Molecular Cartography of a Hawaiian Coral Assemblage
Abstract
1. Introduction
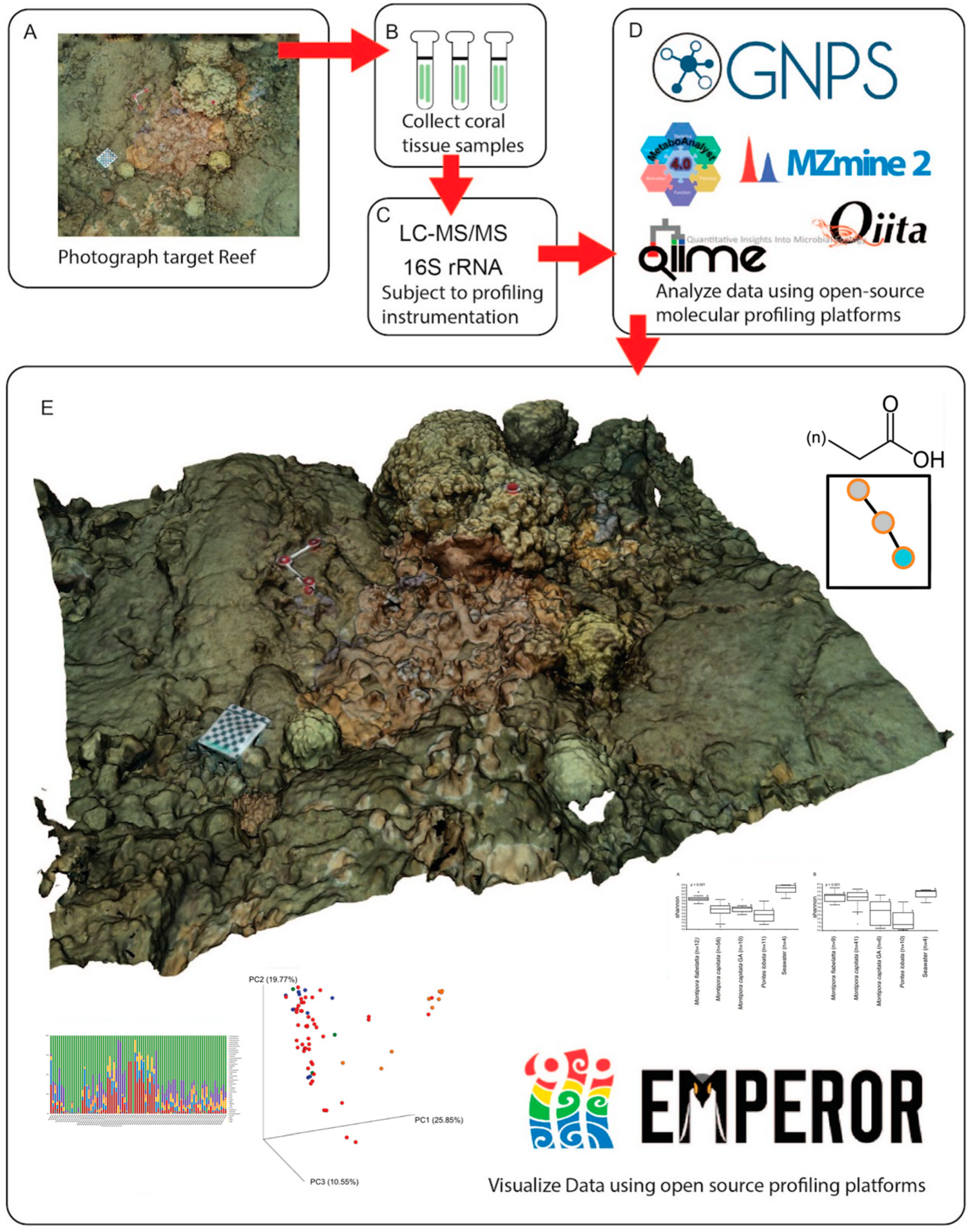
2. Methods
2.1. Coral Tissue Sampling
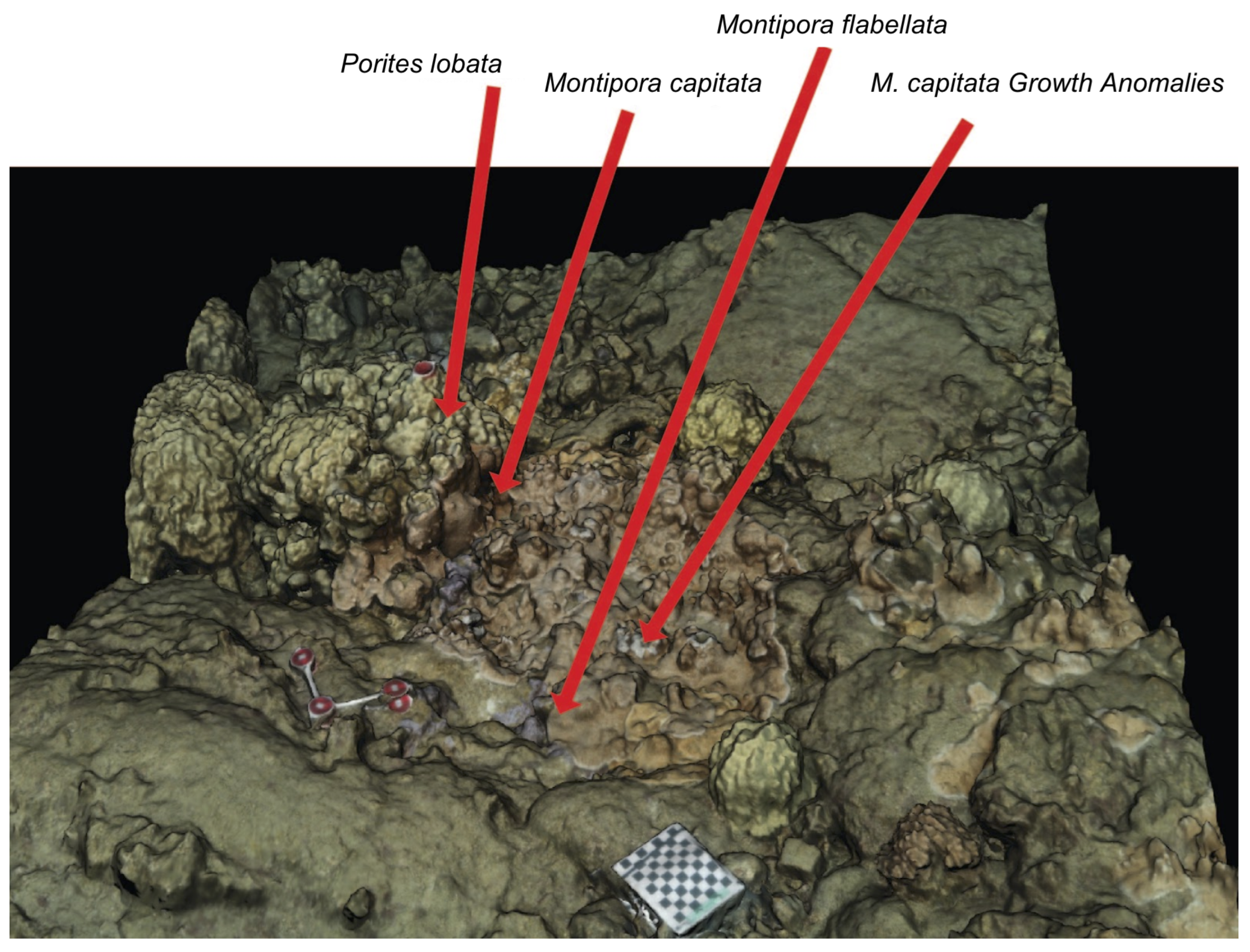
2.2. Three-Dimensional Reconstruction of Coral Assemblage
2.3. Molecular Extraction and LC-MS/MS
2.4. Microbial DNA Extraction and Sequencing
2.5. Data Processing
2.6. Data Analysis
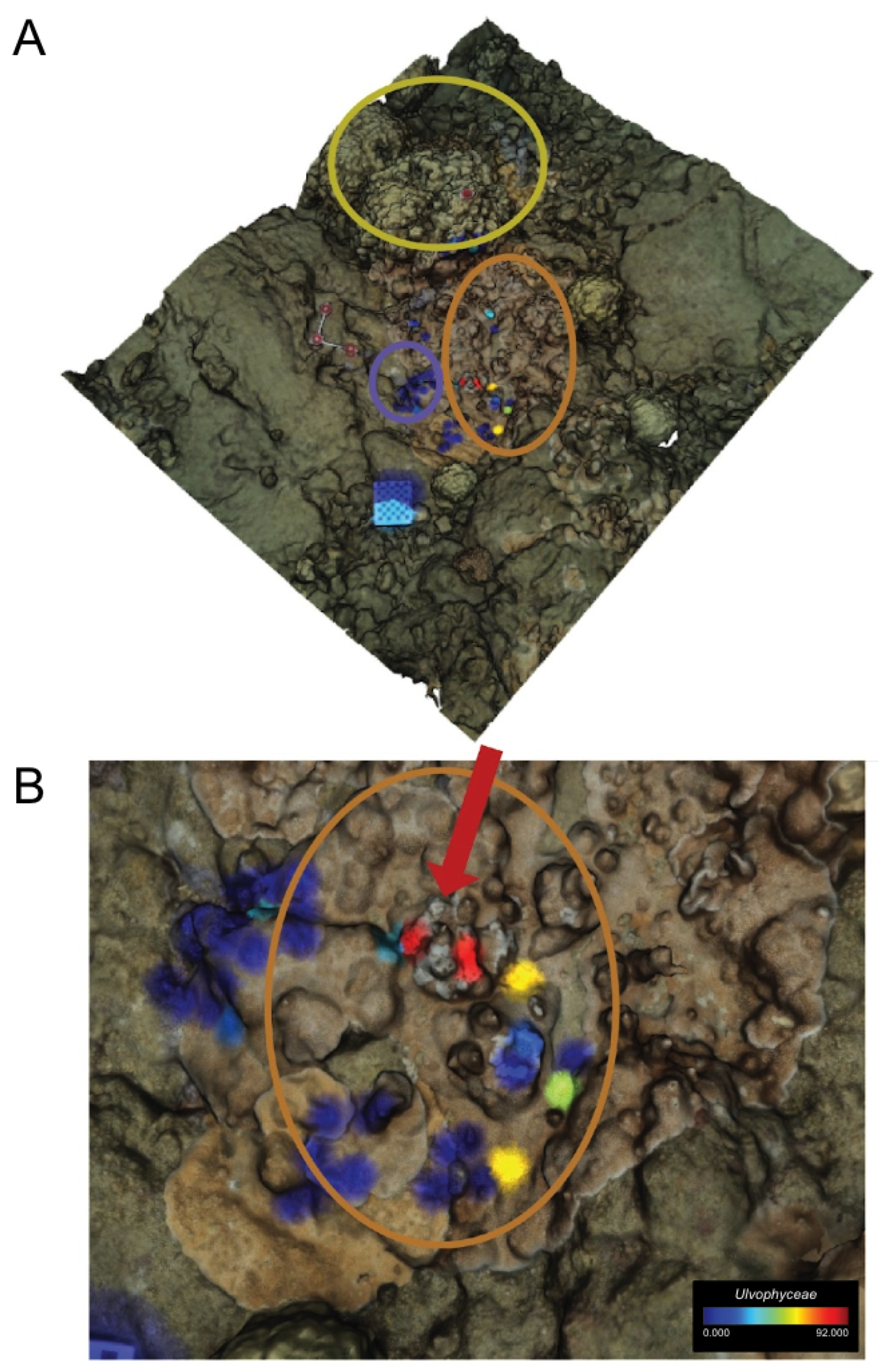
3. Results
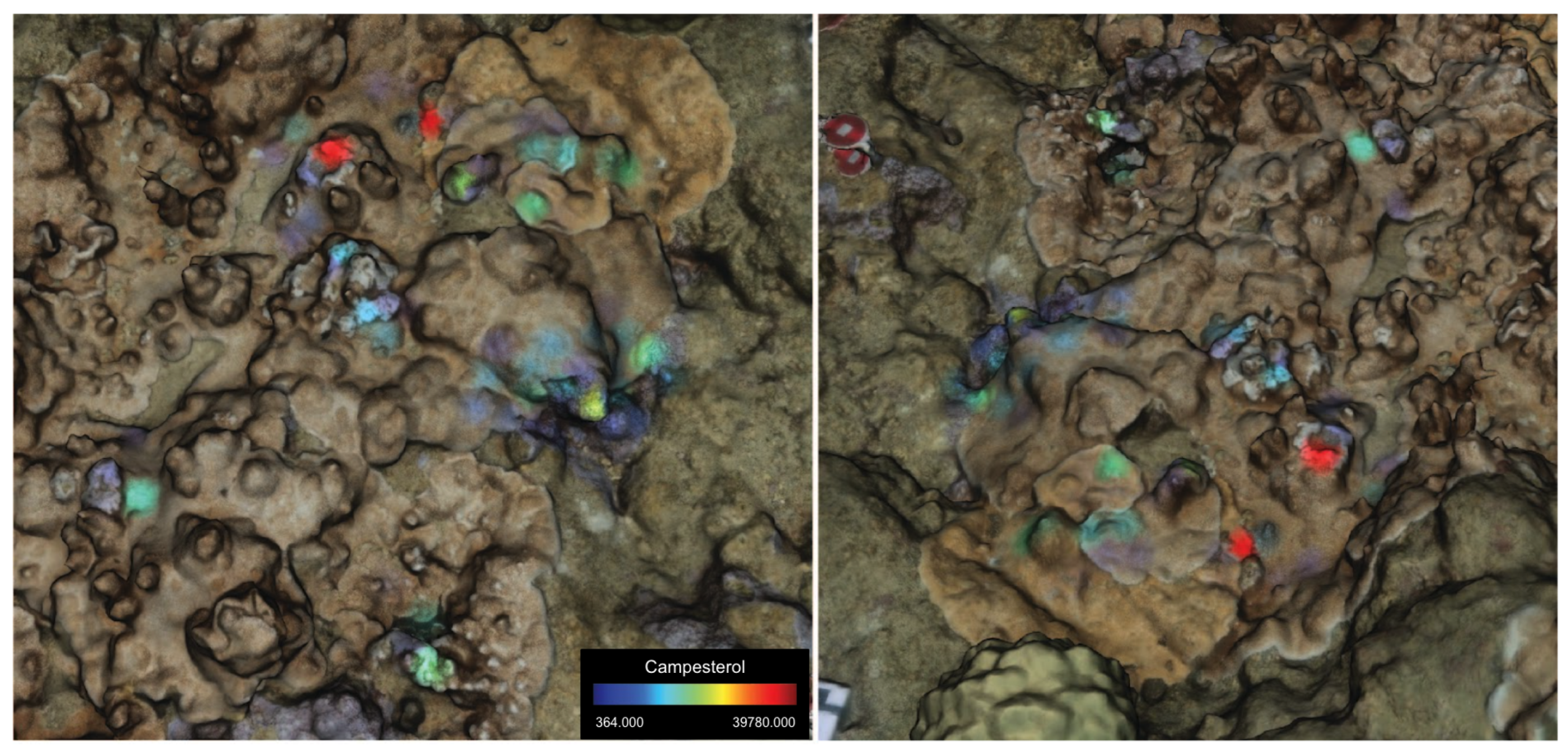
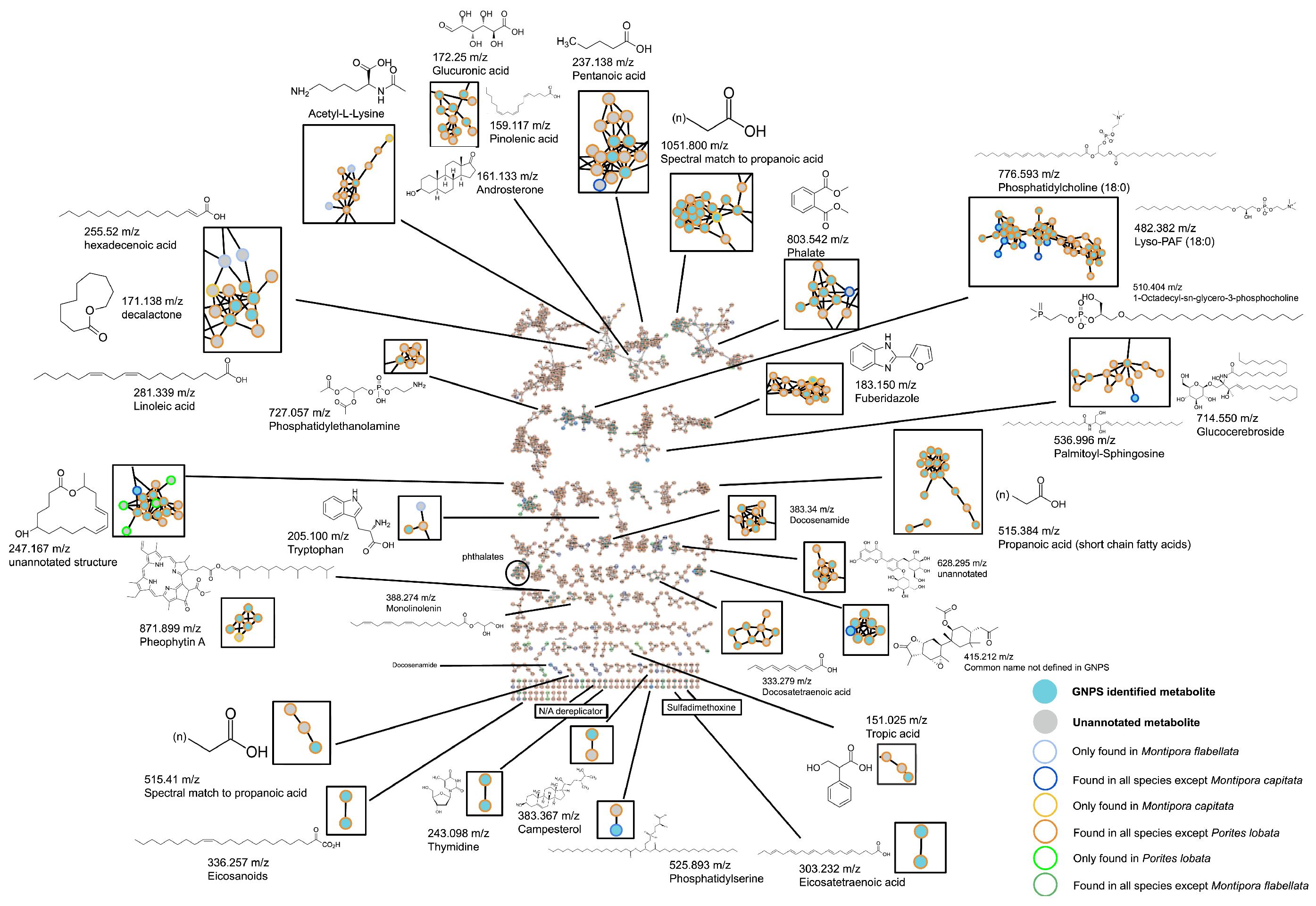
3.1. Metabolomic Profiling
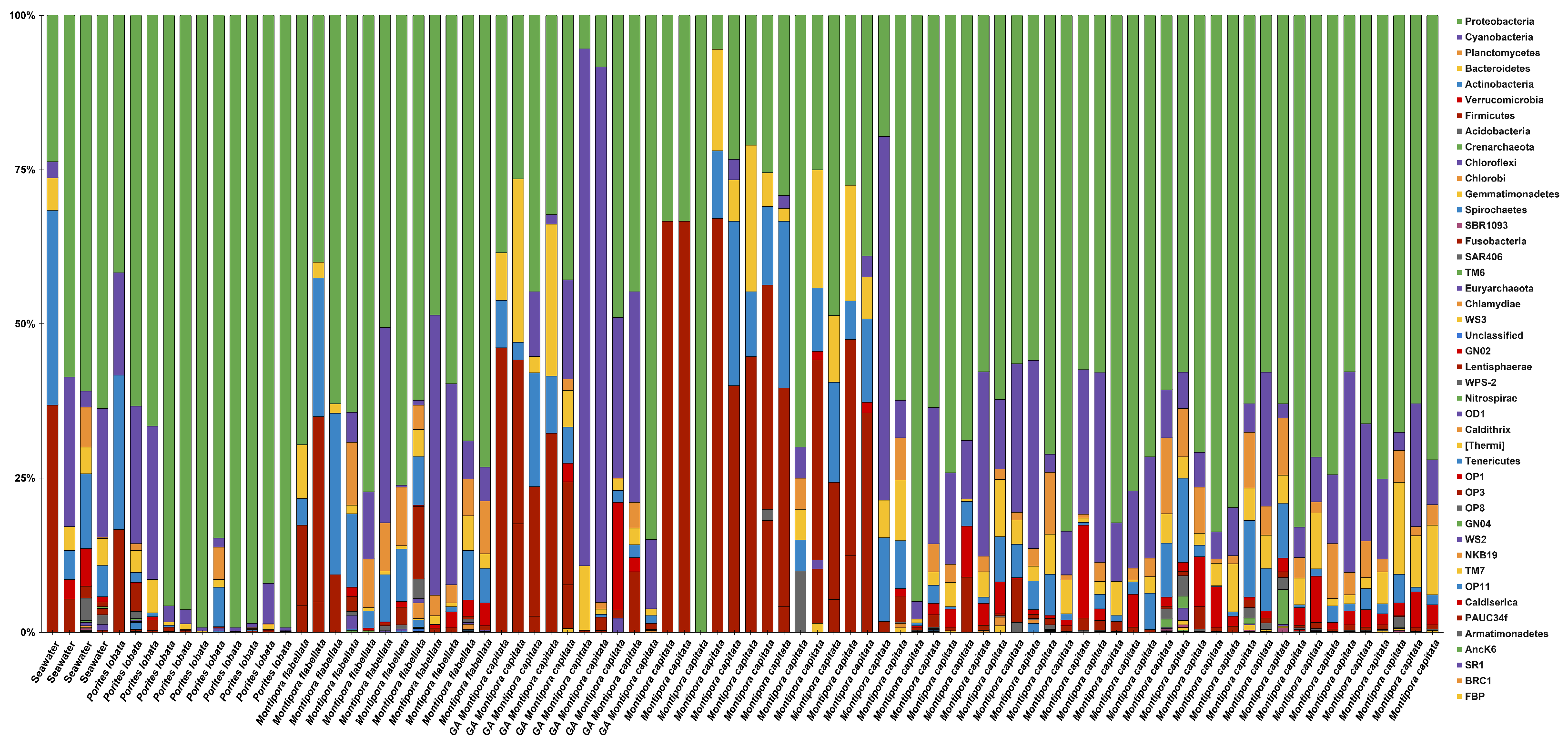
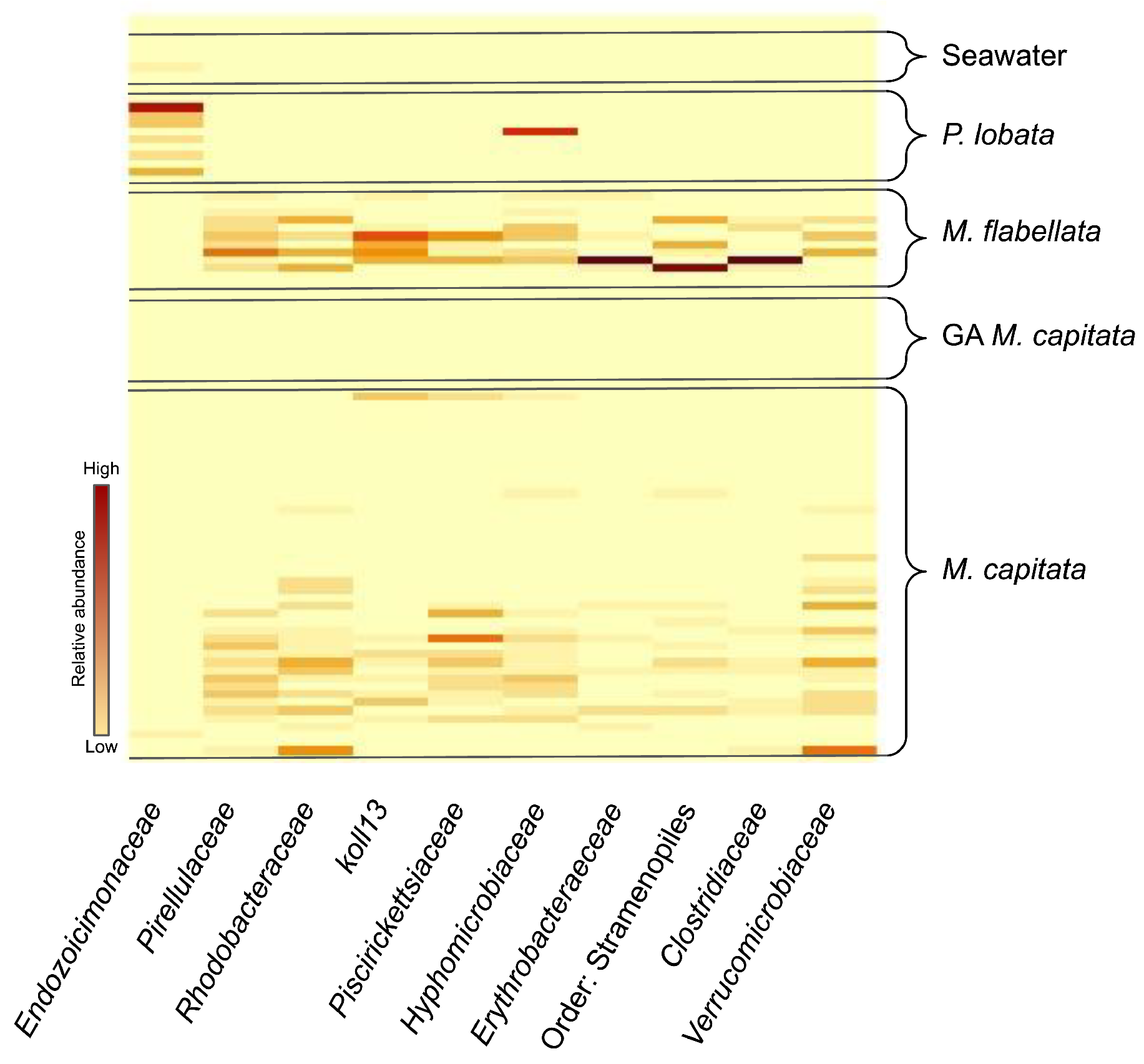
3.2. Microbiome Characterization
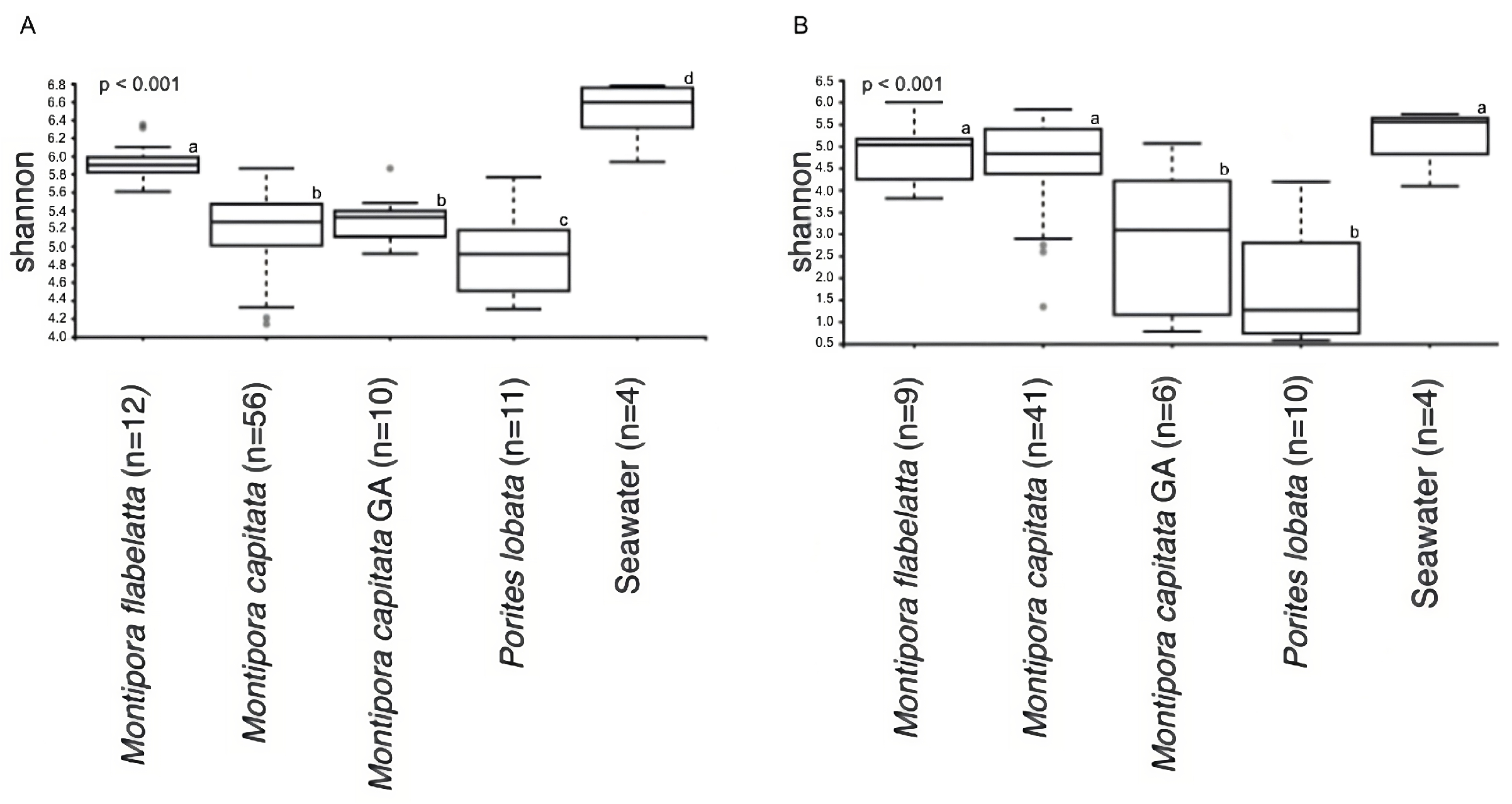
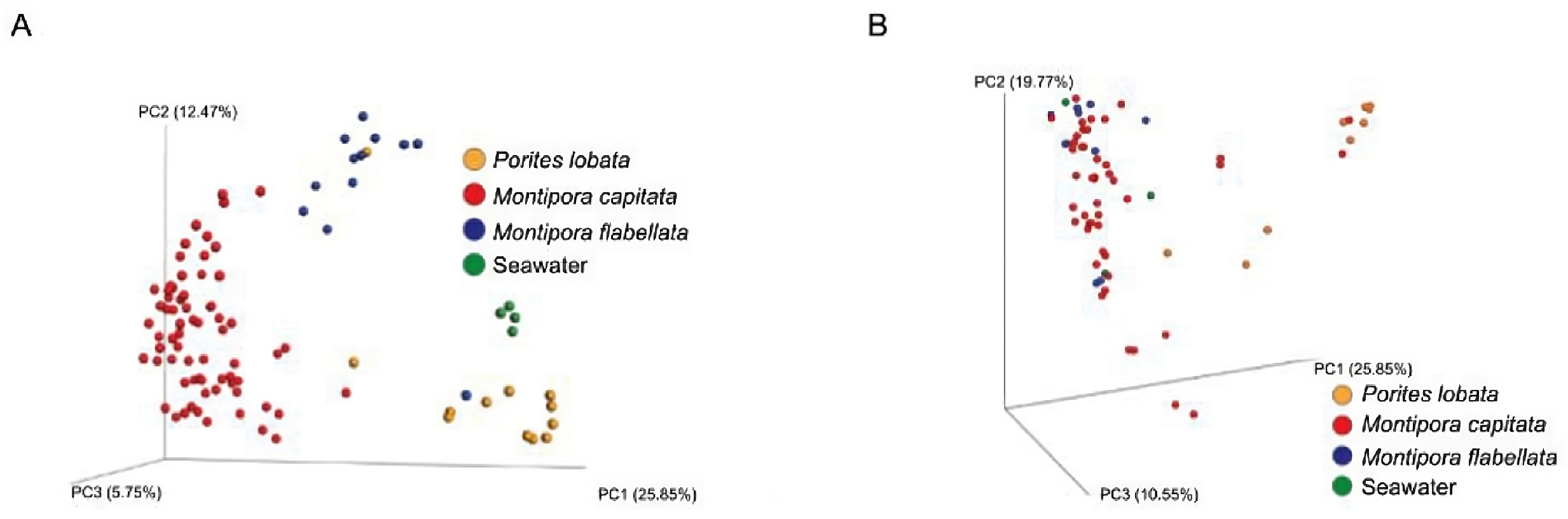
3.3. Metabolomic and Microbial Diversity
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Lamb, J.B.; Willis, B.L.; Fiorenza, E.A.; Couch, C.S.; Howard, R.; Rader, D.N.; True, J.D.; Kelly, L.A.; Ahmad, A.; Jompa, J.; et al. Plastic waste associated with disease on coral reefs. Science 2018, 359, 460–462. [Google Scholar] [CrossRef]
- Roche, R.C.; Williams, G.J.; Turner, J.R. Towards developing a mechanistic understanding of coral reef resilience to thermal stress across multiple scales. Curr. Clim. Change Rep. 2018, 4, 51–64. [Google Scholar] [CrossRef]
- McDevitt-Irwin, J.M.; Baum, J.K.; Garren, M.; Vega Thurber, R.L. Towards developing a mechanistic understanding of coral reef resilience to thermal stress across multiple scales. Front. Mar. Sci. 2017, 4, 262. [Google Scholar] [CrossRef]
- Rohwer, F.; Breitbart, M.; Jara, J.; Azam, F.; Knowlton, N. Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 2001, 20, 85–91. [Google Scholar]
- Rohwer, F.; Seguritan, V.; Azam, F.; Knowlton, N. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 2002, 243, 1–10. [Google Scholar] [CrossRef]
- Bourne, D.G.; Garren, M.; Work, T.M.; Rosenberg, E.; Smith, G.W.; Harvell, C.D. Microbial disease and the coral holobiont. Trends Microbiol. 2009, 17, 554–562. [Google Scholar] [CrossRef]
- Gordon, B.R.; Leggat, W. Symbiodinium-invertebrate symbioses and the role of metabolomics. Mar Drugs 2010, 8, 2546–2568. [Google Scholar] [CrossRef]
- Costa-Lotufo, L.V.; Carnevale-Neto, F.; Trindade-Silva, A.E.; Silva, R.R.; Silva, G.G.Z.; Wilke, D.V.; Pinto, F.C.L.; Sahm, B.D.B.; Jimenez, P.C.; Mendonça, J.N.; et al. Chemical profiling of two congeneric sea mat corals along the Brazilian coast: Adaptive and functional patterns. Chem. Commun. 2018, 54, 1952–1955. [Google Scholar] [CrossRef]
- Sogin, E.M.; Putnam, H.M.; Anderson, P.E.; Gates, R.D. Metabolomic signatures of increases in temperature and ocean acidification from the reef-building coral, Pocillopora damicornis. Metabolomics 2016, 12, 71. [Google Scholar] [CrossRef]
- Hartmann, A.C.; Petras, D.; Quinn, R.A.; Protsyuk, I.; Archer, F.I.; Ransome, E.; Williams, G.J.; Bailey, B.A.; Vermeij, M.J.A.; Alexandrov, T.; et al. Meta-mass shift chemical profiling of metabolomes from coral reefs. Proc. Natl. Acad. Sci. USA 2017, 114, 11685–11690. [Google Scholar] [CrossRef]
- McClanahan, T.R.; Donner, S.D.; Maynard, J.A.; MacNeil, M.A.; Graham, N.A.J.; Maina, J.; Baker, A.C.; Alemu I, J.B.; Beger, M.; Campbell, S.J.; et al. Prioritizing key resilience indicators to support coral reef management in a changing climate. PLoS ONE 2012, 7, e42884. [Google Scholar] [CrossRef]
- Bythell, J.; Pan, P.; Lee, J. Three-dimensional morphometric measurements of reef corals using underwater photogrammetry techniques. Coral Reefs 2001, 20, 193–199. [Google Scholar]
- Leon, J.X.; Roelfsema, C.M.; Saunders, M.I.; Phinn, S.R. Measuring coral reef terrain roughness using ‘Structure-from-Motion’ close-range photogrammetry. Geomorphology 2015, 242, 21–28. [Google Scholar] [CrossRef]
- Burns, J.; Delparte, D.; Gates, R.D.; Takabayashi, M. Integrating structure-from-motion photogrammetry with geospatial software as a novel technique for quantifying 3D ecological characteristics of coral reefs. PeerJ 2015, 3, e1077. [Google Scholar] [CrossRef]
- Burns, J.H.R.; Delparte, D.; Kapono, L.; Belt, M.; Gates, R.D.; Takabayashi, M. End of the chain? Rugosity and fine-scale bathymetry from existing underwater digital imagery using structure-from-motion (SfM) technology. Coral Reefs 2015, 35, 889–894. [Google Scholar]
- Storlazzi, C.D.; Dartnell, P.; Hatcher, G.A.; Gibbs, A.E. Assessing the impact of acute disturbances on the structure and composition of a coral community using innovative 3D reconstruction techniques. Methods Oceanogr. 2015, 15, 49–59. [Google Scholar]
- Protsyuk, I.; Melnik, A.V.; Nothias, L.F.; Rappez, L.; Phapale, P.; Aksenov, A.A.; Bouslimani, A.; Ryazanov, S.; Dorrestein, P.C.; Alexandrov, T. 3D molecular cartography using LC-MS facilitated by Optimus and ‘ili software. Nat. Protoc. 2018, 13, 134–154. [Google Scholar] [CrossRef]
- Townsley, S.J.; Lamarr, T.; Trott, E. A preliminary report of the rehabilitation of the littoral marine community on a new lava flow at Kapoho, Hawai‘i. Ecology 1962, 43, 728–730. [Google Scholar] [CrossRef]
- Burns, J.H.R.; Rozet, N.K.; Takabayashi, M. Morphology, severity, and distribution of growth anomalies in the coral, Montipora capitata, at Wai‘ōpae, Hawai‘i. Coral Reefs 2011, 30, 819–826. [Google Scholar] [CrossRef]
- Burns, J.H.R.; Gregg, T.M.; Takabayashi, M. Does coral disease affect symbiodinium? Investigating the impacts of growth anomaly on symbiont photophysiology. PLoS ONE 2013, 8, e72466. [Google Scholar] [CrossRef]
- Spies, N.P.; Takabayashi, M. Expression of galaxin and oncogene homologs in growth anomaly in the coral Montipora capitata. Dis. Aquat. Organ. 2013, 104, 249–256. [Google Scholar] [CrossRef]
- Burns, J.H.R.; Alexandrov, T.; Ovchinnikova, K.; Gates, R.D.; Takabayashi, M. Data for spatial analysis of growth anomaly lesions on coral colonies using 3D reconstruction techniques. Data Brief 2016, 9, 460–462. [Google Scholar] [CrossRef][Green Version]
- Little, M.; George, E.E.; Arts, M.G.I.; Shivak, J.; Benler, S.; Huckeba, J.; Quinlan, Z.A.; Boscaro, V.; Mueller, B.; Güemes, A.G.C.; et al. Three-dimentional molecular cartography of the Carribean reef-building coral Orbicella faveolata. Front. Mar. Sci. 2021, 8, 627724. [Google Scholar] [CrossRef]
- Li, X.Q.; Chen, Z.A.; Zhang, L.T.; Jia, D. Construction and accuracy test of a 3D model of non-metric camera images using Agisoft PhotoScan. Procedia Environ. Procedia Environ. Sci. 2016, 36, 184–190. [Google Scholar]
- Chen, Y.; Liu, H. Geomagic software for multi-sensor metrology systems. Sens. Rev. 2007, 27, 1–19. [Google Scholar]
- Marotz, C.; Schwartz, T.; Thompson, L.; Humphrey, G.; Gogul, G.; Gaffney, J.; Amir, A.; Knight, R. Earth Microbiome Project (EMP) high throughput (HTP) DNA extraction protocol. BioTechniques 2017, 62, 290–293. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Meyer, F.; Jansson, J.; Gordon, J.; Pace, N.; Tiedje, J.; Ley, R.; Fierer, N.; Field, D.; Kyrpides, N.; et al. The Earth Microbiome Project: Meeting report of the “1st EMP meeting on sample selection and acquisition” at Argonne National Laboratory October 6th 2010. Sens. Rev. 2010, 3, 249–253. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Olivon, F.; Grelier, G.; Roussi, F.; Litaudon, M.; Touboul, D. MZmine 2 Data-Preprocessing to enhance molecular networking reliability. Anal. Chem. 2017, 89, 7836–7840. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience 2013, 2, 16. [Google Scholar] [CrossRef]
- Yeung, N.; Cline, M.S.; Kuchinsky, A.; Smoot, M.E.; Bader, G.D. Exploring biological networks with Cytoscape software. Bioinformatics 2018, 23, 8–13. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bruno, F.; Gallo, A.; De Filippo, F.; Muzzupappa, M.; Petriaggi, B.D.; Caputo, P. 3D documentation and monitoring of the experimental cleaning operations in the underwater archaeological site of Baia (Italy). Digit. Herit. Int. Congr. (DigitalHeritage) 2013, 1, 105–112. [Google Scholar]
- McCarthy, J.; Benjamin, J. Multi-image Photogrammetry for Underwater Archaeological Site Recording: An Accessible, Diver-Based Approach. J. Marit. Archaeol. 2014, 9, 95–114. [Google Scholar] [CrossRef]
- Kersten, T.P.; Lindstaedt, M. Automatic 3D object reconstruction from multiple images for architectural, cultural heritage and archaeological applications using open-source software and web services. Photogramm. Fernerkun. 2012, 727–740. [Google Scholar] [CrossRef]
- Gordon, B.R.; Leggat, W.; Motti, C.A. Extraction protocol for nontargeted NMR and LC-MS metabolomics-based analysis of hard coral and their algal symbionts. Methods Mol. Biol. 2013, 1055, 129–147. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Speiser, J.L.; Miller, M.E.; Tooze, J.; Ip, E. A comparison of random forest variable selection methods for classification prediction modeling. Expert Syst. Appl. 2019, 134, 93–101. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Metabolomic data processing, analysis, and interpretation using MetaboAnalyst. Curr. Protoc. Bioinform. 2011, 34, 4.10.1–14.10.48. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Mohimani, H.; Gurevich, A.; Mikheenko, A.; Garg, N.; Nothias, L.F.; Ninomiya, A.; Takada, K.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of peptidic natural products through database search of mass spectra. Nat. Chem. Biol. 2017, 13, 30–37. [Google Scholar] [CrossRef]
- Walzthoeni, T.; Claassen, M.; Leitner, A.; Herzog, F.; Bohn, S.; Förster, F.; Beck, M.; Aebersold, R. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat. Methods 2012, 9, 901–903. [Google Scholar] [CrossRef]
- Yamashiro, H.; Oku, H.; Onaga, K.; Iwasaki, H.; Takara, K. Coral tumors store reduced level of lipids. J. Exp. Mar. Biol. 2001, 265, 171–179. [Google Scholar] [CrossRef]
- Richardson, L.L. Coral diseases: What is really known? Trends Ecol. Evol. 1998, 13, 438–443. [Google Scholar] [CrossRef]
- Garg, N.; Kapono, C.A.; Lim, Y.W.; Koyama, N.; Vermeij, M.J.; Conrad, D.; Rohwer, F.; Dorrestein, P.C. Mass spectral similarity for untargeted metabolomics data analysis of complex mixtures. Int. J. Mass Spectrom. 2015, 377, 719–727. [Google Scholar] [CrossRef]
- Quinn, R.A.; Vermeij, M.J.; Hartmann, A.C.; Galtier d’Auriac, I.; Benler, S.; Haas, A.; Quistad, S.D.; Lim, Y.W.; Little, M.; Sandin, S.; et al. Metabolomics of reef benthic interactions reveals a bioactive lipid involved in coral defense. Proc. Royal Soc. 2016, 283, 20160469. [Google Scholar]
- Mannochio-Russo, H.; Swift, S.O.I.; Nakayama, K.K.; Wall, C.B.; Gentry, E.C.; Panitchpakdi, M.; Caraballo-Rodriguez, A.M.; Aron, A.T.; Petras, D.; Dorrestein, K.; et al. Microbiomes and metabolomes of dominant coral reef primary producers illustrate a potential role for immunolipids in marine symbioses. Commun. Biol. 2023, 6, 896. [Google Scholar] [CrossRef]
- Speck, M.D.; Conachie, S.P. Widespread Oceanospirillaceae bacteria in Porites spp. J. Mar. Sci. 2012, 2012, 746720. [Google Scholar] [CrossRef]
- Clay, K.; Holah, J. Fungal endophyte symbiosis and plant diversity in successional fields. Science 1999, 285, 1742–1744. [Google Scholar] [CrossRef]
- Chang, J.Y.; Antonopoulos, D.A.; Kalra, A.; Tonelli, A.; Khalife, W.T.; Schmidt, T.M.; Young, V.B. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 2008, 197, 435–438. [Google Scholar] [CrossRef]
- Keesing, F.; Belden, L.K.; Daszak, P.; Dobson, A.; Harvell, C.D.; Holt, R.D.; Hudson, P.; Jolles, A.; Jones, K.E.; Mitchell, C.E.; et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 2010, 468, 647–652. [Google Scholar] [CrossRef]
- Muscatine, L.; Porter, J.W. Reef Corals: Mutualistic Symbioses Adapted to Nutrient-Poor Environments. Bioscience 1977, 3, 454–460. [Google Scholar] [CrossRef]
- Hatcher, B.G. Coral reef primary productivity: A beggar’s banquet. Trends Ecol. Evol. 1988, 27, 106–111. [Google Scholar] [CrossRef]
- Hanshew, A.S.; Mason, C.J.; Raffa, K.F.; Currie, C.R. Minimization of chloroplast contamination in 16S rRNA gene pyrosequencing of insect herbivore bacterial communities. J. Microbiol. Methods 2013, 95, 149–155. [Google Scholar] [CrossRef]
- Massé, A.; Domart-Coulon, I.; Golubic, S.; Duché, D.; Tribollet, A. Early skeletal colonization of the coral holobiont by the microboring Ulvophyceae Ostreobium sp. Sci. Rep. 2018, 8, 2293. [Google Scholar] [CrossRef]
- Claar, D.C.; Takabayashi, M. The effects of growth anomaly on susceptibility of Montipora capitata to turf algal overgrowth. Mar. Freshw. Res. 2015, 67, 666–670. [Google Scholar] [CrossRef]
- Roder, C.; Arif, C.; Bayer, T.; Aranda, M.; Daniels, C.; Shibl, A.; Chavanich, S.; Voolstra, C.R. Bacterial profiling of White Plague Disease in a comparative coral species framework. ISME J. 2014, 8, 31–39. [Google Scholar] [CrossRef]
- Sunagawa, S.; DeSantis, T.Z.; Piceno, Y.M.; Brodie, E.L.; DeSalvo, M.K.; Voolstra, C.R.; Weil, E.; Andersen, G.L.; Medina, M. Bacterial diversity and White Plague Disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 2009, 3, 512–521. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakoa, J.W.P., III; Burns, J.H.R.; Steward, K.; Kapono, L.M.; Kapono, C.A. Molecular Cartography of a Hawaiian Coral Assemblage. Diversity 2023, 15, 1061. https://doi.org/10.3390/d15101061
Nakoa JWP III, Burns JHR, Steward K, Kapono LM, Kapono CA. Molecular Cartography of a Hawaiian Coral Assemblage. Diversity. 2023; 15(10):1061. https://doi.org/10.3390/d15101061
Chicago/Turabian StyleNakoa, Joseph W. P., III, John H. R. Burns, Kanoelani Steward, Lauren M. Kapono, and Clifford A. Kapono. 2023. "Molecular Cartography of a Hawaiian Coral Assemblage" Diversity 15, no. 10: 1061. https://doi.org/10.3390/d15101061
APA StyleNakoa, J. W. P., III, Burns, J. H. R., Steward, K., Kapono, L. M., & Kapono, C. A. (2023). Molecular Cartography of a Hawaiian Coral Assemblage. Diversity, 15(10), 1061. https://doi.org/10.3390/d15101061






