares | Variance Component | Percent Variation | p-Value |
|---|---|---|---|---|---|
| Between rivers | 4 | 42.28 | 0.17 | 3.05 | 0.016 |
| Between landraces within rivers | 39 | 299.16 | 2.19 | 38.84 | 0.001 |
| Within landraces | 44 | 144.43 | 3.28 | 58.11 | 0.001 |
| Total | 87 | 485.88 | 5.65 | 100.00 |
| Marker | Nei’s GST | Hedrick’s G′ST | Jost’s D |
|---|---|---|---|
| GA12 | 0.017 | 0.079 | 0.059 |
| GA13 | 0.045 | 0.063 | 0.007 |
| GA21 | 0.080 | 0.307 | 0.231 |
| GA126 | −0.017 | −0.101 | −0.079 |
| GA127 | 0.016 | 0.098 | 0.079 |
| GA131 | 0.096 | 0.496 | 0.429 |
| GA136 | 0.089 | 0.308 | 0.224 |
| GA161 | 0.017 | 0.089 | 0.070 |
| SSRY32 | 0.036 | 0.064 | 0.020 |
| SSRY46 | 0.117 | 0.156 | 0.017 |
| SSRY70 | 0.119 | 0.411 | 0.311 |
| SSRY83 | 0.001 | 0.001 | 0.000 |
| SSRY169 | 0.082 | 0.114 | 0.015 |
| Global | 0.052 | 0.128 | 0.068 |
| (a) Nei’s GST. | ||||
|---|---|---|---|---|
| Itaya | Nanay | Orosa | Pintuyacu | |
| Nanay | 0.046 | |||
| Orosa | 0.040 | 0.032 | ||
| Pintuyacu | 0.037 | −0.004 | 0.035 | |
| Tahuayo | 0.015 | 0.045 | 0.046 | 0.040 |
| (b) Hedrick’s G’ST. | ||||
| Itaya | Nanay | Orosa | Pintuyacu | |
| Nanay | 0.168 | |||
| Orosa | 0.167 | 0.137 | ||
| Pintuyacu | 0.135 | −0.014 | 0.150 | |
| Tahuayo | 0.057 | 0.161 | 0.188 | 0.141 |
| (c) Jost’s D. | ||||
| Itaya | Nanay | Orosa | Pintuyacu | |
| Nanay | 0.087 | |||
| Orosa | 0.097 | 0.080 | ||
| Pintuyacu | 0.067 | −0.007 | 0.088 | |
| Tahuayo | 0.027 | 0.082 | 0.111 | 0.070 |
| Specimen | Closest Relative | River(s) of Origin | |
|---|---|---|---|
| amarilla 1 | piririca | Itaya | |
| amarilla 2 | arpón | Itaya | |
| amarilla 3 | palo blanco | Orosa | |
| amarilla 4 | palo negro Arimuya | Nanay | |
| añera 1 | lobera colorada | Itaya | |
| añera 2 | cogollo colorado | Pintuyacu | |
| brava (cult.) 1 | morada | Orosa | |
| brava (cult.) 2 | palo negro Antonio (n) | Nanay | |
| indianino 1 | inviernino | Orosa | Same river |
| indianino 2 | palmera | Orosa | Both from same river |
| napino 1 | palmera | Orosa | |
| napino 2 | brava (cult.) 1 | Orosa | |
| piririca 1 | lupuna | Orosa | |
| piririca 2 | arpón | Itaya | |
| señorita 1 | arpón | Itaya | |
| señorita 2 | brava (cult.) 1 | Nanay | |
| umishina 1 | vidrio | Tahuayo | |
| umishina 2 | palo negro Antonio (n) | Nanay | |
| ungurahui 1 | mano de tunche | Orosa | |
| amarilla 5 | lobera 1 | Tahuayo, Orosa | |
| amarilla 6 | palo negro Antonio (n) | Itaya, Nanay | |
| añera 3 | morada | Itaya, Orosa | |
| lobera 1 | vidrio | Orosa, Tahuayo | |
| lobera 2 | amarilla 1 | Orosa, Tahuayo | Each from different river |
| motelillo 1 | morada | Tahuayo, Orosa | Different river |
| motelillo 2 | motelillo 1 | Itaya, Pintuyacu | |
| motelillo 3 | palo negro | Pintuyacu, Nanay | |
| ungurahui 2 | andioca | Nanay, Pintuyacu |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wooding, S.P.; Peña, C.R. Genetic Diversity of Yuca (Manihot esculenta esculenta; Cassava, Manioc), an Indigenous Crop in the Peruvian Amazon. Diversity 2023, 15, 1158. https://doi.org/10.3390/d15121158
Wooding SP, Peña CR. Genetic Diversity of Yuca (Manihot esculenta esculenta; Cassava, Manioc), an Indigenous Crop in the Peruvian Amazon. Diversity. 2023; 15(12):1158. https://doi.org/10.3390/d15121158
Chicago/Turabian StyleWooding, Stephen Park, and César Rubén Peña. 2023. "Genetic Diversity of Yuca (Manihot esculenta esculenta; Cassava, Manioc), an Indigenous Crop in the Peruvian Amazon" Diversity 15, no. 12: 1158. https://doi.org/10.3390/d15121158
APA StyleWooding, S. P., & Peña, C. R. (2023). Genetic Diversity of Yuca (Manihot esculenta esculenta; Cassava, Manioc), an Indigenous Crop in the Peruvian Amazon. Diversity, 15(12), 1158. https://doi.org/10.3390/d15121158






