Abstract
Coccoid cyanobacteria represent an important part of cyanobacterial freshwater diversity, with many studied strains in public databases identified as Synechococcus. This is a diverse genus, both morphologically and ecologically, with a global distribution. However, many of the so-called Synechococcus-like cyanobacteria strains could represent several independent genera that require further studies. In this work, four strains of a Synechococcus-like cyanobacteria isolated from freshwater lakes and terrestrial atmophytic habitats on São Miguel and Flores Islands (Azores archipelago) were studied genetically using the 16S rRNA and 16S–23S rRNA ITS, morphologically with light and transmission electron microscopy, and ecologically. A draft genome was produced from the reference strain by Illumina sequencing, which allowed a more complete phylogenetic study and a deeper taxonomic analysis, revealing a divergent phylogenetic evolution and low ANI and AAI values (69.4% and 66.3%, respectively) to Thermosynechococcus, the closest phylogenetic genus. Although morphologically similar to Synechococcus, the 16S rRNA and genome phylogenetic analysis placed the studied strains in a clade sister to Thermosynechococcus, inside the Thermosynechococcaceae. Thus, Pseudocalidococcus azoricus gen. sp. nov. is described as a new coccoid freshwater genus and species from the Azores archipelago. A detailed comparison with similar morphological taxa is provided, supporting the separation of the new genus. The 16S rRNA with a high genetic similarity to other strains from several continents identified as Synechococcus sp. suggests that the new genus probably has a worldwide distribution. Future studies should be performed to clarify the taxonomic identity of those strains.
Keywords:
AAI; ANI; Azores; coccoid cyanobacteria; DDH; genome; new genus; phylogeny; Synechococcus; 16S rRNA 1. Introduction
Cyanobacteria are one of the most ancient organisms [1], which arose around 3500 million years ago [2]. They are present in a wide diversity of habitats, in terrestrial and aquatic ecosystems [3,4,5], and are common inhabitants of extreme environments [5,6,7]. Recent studies on cyanobacteria diversity in the Azores Islands, a remote oceanic archipelago with a great variety of suitable habitats [6,8], allowed the description of several new taxa [9,10]. Therefore, many cyanobacteria species may still be unknown, and increased sampling efforts should be taken in these remote areas, especially in less-studied habitats such as terrestrial atmophytic habitats.
The morphology of coccoid cyanobacteria is poorly characterized, compromising their taxonomical classification, which was until recently based mainly on morphological characteristics [11,12]. In recent years, a significant effort to improve the coccoid cyanobacteria taxonomy has been made using a polyphasic approach with molecular and morphological data for the description of new taxa or its reassessment [13,14], and more recently using genomic data [11,15].
Synechococcus Nägeli represents a large role, with a recognized polyphyletic nature from the most commonly studied cyanobacteria [16]. However, many strains lack apomorphic features that could aid in the morphologic identification, which often resulted only in the classification of Synechococcus-like cyanobacteria [17]. Synechococcus was traditionally classified as a benthic freshwater rod-like cyanobacteria, according to Nägeli (1849) [18], yet with several described species over the years with an increasing ecological range. Recent molecular studies have confirmed the polyphyletic nature of the genus [11,19], such as Thermosynechococcus Katoh, Itoh, Shen, and Ikeuchi [11,20], which was initially based on its ecophysiological and biochemical features, since all its strains were thermophilic [20], and was recently validated [11].
An important shift in cyanobacteria taxonomy was observed in the twentieth century, with many authors relying more on genetic data [9,12,21,22]. More recently, the fast growth of cyanobacteria genomic data has allowed for a more robust analysis [11,23,24,25], not only for taxonomic resolution [26,27] but also for ecological studies [28,29]. The relevance of this new taxonomical tool is well represented in the last cyanobacterial order and family classification update, with its results based on the available genomic data [23].
In this work, we applied a polyphasic approach to study four Synechococcus-like cyanobacteria strains from BACA (Bank of Algae and Cyanobacteria of the Azores), assigning Pseudocalidococcus azoricus gen. sp. nov. to the Thermosynechococcaceae Komárek, Strunecký, and Johansen, according to its phylogenetic placement. More importantly, Pseudocalidococcus azoricus was defined as a new genus through a combination of molecular data (genomic data, 16S rRNA, and 16S–23S rRNA ITS), morphological characters (by light and transmission electron microscopy), and ecological data. The obtained draft genome and the taxonomic analysis allowed for a better knowledge of the Synechococcus polyphyletic nature. The description of the new taxa followed the International Code of Nomenclature for algae, fungi, and plants [30].
2. Materials and Methods
2.1. Strains and Morphological Characterization
The studied strains (Table 1) were isolated from lakes and a rock wall in São Miguel Island and Flores Island (Azores archipelago), and maintained in BG11 medium at 19 °C and a 14/10 h photoperiod in the BACA culture collection. For the morphological descriptions, at least 50 cells per strain were examined using a Leica DM4 B microscope equipped with a digital camera, the Leica MC 190 HD (Leica, Wetzlar, Germany). Morphological data from the four strains were combined for taxa description.

Table 1.
Strain location of the sampling in the Azores archipelago, Portugal, and GenBank accession codes of the Pseudocalidococcus strains.
The transmission electron microscopy (TEM), biomass was preserved in 2.5% glutaraldehyde and 0.1 M cacodylate buffer, and postfixed with 2% osmium tetroxide; then, it was dehydrated in an acetone series (30%, 50%, 70%, 80%, 90%, 95%, and 100%) and embedded in Spurr’s resin [31]. Ultra-thin sections (70 nm) were placed on formvar-coated grids, contrasted by uranyl acetate and lead citrate, and analyzed with a JEOL JEM 1010 microscope.
2.2. DNA Extraction, Gene Amplification, and Sequencing
The PureLink® Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA) was used for the DNA extraction following the protocol recommended by the manufacturer for Gram-negative bacteria.
For the 16S rRNA and 16S–23S rRNA ITS region amplification, the primers 27F and 23S30R [32,33] were used in a polymerase chain reaction following the protocol described by Luz et al. (2023) [9]. Thermal cycling reactions were carried out in a ProFlex™ 3 × 32-well PCR System (Thermo Fisher, Waltham, Massachusetts, USA) using the same conditions as Taton et al. (2003) [34]. Visualization and purification of the amplified sequences followed Luz et al. (2023) [9], and amplicon sequencing was conducted as a commercial service at MACROGEN (Madrid, Spain) using the 27F, 781F, 781R, CSIF, and 23S30R primers [32,33,35,36,37].
2.3. Genome Sequencing and Assembly
The chosen reference strain (BACA0444) was produced in 50 mL cultures in BG11 medium for three weeks, and biomass was recovered through centrifugation at 7000 RCF. The same kit was used to extract DNA as for the 16S rRNA and 16S–23S rRNA ITS amplicons. However, for the elution, DNAse- and RNAse-free water was used. Sequencing was performed on an Illumina platform in NovoGene (Cambridge, UK) using NovaSeq, producing 1 G of data output.
The draft genome was assembled using the GEN-ERA assembly pipeline 2.0 [38] with SPades v3.15.3 [39] and with the metagenomics option selected. Binning was performed by CONCOCT v1.1.0 [40], and the produced binned genome was assessed for quality using CheckM v1.2.2 [41], BUSCO v5.5.0 [42], and CheckM2 v1.0.2 [43].
2.4. 16S rRNA Phylogenetic Analysis
The sequences of the studied strains were aligned with 96 sequences from other cyanobacteria retrieved from GenBank using BLAST or from the literature. Sequence alignment was carried out using MAFFT v7.520 with the G-INS-i method [44]. The final alignment contained 1076 columns. The best-fit nucleotide model was assessed using ModelFinder [45], with the selection of the GTR + G4 + I + F evolution model according to the Bayesian information criterion.
Phylogenetic trees were constructed using Bayesian inference (BI) with MrBayes v3.2.7a [46] on XSEDE through the CIPRES Science Gateway, and maximum likelihood (ML) using the IQ-Tree online version v1.6.12 [47]. Gloeobacter violaceus PCC 8105 was used as an outgroup. The following conditions were used in the BI analysis: 2.5 × 106 generations, with two runs of four Markov chains, custom parameters (temp = 0.01), sampling every 1000 generations, and a 0.25 burn-in rate (the final average standard deviation of split frequencies was equal to 0.004500). The ML analysis was carried out using the GTR + G4 + I + F model with 1000 ultrafast bootstrap replicates [48]. Trees were visualized in FigTree v1.4.4 [49], and the final composite tree from the maximum likelihood with the addition of the posterior probabilities values from the BI was redrawn with Inkscape v1.3.
2.5. Genome Analysis
Genomes were selected following a literature review and their availability in GenBank. For quality control, an analysis using BUSCO v5.5.0 [42] was performed in all retrieved genomes with the cyanobacteria_odb10 dataset lineage selected (date: 23 February 2021; number_of_BUSCOs: 773) and CheckM v1.2.2. The number of identified genes was analyzed, and genomes with low-conserved genes were removed, along with genomes with fragment genes (5 >; BUSCO analysis) and less than 95% completeness (CheckM analysis). A final dataset of 114 genomes was then used for further analysis.
An adapted and custom python pipeline based on Jamie McGowan (https://github.com/jamiemcg/BUSCO_phylogenomics.git, accessed on 31 August 2023) was used for the phylogenomic analyses, modified to better fit a prokaryote analysis and taking into account updated software, here presented as KABOOM (https://github.com/rubenluz/KABOOM.git, accessed on 31 August 2023). This python pipeline, working in a Conda environment, takes assembled genomes as input (.fasta and .fna), performs BUSCO analyses for the identification of conserved BUSCO genes, trims them, concatenates common and single-copy genes, and performs a phylogenetic analysis based on nucleotides or amino acid sequences. Briefly, common genes to all the selected genomes were retrieved and then aligned using MAFFT v7.520 [44], trimmed using trimAl v1.4.1 [50], and concatenated. Phylogenetic analysis was performed on the final concatenated alignment of 217 genes, with 65,463 columns of amino acids, using IQTREE 2.2.3 [51] with the automatic selection of the LG + F + I + R10 best-fit model according to the Bayesian information criterion by ModelFinder [45] and 1000 ultra-fast bootstrap [48]. The final bootstrap correlation coefficient of split occurrence frequencies was 1 after 102 iterations. The same approach was applied using the nucleotide option for the phylogenomic inference, with a final concatenated alignment of 217 genes with 198,274 columns of nucleotides. The model was selected by ModelFinder [45], with the best-fit model SYM + I + R10 chosen according to Bayesian information criterion. The final bootstrap correlation coefficient of split occurrence frequencies was 0.998 after 200 iterations. Trees were visualized in FigTree v1.4.4 [49] and redrawn with Inkscape v1.3.
The average nucleotide identity (ANI) and the average amino acid identity (AAI) to the closest phylogenetic and morphological taxa were calculated using orthoANI [52] and EzAAI [53], respectively. Digital DNA–DNA hybridization (DDH) was calculated using the genome-to-genome distance calculator [54].
2.6. Analyses of the 16S–23S rRNA ITS Region
For the 16S–23S rRNA ITS secondary-structure identification, M-fold was used, applying the default parameter settings [55]. The D1–D1′, Box-B, and V3 helix sequences were identified after the M-fold results and the published literature. Final structures were redrawn with Inkscape v1.3.
3. Results
Pseudocalidococcus azoricus R.F.S. Luz, J. Kaštovský, V. Gonçalves gen. sp. nov. (Figure 1).
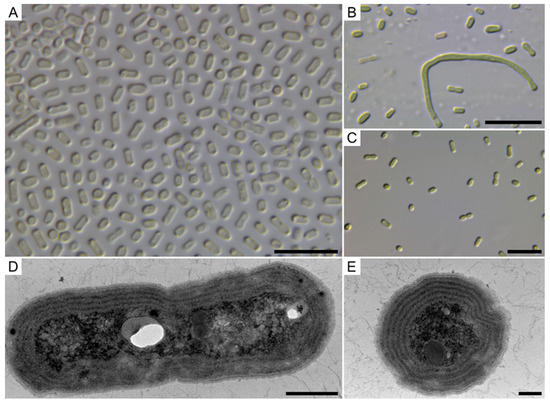
Figure 1.
Morphology of Pseudocalidococcus azoricus BACA0444 under light microscope and TEM. (A) Different cell morphologies in DIC; (B) Normal and elongated cells in DIC; (C) Cells with incomplete binary fission; (D) Transversal cut showing four parietal thylakoids; (E) Longitudinal cut showing five parietal thylakoids in the cell. Scale bars 10 µm (A–C), 500 nm (D), and 200 nm (E).
Diagnosis: Morphologically similar to Synechococcus, but with a distinct phylogenetic placement in the Thermosynechococcaceae by the 16S rRNA phylogeny and phylogenomic analysis. Differs from Thermosynechococcus ecologically, as Pseudocalidococcus is a freshwater and Thermosynechococcus is strictly thermal, and genomically, both in its phylogenetic position and low AAI (66.3%) and ANI (69.4%).
Description: Cells solitary or arranged in small clusters. Without mucilage or any evident envelops. Cells blue-green, cylindrical, rod-shaped (sometimes slightly arcuate) to elongated cylindrical, and occasionally slightly widened at both ends. Cells 1.6–6.5 µm in length (mean = 2.9 µm) and 0.8–2.0 µm wide (mean = 1.4 µm), with a length/width ratio of 1.1–6.3 (mean = 2.2). Observed elongated cells were up to 45 µm in length. Cell division perpendicular to the long axis of the cells. Thylakoids present in a parietal arrangement; up to five.
Holotype: Dried material preserved in a permanently inactive state at Herbário Ruy Telles Palhinha, University of Azores, Portugal, under the AZB 4202 code.
Type locality: Lagoa Comprida, Lajes, Flores Island (Azores archipelago, Portugal); 39°26′26.052″ N 31°13′19.0128″ W, collected by the MONITAIA team project.
Habitat: Aquatic in freshwater lakes and atmophytic.
Etymology: Pseudocalidococcus: Pseudo (fake)—calidum (hot)—coccus: fake thermal cyanobacteria, as it is positioned inside a supposedly thermal family; masculine gender; azoricus: isolated from the Azores archipelago.
Reference strain: BACA0444 (Bank of Algae and Cyanobacteria of the Azores, Azores, Portugal), isolated by Rita Cordeiro.
Gene Sequences: GenBank accession number OM732240 for the 16S rRNA and 16S–23S rRNA ITS genes and GenBank accession number GCA_031729055 for the genome assembly.
3.1. Morphological Analysis
The four strains studied in this work have very similar morphological characteristics (Table 2) despite originating from different ecosystems in the Azores; namely, from a small lake in Furnas village (São Miguel Island), Lake Empadadas Norte (São Miguel Island), Lake Comprida (Flores Island), and an atmophytic site in Ribeira Grande (São Miguel Island). This represents a large geographical distance of separated populations from where the strains of P. azoricus were isolated.

Table 2.
Cell dimensions in the four studied strains, with the minimum, maximum, and arithmetic mean of the length and width in micrometers. The P. azoricus cell dimensions correspond to the combined values of the four strains.
The four strains presented the same morphological cell characteristics, but not all presented the elongated cells, as seen in Figure 1B, with up to five parietally arranged thylakoids (Figure 1E). An attempt was made to compare the morphological characteristics of our strains with the strains that fall within the Pseudocalidococcus phylogenetic clade, but no description was found in the literature.
3.2. 16S rRNA Phylogeny and 16S–23S ITS Secondary Structures
The four Azorean strains were grouped together with other Synechococcus sp. strains (EO68, CHAB TP201738, IPPAS B-1202, PCC 6312, and PCC 6603) in the 16S rRNA phylogenetic analysis (Figure 2), near Thermosynechococcus, with strong support (100 ML, 1 BI), suggesting the position of Pseudocalidococcus in the Thermosynechococcaceae. Furthermore, the Synechococcus sp. strains positioned in the cluster of Pseudocalidococcus azoricus are genetically closely related and must all belong to the genus Pseudocalidococcus. Therefore, Pseudocalidococcus has a wide geographical distribution, being present at least in the United States of America (a freshwater pond in California, PCC 6603 strain) and Kazakhstan (Issyk Lake, IPPAS B-1202 strain), besides the Azores archipelago.
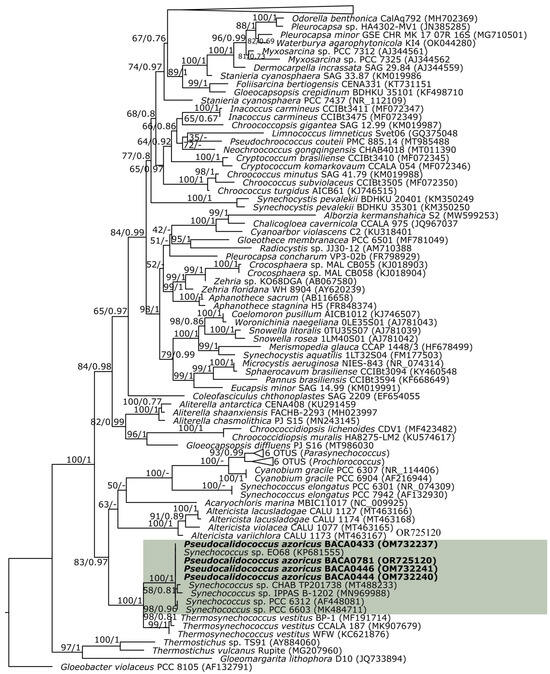
Figure 2.
16S rRNA partial maximum likelihood (ML) phylogenetic tree. Bootstrap values for the maximum likelihood and posterior probabilities for the Bayesian inference are indicated on the tree. The studied strains are in bold font. The new genus is in the green shade.
The secondary structure of the D1-D1′ and Box-B helix of the 16S–23S rRNA ITS is shown in Figure 3. As expected and reinforcing the distance of the Pseudocalidococcus genus to Thermosynechococcus, a large difference was observed between the folded structures in both the sequence and folding. A marked difference can be seen in the formation of the different lateral bulges in both genera in the D1-D1′ helix and in the Box-B helix in the mid-internal loop of Pseudocalidococcus, in contrast with its absence in Thermosynechococcus.
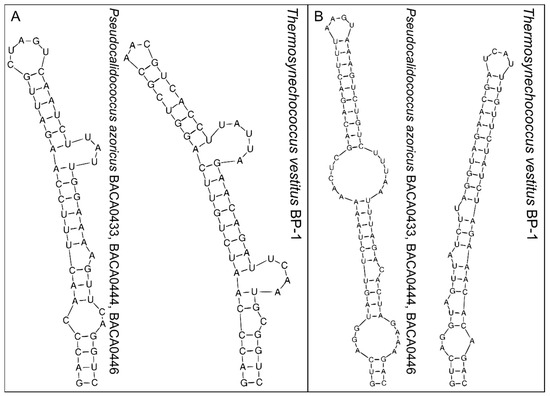
Figure 3.
The 16S–23S rRNA ITS secondary structures of the D1-D1′ (A) and Box-B (B) region of Pseudocalidococcus azoricus BACA0444 and the closest phylogenetic species of Thermosynechococcus vestitus BP-1.
3.3. Genomic Analysis
The produced genome is of high quality, with 34 contigs, a size of 3,463,985 base pairs (Figure 4), and a GC content of 48.7%. Quality control showed a 99.53% completeness and a 0.12% contamination according to CheckM v1.2.2, a 99.8% completeness and a 0.0% contamination according to CheckM2 v1.0.2, and a 98.4% completeness according to BUSCO v5.5.0. Assembly data and annotation statistics to the closest phylogenetic genera are presented in Table 3.
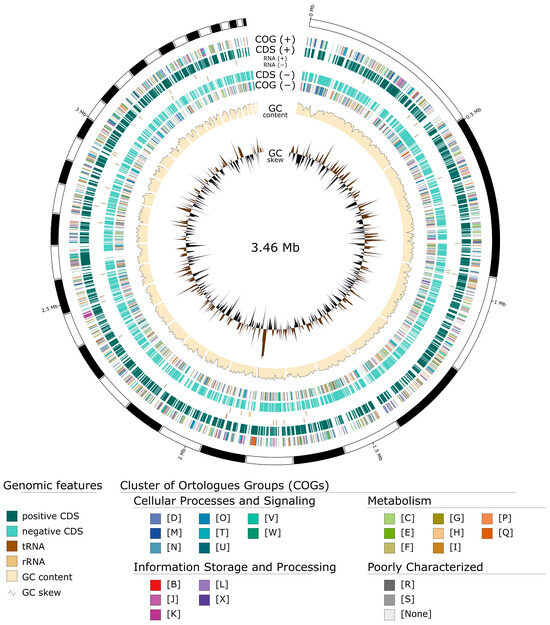
Figure 4.
Circular genome representation of Pseudocalidococcus azoricus BACA0444 using GenoVi [56].

Table 3.
Genome data from Pseudocalidococcus azoricus and the closest phylogenetic genera. Statistical data of assembly were retrieved from CheckM v1.2.2 and Bakta v1.8.2.
The phylogenomic analysis placed the new genus in the same position as the 16S rRNA phylogenetic analysis, confirming its similarity with Thermosynechococcus, with a good bootstrap support of 100 (Figure 5). The ANI and AAI analysis supported the gene separation against Thermosynechococcus, with low values (below 70%). The ANI, AAI, and DDH supported the presence of at least two species in the genus, following the genomic analysis of the available data.
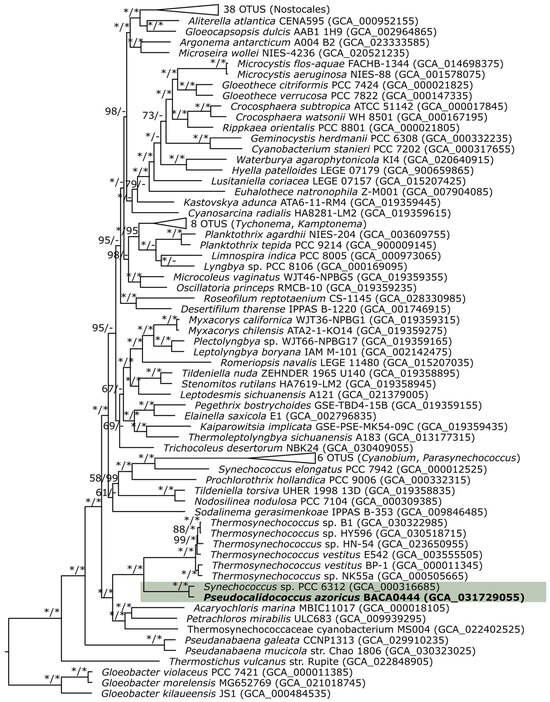
Figure 5.
Amino acid maximum likelihood phylogenomic tree with 114 OTUS of 217 concatenated BUSCO genes. Bootstrap values for the maximum likelihood based on amino acids and nucleotides are indicated on the tree. The new genus is in the green shade. * 100% bootstrap.
4. Discussion
The new genus Pseudocalidococcus is phylogenetically closely related to Thermosynechococcus. However, it is noteworthy that Thermosynechococcus is recognized as a genus strictly associated with thermal environments [11,20], and Pseudocalidococcus strains have been isolated, not only from freshwater lakes, but also from an atmophytic habitat on a rock wall. This habitat distribution suggests that Pseudocalidococcus is primarily thermotolerant rather than thermophilic. Genetically, the 16S rRNA pairwise distance (Table 4) is slightly above (94.8%) the recommended minimum threshold values of 94.5% for the 16S rRNA [57]. However, the combination of the 16S rRNA phylogenetic distance (Figure 1), the genomic analysis, the ANI and AAI values, and the phylogenomic analysis strongly support the creation of the new genus, Pseudocalidococcus.

Table 4.
The 16S pairwise distance similarity, the 16S rRNA ITS pairwise distance similarity, and the ANI, AAI, and DDH (identities/HSP length) percentages.
Compared to Synechococcus, Pseudocalidococcus is morphologically very similar. Pseudocalidococcus present the same type of involution/elongated cells when in culture, as described for the type species Synechococcus elongatus [58,59]. However, phylogenetically, it is distantly placed from S. elongatus PCC 6301, the currently accepted reference strain. The phylogenetic distance provides strong support for recognizing the difference between these genera.
Pseudocalidococcus azoricus falls within the general morphological cell description of Synechococcus nidulans [59]. However, this can be very problematic, as the validity of the latter is questionable. Synechococcus nidulans is a comb., cited in Bourrelly (1970) [60], with the basionym of Lauterbornia nidulans (Richter) Pringsheim [60], and L. nidulans with the basionym of Aphanothece nidulans Richter [61]. Thus, Aphanothece nidulans and Synechococcus nidulans, both currently considered valid, have the same holotype, which is not taxonomically acceptable. In the same year, Komárek (1970) [58] describes Synechococcus leopoliensis comb. nov., arguing that Aphanothece nidulans and Lauterbornia nidulans are different taxa, including the latter as a synonym of Synechococcus leopoliensis [58]. To increase the complexity of the subject, the strain used by Pringsheim (1968) [61] for Lauterbornia description [61] was Kratz-Allen/Bloom 625, which are synonyms of PCC 6301, CCAP 1405/1, and SAG 1402-1 [58,62]. PCC 6301 is the reference strain of Synechococcus [63], and the currently accepted neotype of Synechococcus elongatus [62,64].
Therefore, Synechococcus nidulans is a nom. inval., as it has no valid holotype (it is invalid under the International Code of Nomenclature for algae, fungi, and plants), and only Aphanothece nidulans is still valid, as previously confirmed [58]. Under these terms, Lauterbornia nidulans should be considered as a synonym for Synechococcus elongatus, as they are all based on the same strain, with morphological differences probably related to culture conditions and/or long-term maintenance [65]. This approach allows the separation of Synechococcus leopoliensis, which should be regarded as a synonym of Romeria leopoliensis, as suggested by Komárek and Anagnostidis (2005) [66]. Considering that Synechococcus nidulans is a nom. inval., we disregard this taxon for the proposal of the new species. However, as there are many reports of this taxon in the literature, its validity should be reassessed as soon as possible.
In the Pseudocalidococcus clade, genomic data are only available for the strains P. azoricus BACA0444 and Pseudocalidococcus sp. PCC 6312. The 16S rRNA similarity of these two strains is quite high, with only a 0.2% difference, indicating that these strains belong to the same species, following the criteria of 98.7% for species delimitation in bacteria [67,68]. However, the ANI and AAI values (90.0% and 92.0%, respectively) are well below the recommended 95% threshold for species separation [69,70], supporting the possible separation of these strains in two distinct species. This hypothesis was also supported by the DDH analysis, with a value of 39.5%, considered a good support value for species distinction [71,72]. These contradictory results can be problematic, as the 16S rRNA similarity has been used as a reference for species delimitation [21]. The 98.7% recommended threshold value [67,68] is widely used, and this value is based on DNA–DNA hybridization and the correlation that exists between the 98.7% 16S rRNA values and the 70% DNA–DNA hybridization value, which is the gold standard for microbial species delimitation [71,72]. This pattern was also observed in the Thermosynechococcus strains, as the 16S rRNA similarity (recommended 98.7%) does not match with the ANI and DDH similarities (suggested as 95% and 70%, respectively); instead, a difference of only 0.4–0.7% 16S rRNA similarity corresponds to a more than 10% divergence in the ANI values, and much lower than 70% in the DDH.
To our knowledge, no generic value is accepted for genera distinction using genomic ANI or AAI criteria. Based on the 16S rRNA, a 94.5% similarity is suggested as the threshold for genera separation [57]. In the Cyanophyceae, these values are often not followed, e.g., in Nostocales phylogenetic studies, as different values are applied, resulting in some confusion [73]. The sole use of general genetic threshold values from bacterial broad studies, which normally do not even include cyanobacteria data due to the lack of available genomes, must be avoided. Future cyanobacteria taxonomic studies should adopt a heuristic approach, integrating traditional markers (morphological and amplicons) and genomic data. The 16S rRNA analysis must be complemented with a deep genomic approach, including phylogenomic, ANI, AAI, DDH, and other criteria that might support the new taxa (e.g., GC content, coding density, and number of genes), and through the use of a pangenome analysis [24,26].
5. Conclusions
This work provided a concise description of a new coccoid cyanobacteria, Pseudocalidococcus azoricus gen. sp. nov., using a polyphasic approach. This approach allowed the separation of what would appear to be a Synechococcus nidulans strain to a new and well-defined genus that probably has a global distribution. With the predictable future increase in genomic data, this study provides a new perspective on the values that should be applied in cyanobacteria taxonomy. The growing accessibility of genomic data and the increase in available software or pipelines, such as KABOOM, that facilitate the recovery and use of genomes or metagenomes should be considered in new taxa descriptions, as they bring important insights when discussing closely related taxa with few differentiating morphological characteristics. Our results reinforce the need for deeper studies in cyanobacteria taxonomy, with larger datasets to clarify if the minimum values suggested for species and genera delimitation can be blindly applied. Using such criteria in cyanobacteria may be too conservative and undermine the knowledge of cyanobacterial diversity.
Author Contributions
Conceptualization, R.L. and V.G.; methodology, R.L., R.C. and J.K.; software, R.L.; formal analysis, R.L., J.K. and R.C.; data curation, R.L. and R.C.; writing—original draft preparation, R.L.; writing—review and editing, R.L., R.C., J.K., A.F., R.U., V.V. and V.G; funding acquisition, J.K. and V.G. All authors have read and agreed to the published version of the manuscript.
Funding
Rúben Luz was supported by a Ph.D. grant (M3.1.a/F/002/2020) from the Fundo Regional da Ciência e Tecnologia (FRCT). This work was funded by FEDER funds through the Interreg-MAC 2014-2020 Program under the projects REBECA—Red de excelencia en biotecnología azul (algas) de la región macaronesia (MAC1.1a/060) and REBECA-CCT—Red de Excelencia en Biotecnología Azul de la Región Macaronésica. Consolidación, Certificación y Transferencia (MAC2/1.1b/269), and by Portuguese National Funds, through FCT—Fundação para a Ciência e a Tecnologia, the European Union, QREN, FEDER, and COMPETE, by funding the CIBIO/InBIO (project UIDB/50027/2020 and UIDP/50027/2020). CIIMAR acknowledges funding by the FCT through UIDB/04423/2020 and UIDP/04423/2020. This work was also funded by the Grant Agency of the Czech Republic, 22-06374S.
Data Availability Statement
KABOOM is written in python and is freely available at GitHub (https://github.com/rubenluz/KABOOM.git, accessed on 31 August 2023).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mareš, J.; Hrouzek, P.; Kaňa, R.; Ventura, S.; Strunecký, O.; Komárek, J. The Primitive Thylakoid-Less Cyanobacterium Gloeobacter Is a Common Rock-Dwelling Organism. PLoS ONE 2013, 8, e66323. [Google Scholar] [CrossRef]
- Schirrmeister, B.E.; Gugger, M.; Donoghue, P.C.J. Cyanobacteria and the Great Oxidation Event: Evidence from genes and fossils. Palaeontology 2015, 58, 769–785. [Google Scholar] [CrossRef]
- Whitton, B.A.; Potts, M. Introduction to the Cyanobacteria. In Ecology of Cyanobacteria II: Their Diversity in Space and Time; Springer: Dordrecht, The Netherlands, 2012; pp. 1–13. ISBN 9789400738553. [Google Scholar]
- Scott, J.T.; Marcarelli, A.M. Cyanobacteria in Freshwater Benthic Environments. In Ecology of Cyanobacteria II; Springer: Dordrecht, The Netherlands, 2012; pp. 271–289. [Google Scholar]
- Komárek, J.; Johansen, J.R. Coccoid Cyanobacteria. In Freshwater Algae of North America; Elsevier: Amsterdam, The Netherlands, 2015; pp. 75–133. [Google Scholar]
- Luz, R.; Cordeiro, R.; Fonseca, A.; Raposeiro, P.M.; Gonçalves, V. Distribution and diversity of cyanobacteria in the Azores Archipelago: An annotated checklist. Biodivers. Data J. 2022, 10, e87638. [Google Scholar] [CrossRef]
- Cordeiro, R.; Luz, R.; Vasconcelos, V.; Fonseca, A.; Gonçalves, V. A Critical Review of Cyanobacteria Distribution and Cyanotoxins Occurrence in Atlantic Ocean Islands. Cryptogam. Algol. 2020, 41, 73. [Google Scholar] [CrossRef]
- Luz, R.; Cordeiro, R.; Vilaverde, J.; Raposeiro, P.; Fonseca, A.; Gonçalves, V. Cyanobacteria from freshwater lakes in the Azores archipelago, Portugal: Data from long term phytoplankton monitoring. Biodivers. Data J. 2020, 8, e51928. [Google Scholar] [CrossRef]
- Luz, R.; Cordeiro, R.; Kaštovský, J.; Johansen, J.R.; Dias, E.; Fonseca, A.; Urbatzka, R.; Vasconcelos, V.; Gonçalves, V. New terrestrial cyanobacteria from the Azores Islands: Description of Venetifunis gen. nov. and new species of Albertania, Kovacikia and Pegethrix. Phycologia 2023, 62, 483–498. [Google Scholar] [CrossRef]
- Luz, R.; Cordeiro, R.; Kaštovský, J.; Johansen, J.R.; Dias, E.; Fonseca, A.; Urbatzka, R.; Vasconcelos, V.; Gonçalves, V. Description of Four New Filamentous Cyanobacterial Taxa from Freshwater Habitats in the Azores Archipelago. J. Phycol. 2023. [Google Scholar] [CrossRef]
- Komárek, J.; Johansen, J.R.; Šmarda, J.; Strunecký, O. Phylogeny and taxonomy of Synechococcus-like cyanobacteria. Fottea 2020, 20, 171–191. [Google Scholar] [CrossRef]
- Mareš, J.; Johansen, J.R.; Hauer, T.; Zima, J.; Ventura, S.; Cuzman, O.; Tiribilli, B.; Kaštovský, J. Taxonomic resolution of the genus Cyanothece (Chroococcales, Cyanobacteria), with a treatment on Gloeothece and three new genera, Crocosphaera, Rippkaea, and Zehria. J. Phycol. 2019, 55, 578–610. [Google Scholar] [CrossRef]
- Jung, P.; Azua-Bustos, A.; Gonzalez-Silva, C.; Mikhailyuk, T.; Zabicki, D.; Holzinger, A.; Lakatos, M.; Büdel, B. Emendation of the coccoid cyanobacterial genus Gloeocapsopsis and description of the new species Gloeocapsopsis diffluens sp. nov. and Gloeocapsopsis dulcis sp. nov. isolated from the coastal range of the Atacama Desert (Chile). Front. Microbiol. 2021, 12, 671742. [Google Scholar] [CrossRef]
- Pokorný, J.; Štenclová, L.; Kaštovský, J. Unsuspected findings about phylogeny and ultrastructure of the enigmatic cyanobacterium Microcrocis geminata resulted in its epitypification and novel placement in Geminocystaceae. Fottea 2023, 23, 110–121. [Google Scholar] [CrossRef]
- Pessi, I.S.; Popin, R.V.; Durieu, B.; Lara, Y.; Tytgat, B.; Savaglia, V.; Roncero-Ramos, B.; Hultman, J.; Verleyen, E.; Vyverman, W.; et al. Novel diversity of polar Cyanobacteria revealed by genome-resolved metagenomics. Microb. Genom. 2023, 9, 001056. [Google Scholar] [CrossRef] [PubMed]
- Dvořák, P.; Casamatta, D.A.; Poulíčková, A.; Hašler, P.; Ondřej, V.; Sanges, R. Synechococcus: 3 billion years of global dominance. Mol. Ecol. 2014, 23, 5538–5551. [Google Scholar] [CrossRef] [PubMed]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Nägeli, C. Gattungen Einzelliger Algen: Physiologisch und Systematisch Bearbeitet; Friedrich Schulthess: Zurich, Switzerland, 1849; Volume 10. [Google Scholar]
- Callieri, C. Synechococcus plasticity under environmental changes. FEMS Microbiol. Lett. 2017, 364, fnx229. [Google Scholar]
- Katoh, H.; Itoh, S.; Shen, J.-R.; Ikeuchi, M. Functional Analysis of psbV and a Novel c-type Cytochrome Gene psbV2 of the Thermophilic Cyanobacterium Thermosynechococcus elongatus Strain BP-1. Plant Cell Physiol. 2001, 42, 599–607. [Google Scholar]
- Mai, T.; Johansen, J.R.; Pietrasiak, N.; Bohunická, M.; Martin, M.P. Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. Phytotaxa 2018, 365, 1–59. [Google Scholar] [CrossRef]
- Kaštovský, J.; Johansen, J.R.; Hauerová, R.; Akagha, M.U. Hot Is Rich—An Enormous Diversity of Simple Trichal Cyanobacteria from Yellowstone Hot Springs. Diversity 2023, 15, 975. [Google Scholar] [CrossRef]
- Strunecký, O.; Ivanova, A.P.; Mareš, J. An updated classification of cyanobacterial orders and families based on phylogenomic and polyphasic analysis. J. Phycol. 2022, 59, 12–51. [Google Scholar] [CrossRef]
- Stanojković, A.; Skoupý, S.; Škaloud, P.; Dvořák, P. High genomic differentiation and limited gene flow indicate recent cryptic speciation within the genus Laspinema (cyanobacteria). Front. Microbiol. 2022, 13, 977454. [Google Scholar] [CrossRef]
- Cai, H.; Mclimans, C.J.; Beyer, J.E.; Krumholz, L.R.; David Hambright, K. Microcystis pangenome reveals cryptic diversity within and across morphospecies. Sci. Adv. 2023, 9, eadd3783. [Google Scholar] [CrossRef] [PubMed]
- Dvořák, P.; Jahodářová, E.; Stanojković, A.; Skoupý, S.; Casamatta, D.A. Population genomics meets the taxonomy of cyanobacteria. Algal Res. 2023, 72, 103128. [Google Scholar]
- Willis, A.; Woodhouse, J.N. Defining Cyanobacterial Species: Diversity and Description Through Genomics. CRC Crit. Rev. Plant Sci. 2020, 39, 101–124. [Google Scholar] [CrossRef]
- Dick, G.J.; Duhaime, M.B.; Evans, J.T.; Errera, R.M.; Godwin, C.M.; Kharbush, J.J.; Nitschky, H.S.; Powers, M.A.; Vanderploeg, H.A.; Schmidt, K.C.; et al. The genetic and ecophysiological diversity of Microcystis. Environ. Microbiol. 2021, 23, 7278–7313. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.Y.; Teng, W.K.; Zhao, L.; Hu, C.X.; Zhou, Y.K.; Han, B.P.; Song, L.R.; Shu, W.S. Comparative genomics reveals insights into cyanobacterial evolution and habitat adaptation. ISME J. 2021, 15, 211–227. [Google Scholar] [CrossRef]
- Turland, N.; Wiersema, J.; Barrie, F.; Greuter, W.; Hawksworth, D.; Herendeen, P.; Knapp, S.; Kusber, W.-H.; Li, D.-Z.; Marhold, K.; et al. International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code): Adopted by the Nineteenth International Botanical Congress, Shenzhen, China; Koeltz Botanical Books: Oberreifenberg, Germany, 2017; ISBN 9783946583165. [Google Scholar]
- Spurr, A.R. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 1969, 26, 31–43. [Google Scholar] [CrossRef]
- Neilan, B.A.; Jacobs, D.; Del Dot, T.; Blackall, L.L.; Hawkins, P.R.; Cox, P.T.; Goodman, A.E. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int. J. Syst. Bacteriol. 1997, 47, 693–697. [Google Scholar] [CrossRef]
- Lepère, C.; Wilmotte, A.; Meyer, B. Molecular diversity of Microcystis strains (Cyanophyceae, Chroococcales) based on 16S rDNA sequences. Syst. Geogr. Plants 2000, 70, 275–283. [Google Scholar] [CrossRef]
- Taton, A.; Grubisic, S.; Brambilla, E.; De Wit, R.; Wilmotte, A. Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): A morphological and molecular approach. Appl. Environ. Microbiol. 2003, 69, 5157–5169. [Google Scholar] [CrossRef]
- Nübel, U.; Garcia-Pichel, F.; Muyzer, G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 1997, 63, 3327–3332. [Google Scholar] [CrossRef]
- Cordeiro, R.; Azevedo, J.; Luz, R.; Vasconcelos, V.; Gonçalves, V.; Fonseca, A. Cyanotoxin Screening in BACA Culture Collection: Identification of New Cylindrospermopsin Producing Cyanobacteria. Toxins 2021, 13, 258. [Google Scholar] [CrossRef] [PubMed]
- Janse, I.; Kardinaal, W.E.A.; Meima, M.; Fastner, J.; Visser, P.M.; Zwart, G. Toxic and nontoxic Microcystis colonies in natural populations can be differentiated on the basis of rRNA gene internal transcribed spacer diversity. Appl. Environ. Microbiol. 2004, 70, 3979–3987. [Google Scholar] [CrossRef] [PubMed]
- Cornet, L.; Durieu, B.; Baert, F.; D’hooge, E.; Colignon, D.; Meunier, L.; Lupo, V.; Cleenwerck, I.; Daniel, H.M.; Rigouts, L.; et al. The GEN-ERA toolbox: Unified and reproducible workflows for research in microbial genomics. Gigascience 2023, 12, giad022. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Chklovski, A.; Parks, D.H.; Woodcroft, B.J.; Tyson, G.W. CheckM2: A rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. Nat. Methods 2023, 20, 1203–1212. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Sy Vinh, L.; Rosenberg, M.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2017, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v1.4. Molecular Evolution, Phylogenetics and Epidemiology 2012. Available online: https://github.com/rambaut/figtree (accessed on 30 August 2023).
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R.; Teeling, E. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Lee, I.; Kim, Y.O.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Kim, D.; Park, S.; Chun, J. Introducing EzAAI: A pipeline for high throughput calculations of prokaryotic average amino acid identity. J. Microbiol. 2021, 59, 476–480. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Cumsille, A.; Durán, R.E.; Rodríguez-Delherbe, A.; Saona-Urmeneta, V.; Cámara, B.; Seeger, M.; Araya, M.; Jara, N.; Buil-Aranda, C. GenoVi, an open-source automated circular genome visualizer for bacteria and archaea. PLoS Comput. Biol. 2023, 19, e1010998. [Google Scholar] [CrossRef]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Komárek, J. Generic identity of the “Anacystis nidulans” strain KRATZ-ALLEN/Bloom. 625 with Synechococcus NÄG. 1849. Arch Protistenkd. 1970, 112, 343–364. [Google Scholar]
- Komárek, J.; Anagnostidis, K. Süßwasserflora von Mitteleuropa: Cyanoprokaryota 1; Teil: Chroococcales; Ettl, H., Gerloff, J., Heynig, H., Mollenhauer, D., Eds.; Spektrum Akademischer Verlag: Heidelberg, Germany, 2000. [Google Scholar]
- Bourrelly, P. Les Algues d’Eau Douce, Initiation à La Systématique, Tome III: Les Algues Bleues et Rouges, Les Eugléniens, Peridiniens et Cryptomonadines; Boubée N. et Cie: Paris, France, 1970. [Google Scholar]
- Pringsheim, E.G. Kleine Mitteilungen über Flagellaten und Algen. XVI. Lauterbornia (Anacystis) nidulans (Richter) nov. gen. comb. Cyanophyceae. Arch. Mikrobiol. 1968, 63, 1–6. [Google Scholar] [CrossRef]
- Rippka, R.; Cohen-Bazire, G. The cyanobacteriales: A legitimate order based on the type strain Cyanobacterium stanieri? Ann. Microbiol. 1983, 134, 21–36. [Google Scholar] [CrossRef] [PubMed]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Stanier, R.Y.; Kunisawa, R.; Mandel, M.; Cohen-Bazire, G. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 1971, 35, 171–205. [Google Scholar] [CrossRef]
- Lopez-Igual, R.; Mazel, D.; Gugger, M.; Golden, S.S.; Adomako, M.; Ernst, D.; Simkovsky, R.; Chao, Y.Y.; Wang, J.; Fang, M.; et al. Comparative Genomics of Synechococcus elongatus Explains the Phenotypic Diversity of the Strains. mBio 2022, 13, e00862-22. [Google Scholar] [CrossRef]
- Komárek, J.; Anagnostidis, K. Süßwasserflora von Mitteleuropa, Bd. 19/2: Cyanoprokaryota: Bd. 2/Part 2: Oscillatoriales, 1st ed.; Spektrum Akademischer Verlag: Heidelberg, Germany, 2005. [Google Scholar]
- Stackebrandt, E.; Ebers, J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 2006, 33, 152–155. [Google Scholar]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Varghese, N.J.; Mukherjee, S.; Ivanova, N.; Konstantinidis, K.T.; Mavrommatis, K.; Kyrpides, N.C.; Pati, A. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015, 43, 6761–6771. [Google Scholar] [CrossRef] [PubMed]
- Tindall, B.J.; Rosselló-Móra, R.; Busse, H.J.; Ludwig, W.; Kämpfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010, 60, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Kabirnataj, S.; Nematzadeh, G.A.; Talebi, A.F.; Saraf, A.; Suradkar, A.; Tabatabaei, M.; Singh, P. Description of novel species of Aliinostoc, Desikacharya and Desmonostoc using a polyphasic approach. Int. J. Syst. Evol. Microbiol. 2020, 70, 3413–3426. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).