Differences in the Composition of Akkermansia Species and Families of Christensenellaceae and Ruminococcaceae Bacteria in the Gut Microbiota of Healthy Polish Women following a Typical Western Diet
Abstract
:1. Introduction
2. Material and Methods
2.1. Participants and Biological Material
- -
- part A: Eating habits;
- -
- part B: Frequency of food consumption;
- -
- part C: Views on food and nutrition;
- -
- part D: Lifestyle and personal data.
2.2. Genetic Library Construction
2.3. Statistical Analysis
2.4. Ethical Considerations
3. Results
3.1. Participants
3.2. Alpha and Beta Biodiversity
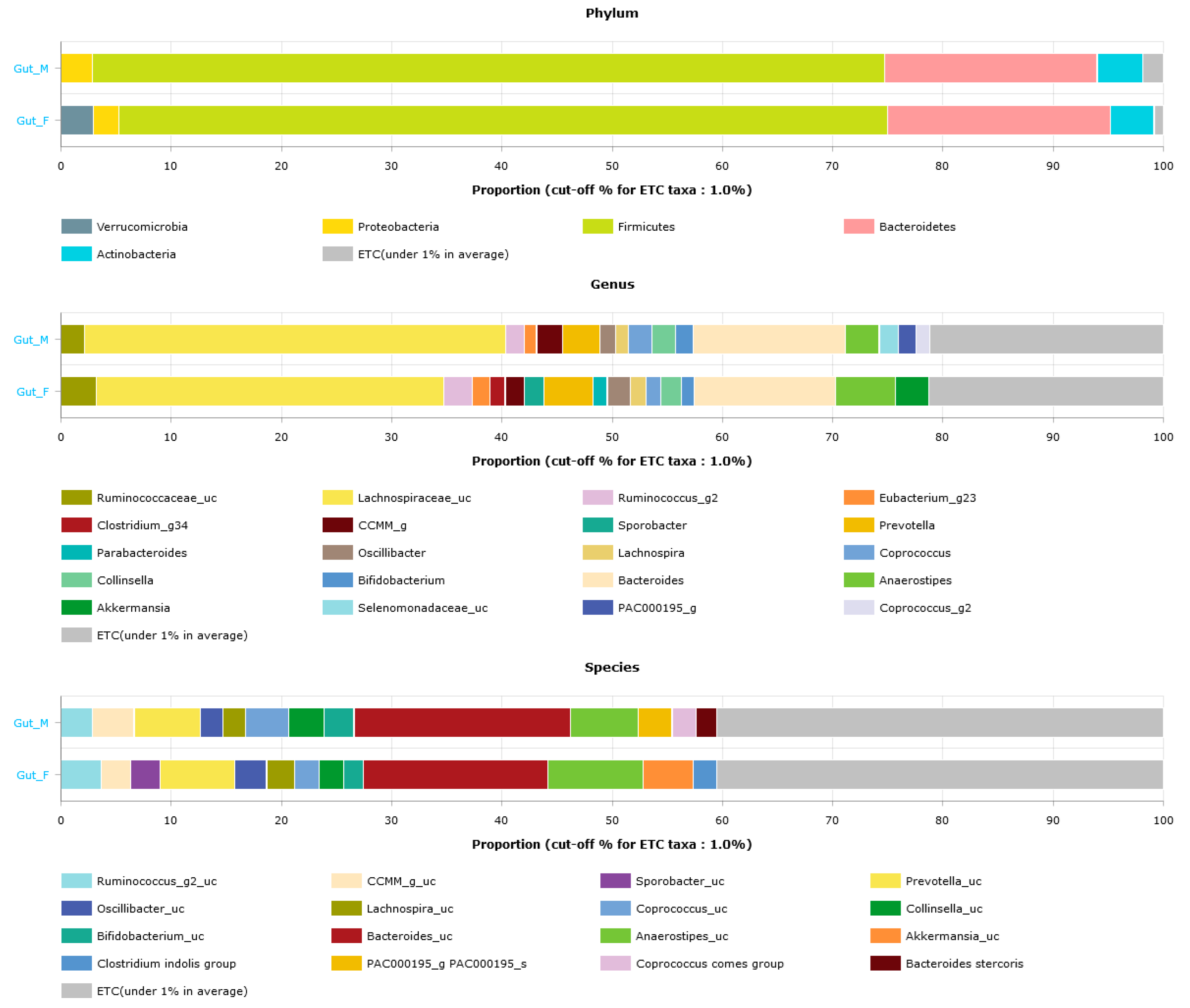
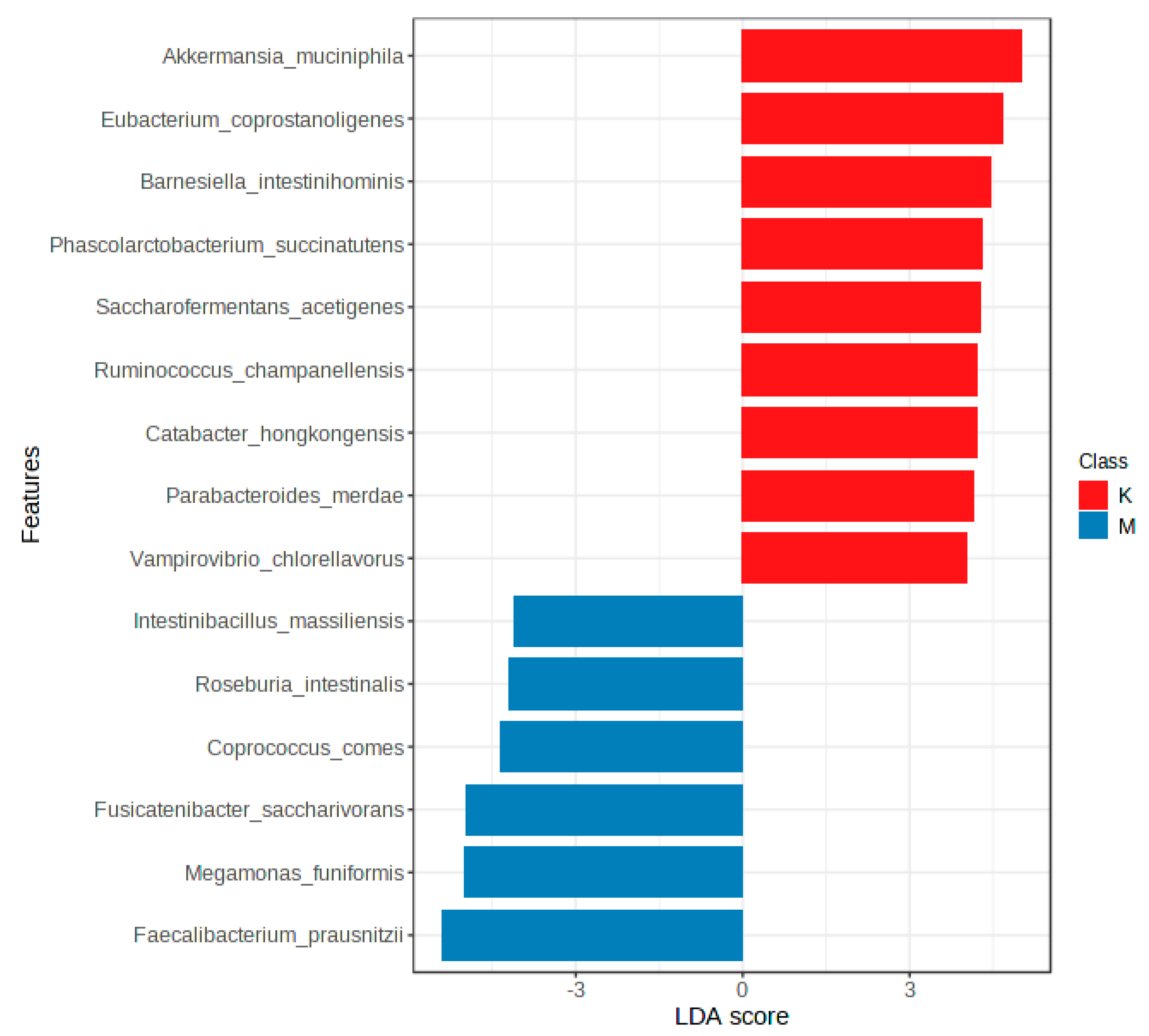
3.3. Gut Microbiota Profiles
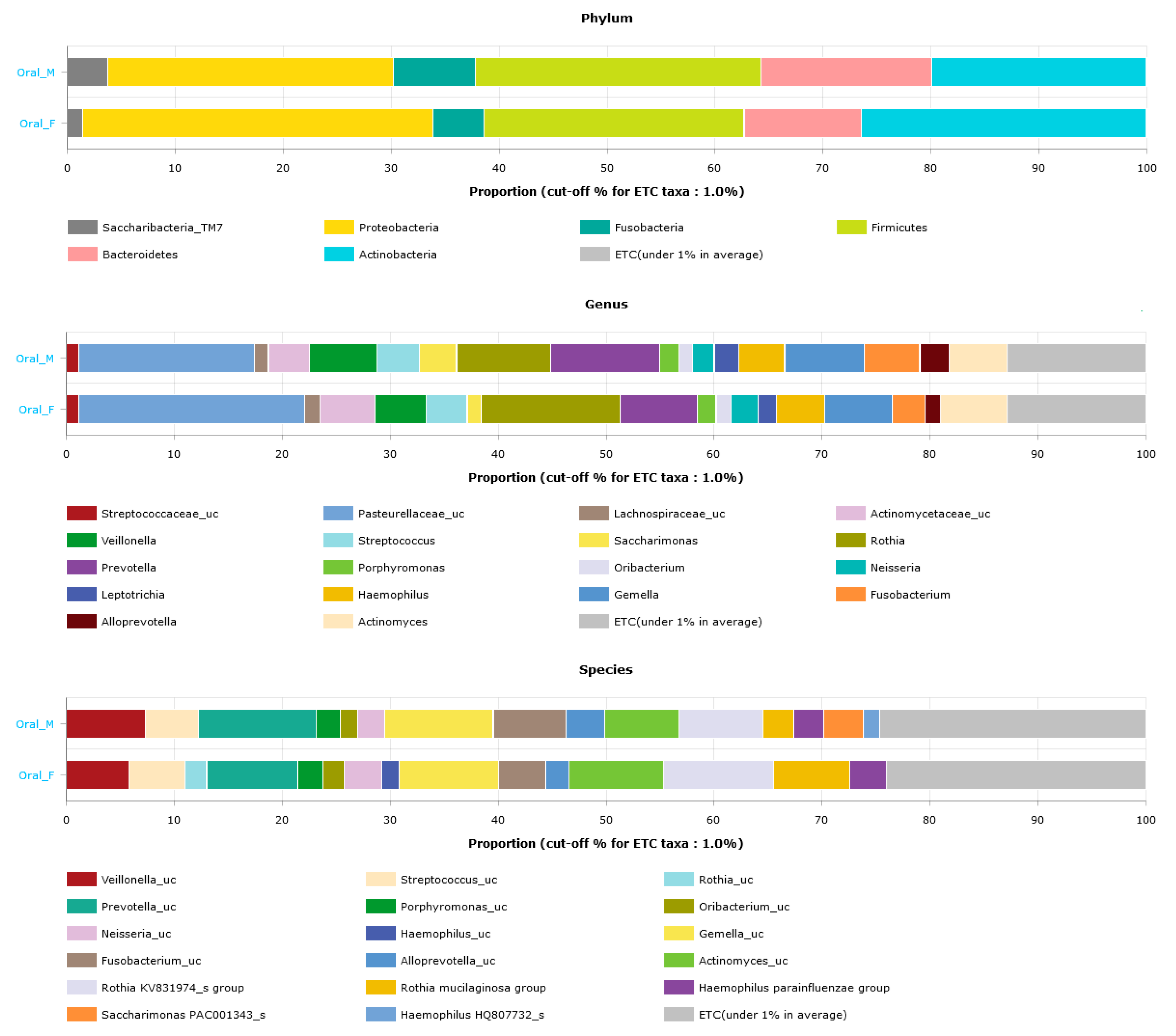
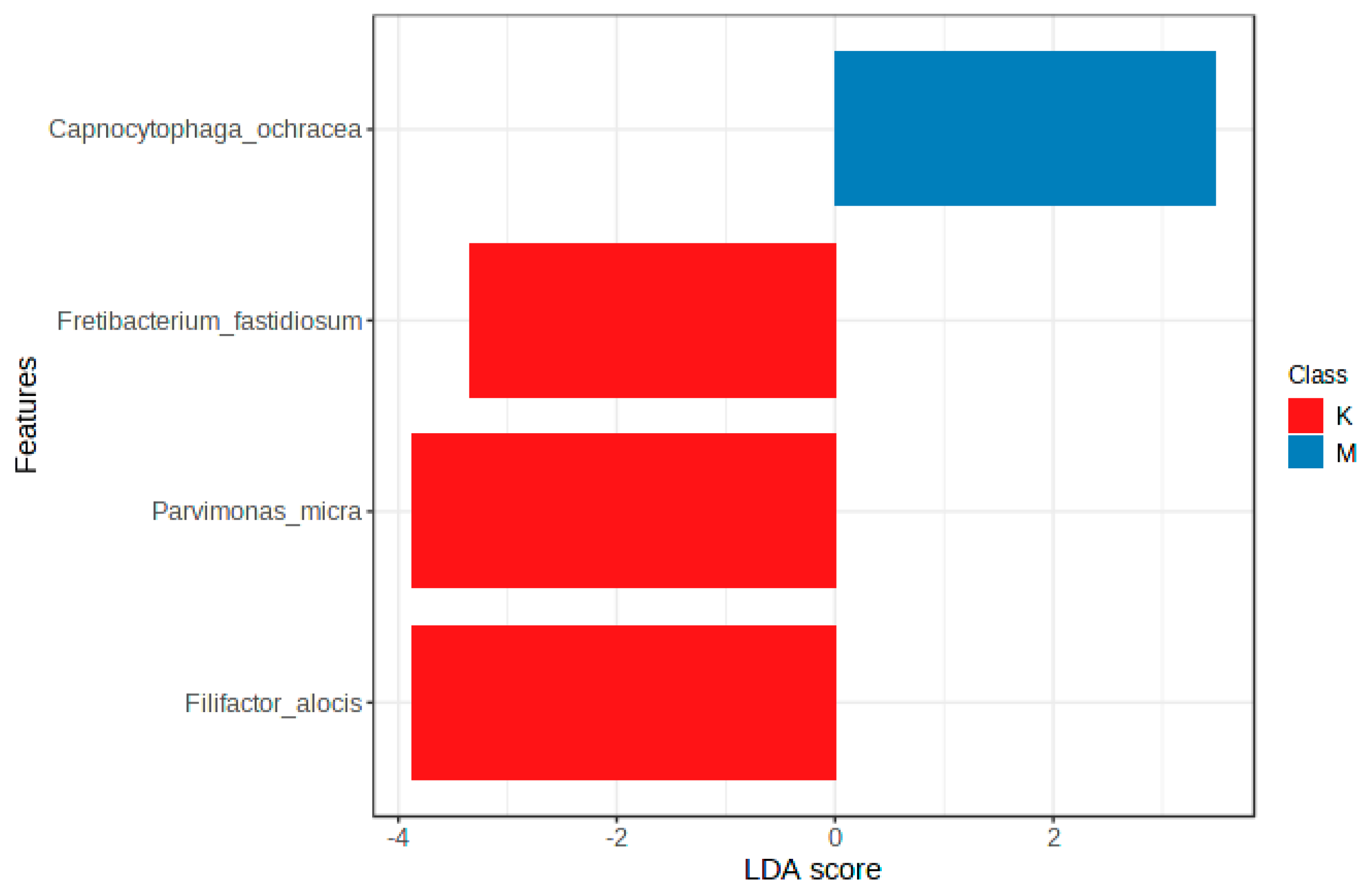
3.4. Oral Microbiota Profiles
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, X.; Fan, X.; Ying, J.; Chen, S. Emerging trends and research foci in gastrointestinal microbiome. J. Transl. Med. 2019, 17, 67. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Boccuto, L.; Tack, J.; Ianiro, G.; Abenavoli, L.; Scarpellini, E. Human Genes Involved in the Interaction between Host and Gut Microbiome: Regulation and Pathogenic Mechanisms. Genes 2023, 14, 857. [Google Scholar] [CrossRef]
- Sakkas, H.; Bozidis, P.; Touzios, C.; Kolios, D.; Athanasiou, G.; Athanasopoulou, E.; Gerou, I.; Gartzonika, C. Nutritional Status and the Influence of the Vegan Diet on the Gut Microbiota and Human Health. Medicina 2020, 56, 88. [Google Scholar] [CrossRef] [PubMed]
- Klement, R.J.; Pazienza, V. Impact of Different Types of Diet on Gut Microbiota Profiles and Cancer Prevention and Treatment. Medicina 2019, 55, 84. [Google Scholar] [CrossRef] [PubMed]
- Saini, A.; Dalal, P.; Sharma, D. Deciphering the interdependent labyrinth between gut microbiota and the immune system. Lett. Appl. Microbiol. 2022, 75, 1122–1135. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Abu-Ali, G.; Huttenhower, C. The healthy human microbiome. Genome Med. 2016, 8, 51. [Google Scholar] [CrossRef]
- Mentella, M.C.; Scaldaferri, F.; Pizzoferrato, M.; Gasbarrini, A.; Miggiano, G.A.D. Nutrition, IBD and Gut Microbiota: A Review. Nutrients 2020, 12, 944. [Google Scholar] [CrossRef]
- El-Salhy, M.; Hatlebakk, J.G.; Hausken, T. Diet in Irritable Bowel Syndrome (IBS): Interaction with Gut Microbiota and Gut Hormones. Nutrients 2019, 11, 1824. [Google Scholar] [CrossRef]
- Caio, G.; Lungaro, L.; Segata, N.; Guarino, M.; Zoli, G.; Volta, U.; De Giorgio, R. Effect of Gluten-Free Diet on Gut Microbiota Composition in Patients with Celiac Disease and Non-Celiac Gluten/Wheat Sensitivity. Nutrients 2020, 12, 1832. [Google Scholar] [CrossRef]
- Quesada-Vázquez, S.; Aragonès, G.; Del Bas, J.M.; Escoté, X. Diet, Gut Microbiota and Non-Alcoholic Fatty Liver Disease: Three Parts of the Same Axis. Cells 2020, 9, 176. [Google Scholar] [CrossRef] [PubMed]
- Pluta, R.; Ułamek-Kozioł, M.; Januszewski, S.; Czuczwar, S.J. Gut microbiota and pro/prebiotics in Alzheimer’s disease. Aging 2020, 12, 5539–5550. [Google Scholar] [CrossRef]
- Gentile, F.; Doneddu, P.E.; Riva, N.; Nobile-Orazio, E.; Quattrini, A. Diet, Microbiota and Brain Health: Unraveling the Network Intersecting Metabolism and Neurodegeneration. Int. J. Mol. Sci. 2020, 21, 7471. [Google Scholar] [CrossRef]
- Gubert, C.; Kong, G.; Renoir, T.; Hannan, A.J. Exercise, diet and stress as modulators of gut microbiota: Implications for neurodegenerative diseases. Neurobiol. Dis. 2020, 134, 104621. [Google Scholar] [CrossRef] [PubMed]
- Conte, L.; Toraldo, D.M. Targeting the gut-lung microbiota axis by means of a high-fibre diet and probiotics may have anti-inflammatory effects in COVID-19 infection. Ther. Adv. Respir. Dis. 2020, 14, 1753466620937170. [Google Scholar] [CrossRef] [PubMed]
- Illiano, P.; Brambilla, R.; Parolini, C. The mutual interplay of gut microbiota, diet and human disease. FEBS J. 2020, 287, 833–855. [Google Scholar] [CrossRef] [PubMed]
- Donald, K.; Finlay, B.B. Early-life interactions between the microbiota and immune system: Impact on immune system development and atopic disease. Nat. Rev. Immunol. 2023, 23, 735–748. [Google Scholar] [CrossRef]
- Salazar, N.; González, S.; Nogacka, A.M.; Rios-Covián, D.; Arboleya, S.; Gueimonde, M.; Reyes-Gavilán, C.G.L. Microbiome: Effects of Ageing and Diet. Curr. Issues Mol. Biol. 2020, 36, 33–62. [Google Scholar] [CrossRef]
- Alsharairi, N.A. The Role of Short-Chain Fatty Acids in Mediating Very Low-Calorie Ketogenic Diet-Infant Gut Microbiota Relationships and Its Therapeutic Potential in Obesity. Nutrients 2021, 13, 3702. [Google Scholar] [CrossRef]
- Redondo-Useros, N.; Nova, E.; González-Zancada, N.; Díaz, L.E.; Gómez-Martínez, S.; Marcos, A. Microbiota and Lifestyle: A Special Focus on Diet. Nutrients 2020, 12, 1776. [Google Scholar] [CrossRef]
- Mohr, A.E.; Jäger, R.; Carpenter, K.C.; Kerksick, C.M.; Purpura, M.; Townsend, J.R.; West, N.P.; Black, K.; Gleeson, M.; Pyne, D.B.; et al. The athletic gut microbiota. J. Int. Soc. Sports Nutr. 2020, 17, 24. [Google Scholar] [CrossRef] [PubMed]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L.; et al. The long-term stability of the human gut microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, F.; Ghosh, T.S.; O’Toole, P.W. The Healthy Microbiome-What Is the Definition of a Healthy Gut Microbiome? Gastroenterology 2021, 160, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Tuominen, H.; Rautava, J. Oral Microbiota and Cancer Development. Pathobiology 2021, 88, 116–126. [Google Scholar] [CrossRef]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.; Yu, W.H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef]
- Zapała, B.; Stefura, T.; Milewicz, T.; Wątor, J.; Piwowar, M.; Wójcik-Pędziwiatr, M.; Doręgowska, M.; Dudek, A.; Jania, Z.; Rudzińska-Bar, M. The Role of the Western Diet and Oral Microbiota in Parkinson’s Disease. Nutrients 2022, 14, 355. [Google Scholar] [CrossRef]
- Wade, W.G. The oral microbiome in health and disease. Pharmacol. Res. 2013, 69, 137–143. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Xie, H.; Guo, R.; Zhong, H.; Feng, Q.; Lan, Z.; Qin, B.; Ward, K.J.; Jackson, M.A.; Xia, Y.; Chen, X.; et al. Shotgun Metagenomics of 250 Adult Twins Reveals Genetic and Environmental Impacts on the Gut Microbiome. Cell Syst. 2016, 3, 572–584.e3. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Xiao, Z.; Wu, Y.; Ge, C. Diet and Skin Aging-From the Perspective of Food Nutrition. Nutrients 2020, 12, 870. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef]
- Ponzo, V.; Fedele, D.; Goitre, I.; Leone, F.; Lezo, A.; Monzeglio, C.; Finocchiaro, C.; Ghigo, E.; Bo, S. Diet-Gut Microbiota Interactions and Gestational Diabetes Mellitus (GDM). Nutrients 2019, 11, 330. [Google Scholar] [CrossRef] [PubMed]
- Christ, A.; Lauterbach, M.; Latz, E. Western Diet and the Immune System: An Inflammatory Connection. Immunity 2019, 51, 794–811. [Google Scholar] [CrossRef]
- Malinowska, A.M.; Kok, D.E.; Steegenga, W.T.; Hooiveld, G.J.E.J.; Chmurzynska, A. Human gut microbiota composition and its predicted functional properties in people with western and healthy dietary patterns. Eur. J. Nutr. 2022, 61, 3887–3903. [Google Scholar] [CrossRef]
- Paoli, A.; Mancin, L.; Bianco, A.; Thomas, E.; Mota, J.F.; Piccini, F. Ketogenic Diet and Microbiota: Friends or Enemies? Genes 2019, 10, 534. [Google Scholar] [CrossRef]
- Bibbò, S.; Ianiro, G.; Giorgio, V.; Scaldaferri, F.; Masucci, L.; Gasbarrini, A.; Cammarota, G. The role of diet on gut microbiota composition. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4742–4749. [Google Scholar]
- Bourdeau-Julien, I.; Castonguay-Paradis, S.; Rochefort, G.; Perron, J.; Lamarche, B.; Flamand, N.; Di Marzo, V.; Veilleux, A.; Raymond, F. The diet rapidly and differentially affects the gut microbiota and host lipid mediators in a healthy population. Microbiome 2023, 11, 26. [Google Scholar] [CrossRef]
- Agans, R.; Gordon, A.; Kramer, D.L.; Perez-Burillo, S.; Rufián-Henares, J.A.; Paliy, O. Dietary Fatty Acids Sustain the Growth of the Human Gut Microbiota. Appl. Environ. Microbiol. 2018, 84, e01525-18. [Google Scholar] [CrossRef]
- Vinelli, V.; Biscotti, P.; Martini, D.; Del Bo, C.; Marino, M.; Meroño, T.; Nikoloudaki, O.; Calabrese, F.M.; Turroni, S.; Taverniti, V.; et al. Effects of Dietary Fibers on Short-Chain Fatty Acids and Gut Microbiota Composition in Healthy Adults: A Systematic Review. Nutrients 2022, 14, 2559. [Google Scholar] [CrossRef] [PubMed]
- Tomasello, G.; Mazzola, M.; Leone, A.; Sinagra, E.; Zummo, G.; Farina, F.; Damiani, P.; Cappello, F.; Gerges Geagea, A.; Jurjus, A.; et al. Nutrition, oxidative stress and intestinal dysbiosis: Influence of diet on gut microbiota in inflammatory bowel diseases. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2016, 160, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Moszak, M.; Szulińska, M.; Bogdański, P. You Are What You Eat-The Relationship between Diet, Microbiota, and Metabolic Disorders-A Review. Nutrients 2020, 12, 1096. [Google Scholar] [CrossRef] [PubMed]
- Bull, M.J.; Plummer, N.T. Part 1: The Human Gut Microbiome in Health and Disease. Integr. Med. (Encinitas). 2014, 13, 17–22. [Google Scholar] [PubMed]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef]
- Jeżewska-Zychowicz, M.; Gawęcki, J.; Wądołowska, L. I wsp. KomPAN® Kwestionariusz do Badania Poglądów i Zwyczajów Żywieniowych dla osób w Wieku od 16 do 65 lat, Wersja 1.2—Kwestionariusz do Samodzielnego Wypełnienia Przez Respondenta. Rozdz. 2. In KomPAN® Kwestionariusz do Badania Poglądów i Zwyczajów Żywieniowych Oraz Procedura Opracowania Danych; Red Gawęcki, J., Ed.; Komitetu Nauki o Żywieniu Człowieka Polskiej Akademii Nauk: Warszawa, Poland, 2020; pp. 22–34. Available online: http://www.knozc.pan.pl/ (accessed on 15 October 2023).
- Wądołowska, L.; Krusińska, B. Procedura Opracowania Danych Żywieniowych z Kwestionariusza KomPAN. Rozdz. 3. In Kwestionariusz do Badania Poglądów i Zwyczajów Żywieniowych Oraz Procedura Opracowania Danych; Red Gawęcki, J., Ed.; Komitetu Nauki o Żywieniu Człowieka Polskiej Akademii Nauk: Warszawa, Poland, 2014; pp. 34–51. Available online: http://www.knozc.pan.pl/ (accessed on 15 October 2023).
- Kowalkowska, J.; Wadolowska, L.; Czarnocinska, J.; Czlapka-Matyasik, M.; Galinski, G.; Jezewska-Zychowicz, M.; Bronkowska, M.; Dlugosz, A.; Loboda, D.; Wyka, J. Reproducibility of a Questionnaire for Dietary Habits, Lifestyle and Nutrition Knowledge Assessment (KomPAN) in Polish Adolescents and Adults. Nutrients 2018, 10, 1845. [Google Scholar] [CrossRef]
- Herbowski, P.; Lorkiewicz-Muszyńska, D. Rola ekspertyzy antropologicznej podczas weryfikacji wyników okazania osób. Probl. Kryminal. 2017, 297, 5–13. [Google Scholar]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Good, I.J. The population frequencies of species and the estimation of population parameters. Biometrika 1953, 40, 237–264. [Google Scholar] [CrossRef]
- Schwenger, K.J.; Clermont-Dejean, N.; Allard, J.P. The role of the gut microbiome in chronic liver disease: The clinical evidence revised. JHEP Rep. 2019, 1, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Dhar, D.; Mohanty, A. Gut microbiota and COVID-19- possible link and implications. Virus Res. 2020, 285, 198018. [Google Scholar] [CrossRef] [PubMed]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, S.J.; Chung, D.; Mahmoud, N.; Sepulveda, A.R.; Manafe, M.; Arch, J.; Adada, H.; van der Merwe, T. Why do African Americans get more colon cancer than Native Africans? J. Nutr. 2007, 137 (Suppl. 1), 175S–182S. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.; Carbonero, F.; Zoetendal, E.G.; DeLany, J.P.; Wang, M.; Newton, K.; Gaskins, H.R.; O’Keefe, S.J. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am. J. Clin. Nutr. 2013, 98, 111–120. [Google Scholar] [CrossRef]
- Gomez, A.; Petrzelkova, K.J.; Burns, M.B.; Yeoman, C.J.; Amato, K.R.; Vlckova, K.; Modry, D.; Todd, A.; Jost Robinson, C.A.; Remis, M.J.; et al. Gut Microbiome of Coexisting BaAka Pygmies and Bantu Reflects Gradients of Traditional Subsistence Patterns. Cell Rep. 2016, 14, 2142–2153. [Google Scholar] [CrossRef]
- Schnorr, S.L.; Candela, M.; Rampelli, S.; Centanni, M.; Consolandi, C.; Basaglia, G.; Turroni, S.; Biagi, E.; Peano, C.; Severgnini, M.; et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014, 5, 3654. [Google Scholar] [CrossRef]
- Obregon-Tito, A.J.; Tito, R.Y.; Metcalf, J.; Sankaranarayanan, K.; Clemente, J.C.; Ursell, L.K.; Zech Xu, Z.; Van Treuren, W.; Knight, R.; Gaffney, P.M.; et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat. Commun. 2015, 6, 6505. [Google Scholar] [CrossRef]
- Wang, P.; Ding, S.; Sun, L.; Feng, Y.; Guo, K.; Zhu, Y.; Huang, D.; Ruan, S. Characteristics and differences of gut microbiota in patients with different Traditional Chinese Medicine Syndromes of Colorectal Cancer and normal population. J. Cancer 2020, 11, 7357–7367. [Google Scholar] [CrossRef]
- Gao, L.; Xu, T.; Huang, G.; Jiang, S.; Gu, Y.; Chen, F. Oral microbiomes: More and more importance in oral cavity and whole body. Protein Cell. 2018, 9, 488–500. [Google Scholar] [CrossRef]
- Cresci, G.A.; Bawden, E. Gut Microbiome: What We Do and Don’t Know. Nutr. Clin. Pract. 2015, 30, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.C.; Dinsmoor, A.M.; Wang, M.; Donovan, S.M. Microbiome Composition in Pediatric Populations from Birth to Adolescence: Impact of Diet and Prebiotic and Probiotic Interventions. Dig. Dis. Sci. 2020, 65, 706–722. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Van Hul, M. Mediterranean diet, gut microbiota and health: When age and calories do not add up! Gut 2020, 69, 1167–1168. [Google Scholar] [CrossRef] [PubMed]
- Hills, R.D., Jr.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut Microbiome: Profound Implications for Diet and Disease. Nutrients 2019, 11, 1613. [Google Scholar] [CrossRef]
- Rinninella, E.; Cintoni, M.; Raoul, P.; Lopetuso, L.R.; Scaldaferri, F.; Pulcini, G.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. Food Components and Dietary Habits: Keys for a Healthy Gut Microbiota Composition. Nutrients 2019, 11, 2393. [Google Scholar] [CrossRef]
- Krishnan, K.; Chen, T.; Paster, B.J. A practical guide to the oral microbiome and its relation to health and disease. Oral Dis. 2017, 23, 276–286. [Google Scholar] [CrossRef]
- Keshavarz Azizi Raftar, S.; Hoseini Tavassol, Z.; Amiri, M.; Ejtahed, H.S.; Zangeneh, M.; Sadeghi, S.; Ashrafian, F.; Kariman, A.; Khatami, S.; Siadat, S.D. Assessment of fecal Akkermansia muciniphila in patients with osteoporosis and osteopenia: A pilot study. J. Diabetes Metab. Disord. 2021, 20, 279–284. [Google Scholar] [CrossRef]
- de Moraes, A.C.; Fernandes, G.R.; da Silva, I.T.; Almeida-Pititto, B.; Gomes, E.P.; Pereira, A.D.; Ferreira, S.R. Enterotype May Drive the Dietary-Associated Cardiometabolic Risk Factors. Front. Cell Infect. Microbiol. 2017, 7, 47. [Google Scholar] [CrossRef]
- Chang, Y.; Woo, H.G.; Jeong, J.H.; Kim, G.H.; Park, K.D.; Song, T.J. Microbiota dysbiosis and functional outcome in acute ischemic stroke patients. Sci. Rep. 2021, 11, 10977. [Google Scholar] [CrossRef]
- Li, N.; Wang, X.; Sun, C.; Wu, X.; Lu, M.; Si, Y.; Ye, X.; Wang, T.; Yu, X.; Zhao, X.; et al. Change of intestinal microbiota in cerebral ischemic stroke patients. BMC Microbiol. 2019, 19, 191. [Google Scholar] [CrossRef]
- Li, X.; Li, Z.; He, Y.; Li, P.; Zhou, H.; Zeng, N. Regional distribution of Christensenellaceae and its associations with metabolic syndrome based on a population-level analysis. PeerJ 2020, 8, e9591. [Google Scholar] [CrossRef] [PubMed]
- Brooks, A.W.; Priya, S.; Blekhman, R.; Bordenstein, S.R. Gut microbiota diversity across ethnicities in the United States. PLoS Biol. 2018, 16, e2006842. [Google Scholar] [CrossRef] [PubMed]
- Wallis, K.F.; Melnyk, S.B.; Miousse, I.R. Sex-Specific Effects of Dietary Methionine Restriction on the Intestinal Microbiome. Nutrients 2020, 12, 781. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Burrows, M.; Khan, A.A.; Graham, L.; Volchkov, P.; Becker, L.; Antonopoulos, D.; Umesaki, Y.; Chervonsky, A.V. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013, 39, 400–412. [Google Scholar] [CrossRef]
- Markle, J.G.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef]
- Org, E.; Mehrabian, M.; Parks, B.W.; Shipkova, P.; Liu, X.; Drake, T.A.; Lusis, A.J. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes 2016, 7, 313–322. [Google Scholar] [CrossRef]
- Fushuku, S.; Fukuda, K. Gender difference in the composition of fecal flora in laboratory mice, as detected by denaturing gradient gel electrophoresis (DGGE). Exp. Anim. 2008, 57, 489–493. [Google Scholar] [CrossRef]
- Elderman, M.; Hugenholtz, F.; Belzer, C.; Boekschoten, M.; van Beek, A.; de Haan, B.; Savelkoul, H.; de Vos, P.; Faas, M. Sex and strain dependent differences in mucosal immunology and microbiota composition in mice. Biol. Sex Differ. 2018, 9, 26. [Google Scholar] [CrossRef]
- Fransen, F.; van Beek, A.A.; Borghuis, T.; Meijer, B.; Hugenholtz, F.; van der Gaast-de Jongh, C.; Savelkoul, H.F.; de Jonge, M.I.; Faas, M.M.; Boekschoten, M.V.; et al. The impact of gut microbiota on gender-specific differences in immunity. Front. Immunol. 2017, 8, 754. [Google Scholar] [CrossRef]
- Mueller, S.; Saunier, K.; Hanisch, C.; Norin, E.; Alm, L.; Midtvedt, T.; Cresci, A.; Silvi, S.; Orpianesi, C.; Verdenelli, M.C.; et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: A cross-sectional study. Appl. Environ. Microbiol. 2006, 72, 1027–1033. [Google Scholar] [CrossRef]
- Li, M.; Wang, B.; Zhang, M.; Rantalainen, M.; Wang, S.; Zhou, H.; Zhang, Y.; Shen, J.; Pang, X.; Zhang, M.; et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. USA 2008, 105, 2117–2122. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Schloss, P.D. Dynamics and associations of microbial community types across the human body. Nature 2014, 509, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Dominianni, C.; Sinha, R.; Goedert, J.J.; Pei, Z.; Yang, L.; Hayes, R.B.; Ahn, J. Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS ONE 2015, 10, e0124599. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Manning, S.D. Impact of age and sex on the composition and abundance of the intestinal microbiota in individuals with and without enteric infections. Ann. Epidemiol. 2016, 26, 380–385. [Google Scholar] [CrossRef]
- Haro, C.; Rangel-Zúñiga, O.A.; Alcalá-Díaz, J.F.; Gómez-Delgado, F.; Pérez-Martínez, P.; Delgado-Lista, J.; Quintana-Navarro, G.M.; Landa, B.B.; Navas-Cortés, J.A.; Tena-Sempere, M.; et al. Intestinal microbiota is influenced by gender and body mass index. PLoS ONE 2016, 11, e0154090. [Google Scholar] [CrossRef]
- Borgo, F.; Garbossa, S.; Riva, A.; Severgnini, M.; Luigiano, C.; Benetti, A.; Pontiroli, A.E.; Morace, G.; Borghi, E. Body mass index and sex affect diverse microbial niches within the gut. Front. Microbiol. 2018, 9, 213. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, M.; Xue, J.; Huang, J.; Zhuang, R.; Zhou, X.; Zhang, H.; Fu, Q.; Hao, Y. Body mass index differences in the gut microbiota are gender specific. Front. Microbiol. 2018, 9, 1250. [Google Scholar] [CrossRef]
- Takagi, T.; Naito, Y.; Inoue, R.; Kashiwagi, S.; Uchiyama, K.; Mizushima, K.; Tsuchiya, S.; Dohi, O.; Yoshida, N.; Kamada, K.; et al. Differences in gut microbiota associated with age, sex, and stool consistency in healthy Japanese subjects. J. Gastroenterol. 2019, 54, 53–63. [Google Scholar] [CrossRef]
- Sinha, T.; Vich Vila, A.; Garmaeva, S.; Jankipersadsing, S.A.; Imhann, F.; Collij, V.; Bonder, M.J.; Jiang, X.; Gurry, T.; Alm, E.J.; et al. Analysis of 1135 gut metagenomes identifies sex-specific resistome profiles. Gut Microbes 2019, 10, 358–366. [Google Scholar] [CrossRef]
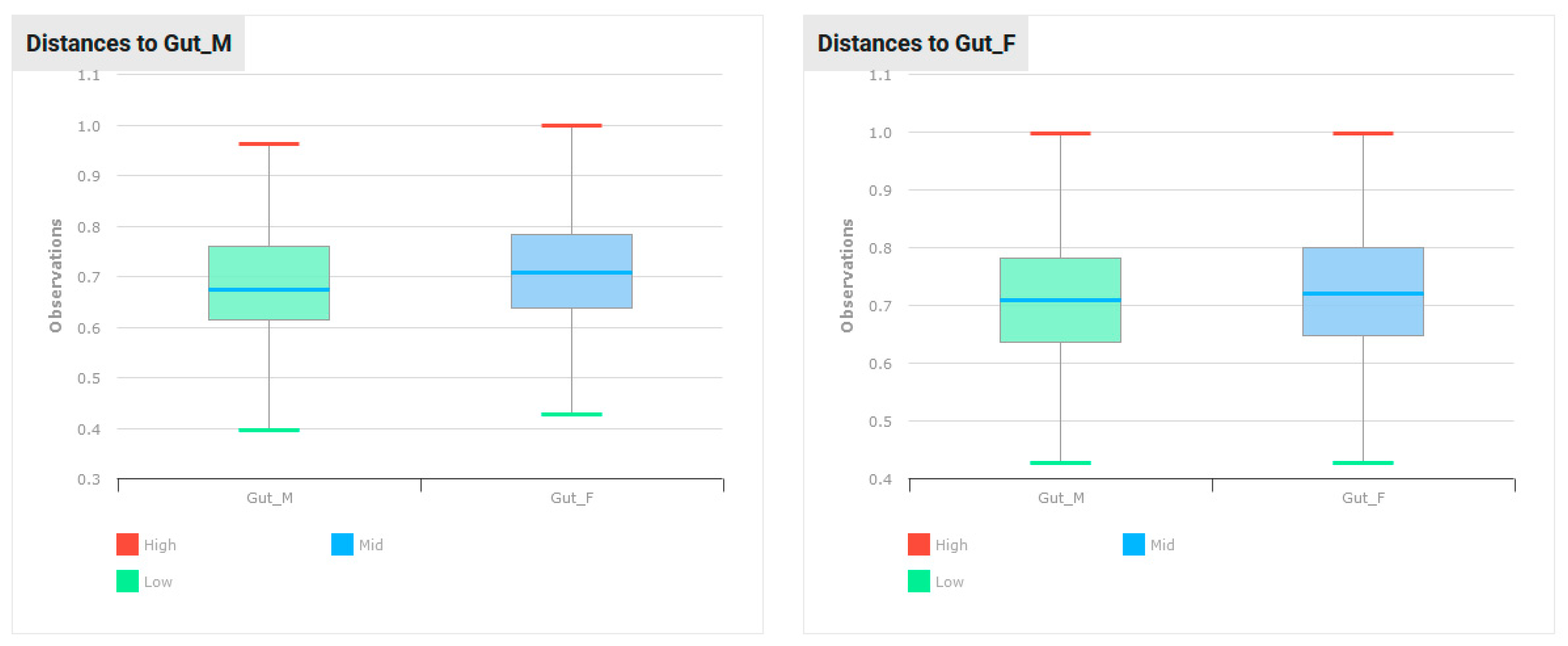
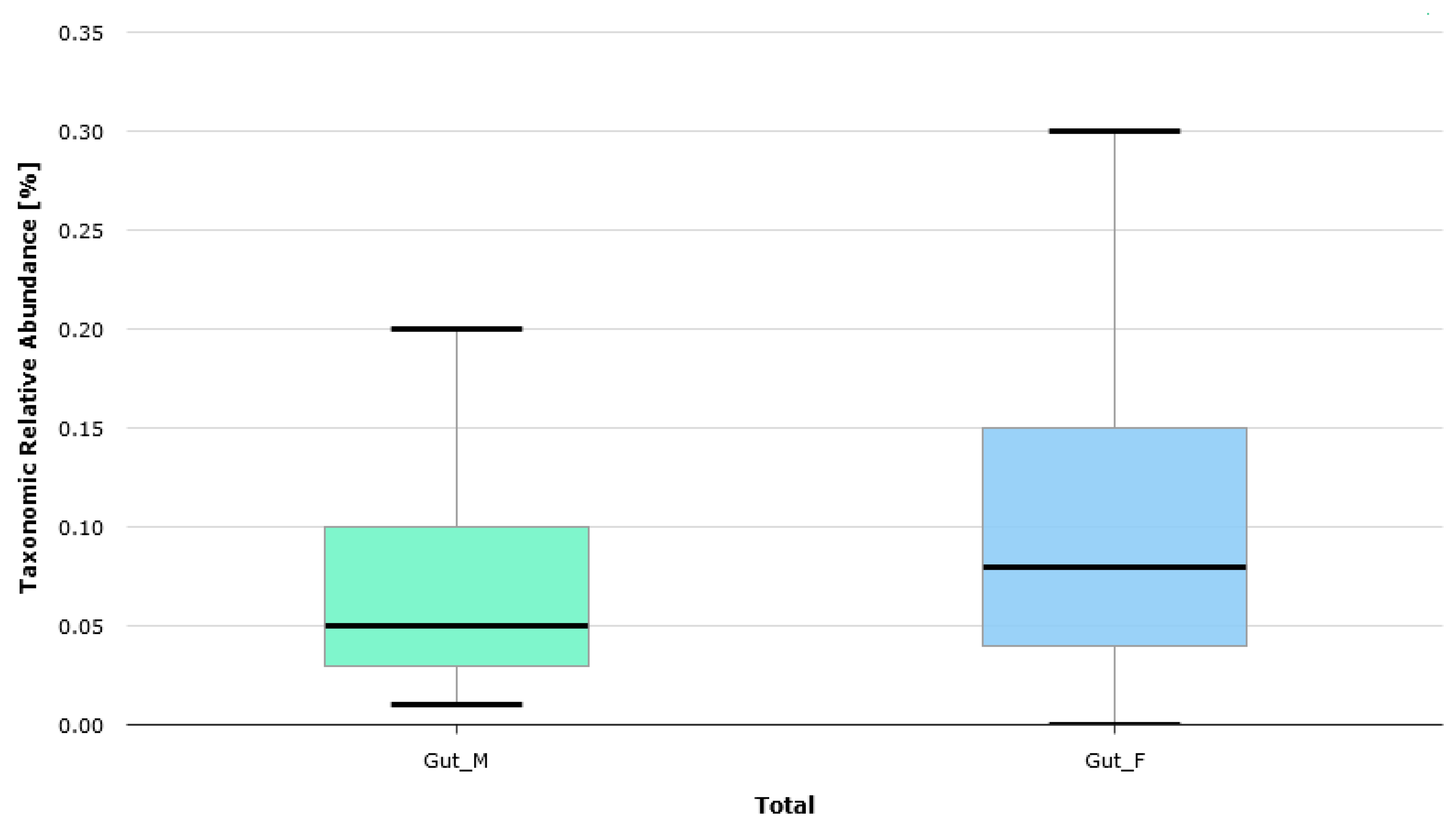

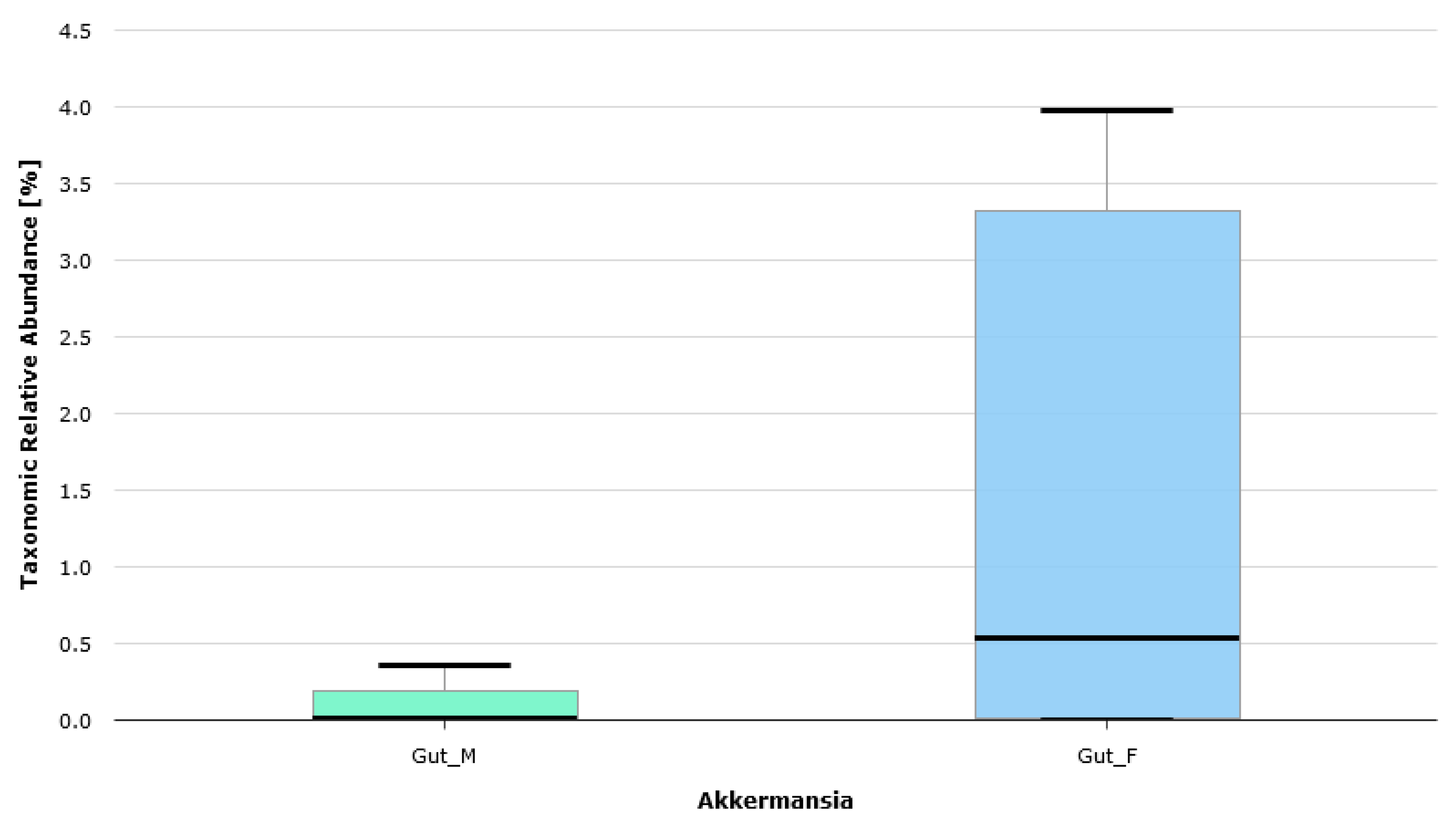
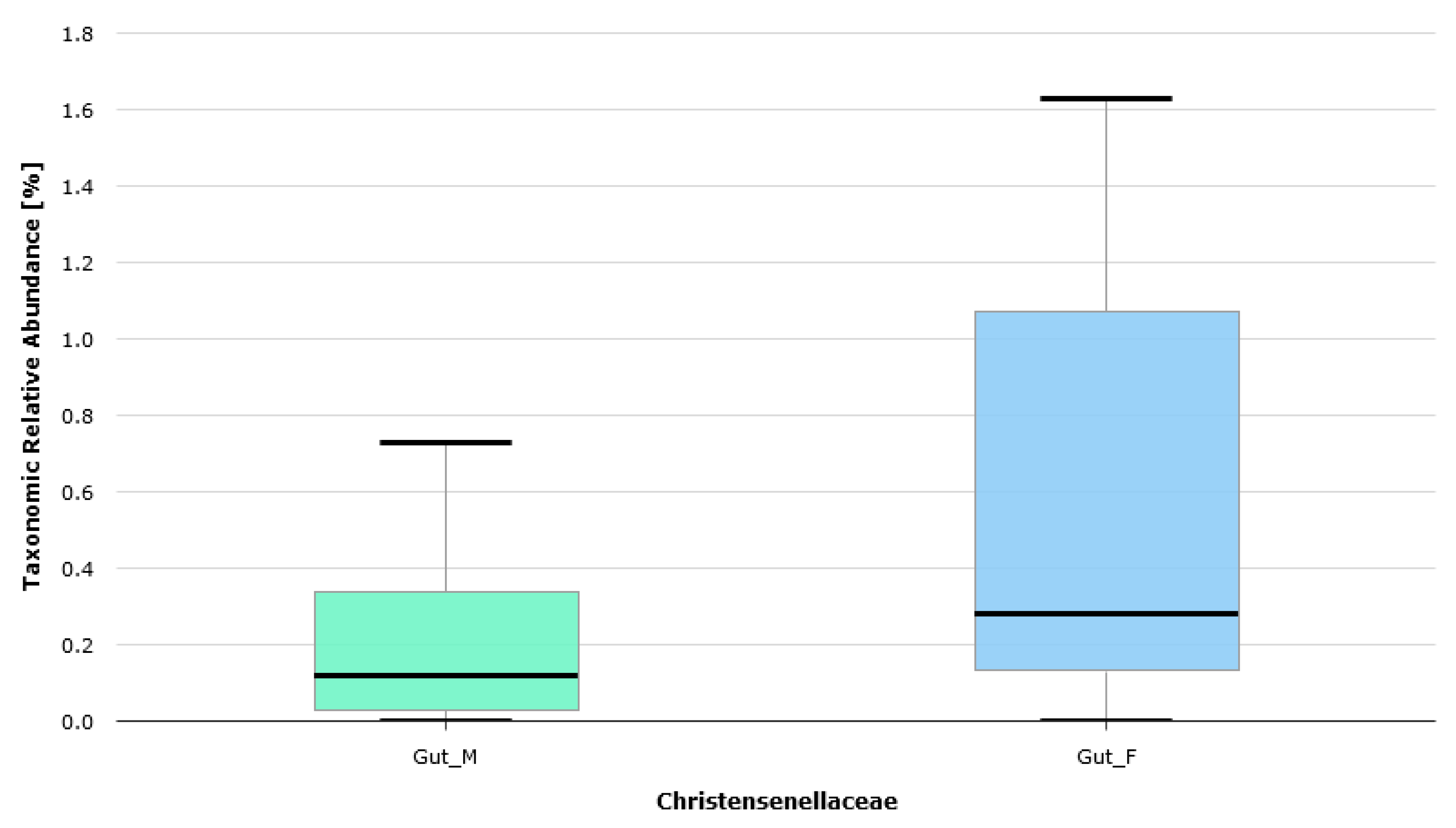

| BMI [kg/m2] | BMI Interpretation |
|---|---|
| <16.00 | Underweight (Severe thinness) |
| 16.00–16.99 | Underweight (Moderate thinness) |
| 17.00–18.49 | Underweight (Mild thinness) |
| 18.50–24.99 | Normal range |
| 25.00–29.99 | Overweight (Pre-obese) |
| 30.00–34.99 | Obese (Class I) |
| 35.00–39.99 | Obese (Class II) |
| ≥40.00 | Obese (Class III) |
| M | SD | Min | Max | Q1 | Me | Q3 | |
|---|---|---|---|---|---|---|---|
| Age (years) | 39.90 | 10.51 | 22.00 | 56.00 | 31.50 | 40.50 | 47.75 |
| Weight (kg) | 64.10 | 13.04 | 47.00 | 96.00 | 55.75 | 59.50 | 67.75 |
| Height (m) | 1.67 | 0.10 | 1.55 | 1.86 | 1.59 | 1.65 | 1.76 |
| BMI (kg/m2) | 22.73 | 2.80 | 18.00 | 28.00 | 21.00 | 22.00 | 25.00 |
| Question | Option 1 and Response Percentage | Option 2 and Response Percentage | Option 3 and Response Percentage |
|---|---|---|---|
| Education | University—77% | Secondary—23% | - |
| Place of residence | City—82% | Countryside—18% | - |
| Physical activity at work | Little—100% | - | - |
| Leisure time physical activity | Little—59% | Moderate—27% | High—14% |
| Following a specific diet | No—100% | - | - |
| Smoking status | Non-smoker—100% | - | - |
| Self-assessment of health in comparison with peers | Same as peers—80% | Better than peers—20% | - |
| The number of meals during the day | Three meals—43% | Four meals—41% | Five meals—16% |
| Eating meals at regular times | Yes, but only some of them—77% | No—23% | - |
| Frequency of consumption of white bread | Several times a day—57% | Once a week—23% | Once a day—20% |
| Frequency of consumption of whole meal bread | 1–3 times a month—45% | Once a day—30% | Once a week—25% |
| Frequency of consumption of refined grain products | Several times a week—55% | Once a week—27% | 1–3 times a month—18% |
| Frequency of consumption of whole grain products | 1–3 times a month—70% | Several times a week—30% | - |
| Frequency of consumption of cottage cheese | 1–3 times a month—50% | Once a week—32% | Several times a week—18% |
| Frequency of consumption of cheese | Several times a week—52% | 1–3 times a month—25% | Once a day—23% |
| Frequency of consumption of ready-to-eat meat products | Several times a week—57% | Once a day—25% | 1–3 times a month—18% |
| Frequency of consumption of red meat | Once a week—59% | Several times a week—21% | 1–3 times a month—20% |
| Frequency of consumption of white meat | Several times a week—77% | 1–3 times a month—23% | - |
| Frequency of consumption of fish | Once a week—52% | 1–3 times a month—48% | - |
| Frequency of consumption of egg | Once a week—52% | Several times a week—25% | 1–3 times a month—23% |
| Frequency of consumption of legumes | 1–3 times a month—100% | - | - |
| Frequency of consumption of fruit | Several times a week—59% | Once a day—21% | Several times a day—20% |
| Frequency of consumption of vegetable | Several times a day—50% | Once a day—39% | Once a week—11% |
| Salting ready meals | No—68% | Yes, but only sometimes—32% | - |
| The type of frying fat used | Plant oil—100% | - | - |
| Type of milk consumed | With reduced fat content—100% | - | - |
| Frequency of consumption of fermented milk drinks | Several times a week—80% | Once a day—20% | - |
| Sweetening hot beverages | Yes, one teaspoon of sugar or honey—64% | No—36% | - |
| Frequency of drinking water | Several times a day—100% | - | - |
| Stool Samples | Oral Swabs | |
|---|---|---|
| Total read counts | 3,230,199 | 1,345,530 |
| Average counts per sample | 78,785 | 31,291 |
| Maximum counts per sample | 110,589 | 53,060 |
| Minimum counts per sample | 48,673 | 5433 |
| Indicator | p-Value (Gut Microbiota) | p-Value (Oral Microbiota) |
|---|---|---|
| ACE | 0.559 | 0.492 |
| Chao1 | 0.711 | 0.507 |
| Shannon | 0.156 | 0.645 |
| Simpson | 0.124 | 0.937 |
| Phylogenetic Diversity | 0.127 | 0.796 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zapała, B.; Pustelnik, J.; Dudek, A.; Milewicz, T. Differences in the Composition of Akkermansia Species and Families of Christensenellaceae and Ruminococcaceae Bacteria in the Gut Microbiota of Healthy Polish Women following a Typical Western Diet. Diversity 2023, 15, 1103. https://doi.org/10.3390/d15101103
Zapała B, Pustelnik J, Dudek A, Milewicz T. Differences in the Composition of Akkermansia Species and Families of Christensenellaceae and Ruminococcaceae Bacteria in the Gut Microbiota of Healthy Polish Women following a Typical Western Diet. Diversity. 2023; 15(10):1103. https://doi.org/10.3390/d15101103
Chicago/Turabian StyleZapała, Barbara, Justyna Pustelnik, Alicja Dudek, and Tomasz Milewicz. 2023. "Differences in the Composition of Akkermansia Species and Families of Christensenellaceae and Ruminococcaceae Bacteria in the Gut Microbiota of Healthy Polish Women following a Typical Western Diet" Diversity 15, no. 10: 1103. https://doi.org/10.3390/d15101103
APA StyleZapała, B., Pustelnik, J., Dudek, A., & Milewicz, T. (2023). Differences in the Composition of Akkermansia Species and Families of Christensenellaceae and Ruminococcaceae Bacteria in the Gut Microbiota of Healthy Polish Women following a Typical Western Diet. Diversity, 15(10), 1103. https://doi.org/10.3390/d15101103





