The Evolution of Collembola Higher Taxa (Arthropoda, Hexapoda) Based on Mitogenome Data †
Abstract
1. Introduction
2. Materials and Methods
2.1. Species and Mitogenomic Data Acquisition
2.2. Phylogenetic Analyses
2.3. Tree Topology Tests
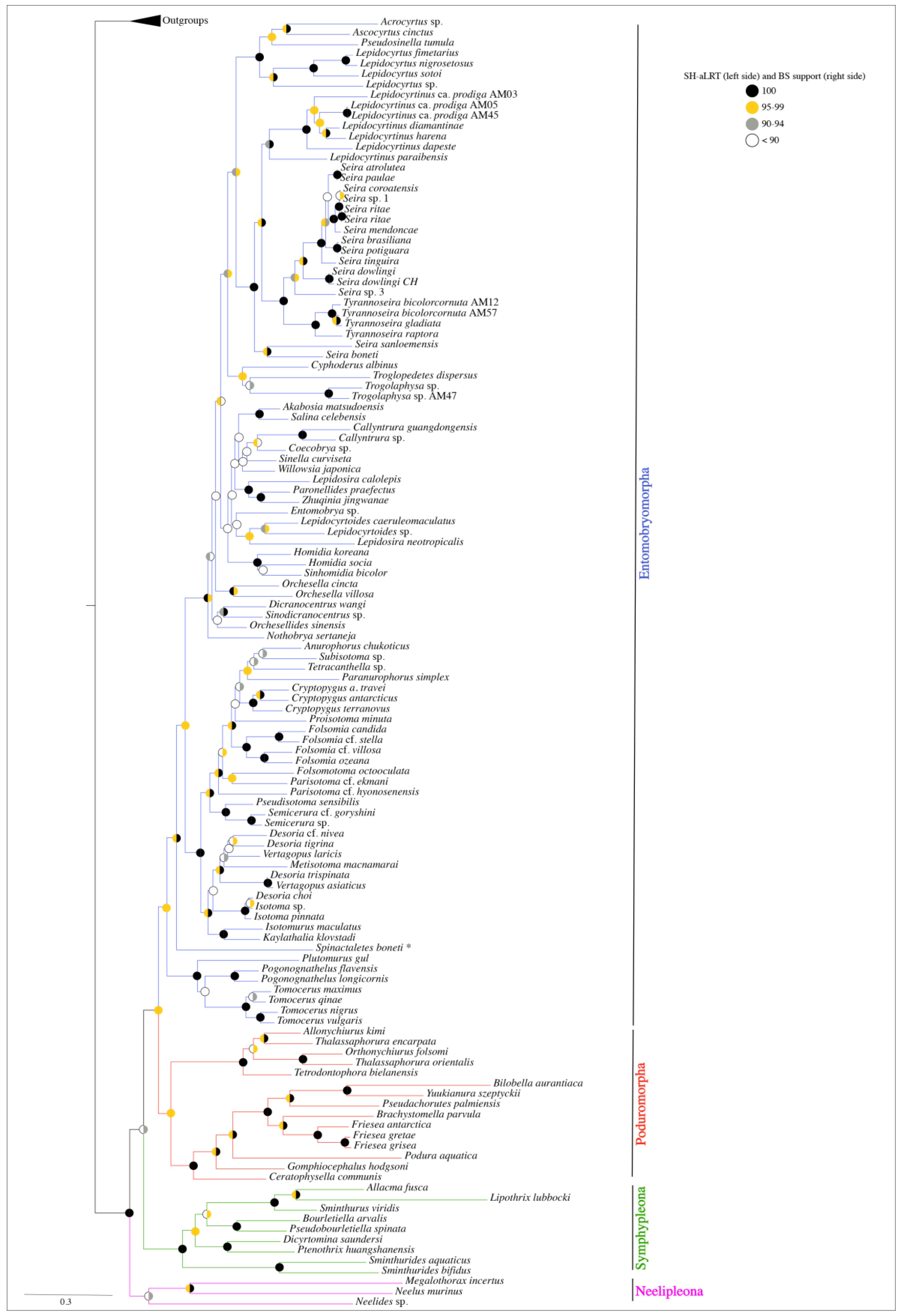
3. Results
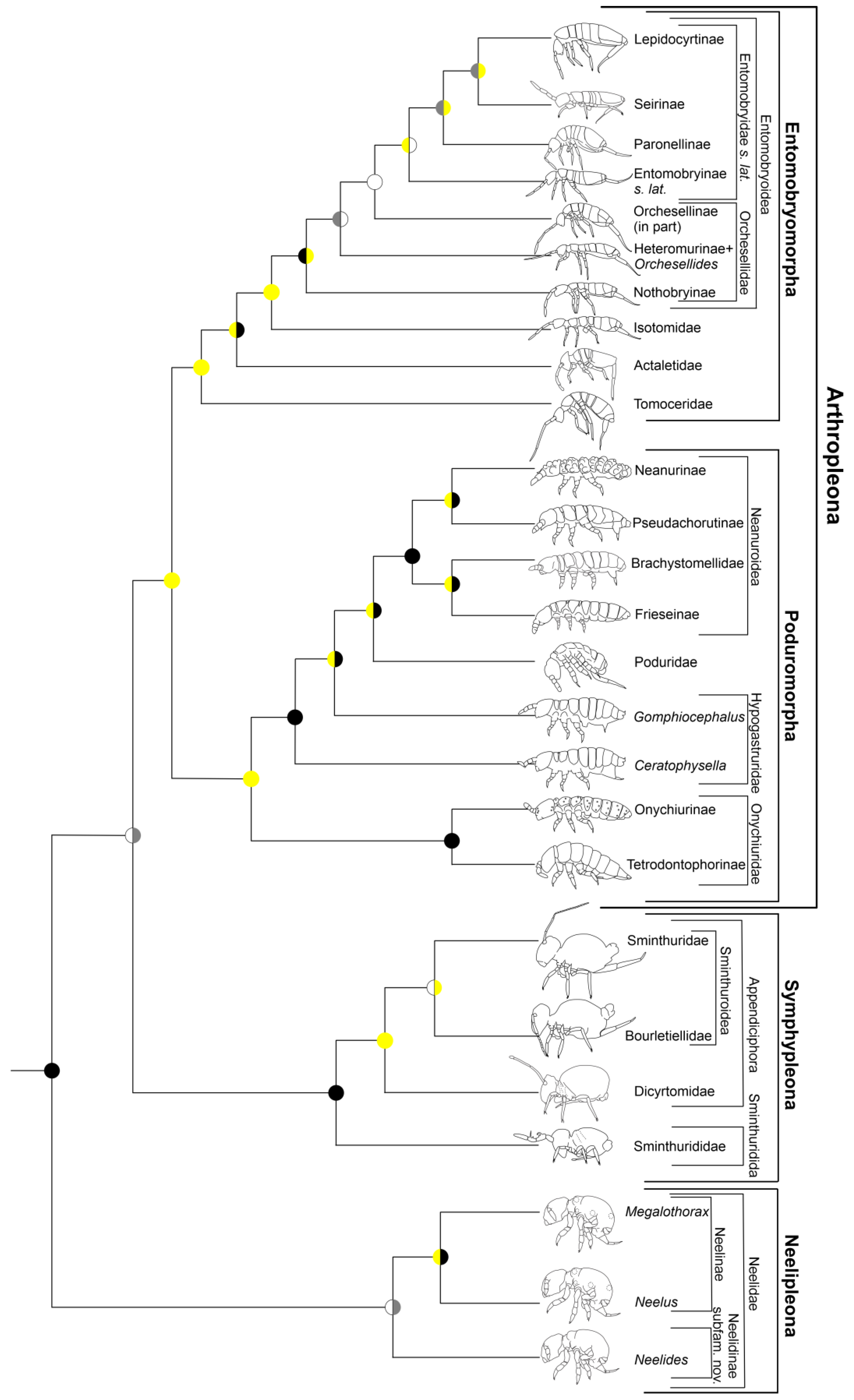
3.1. Matrices and Trees
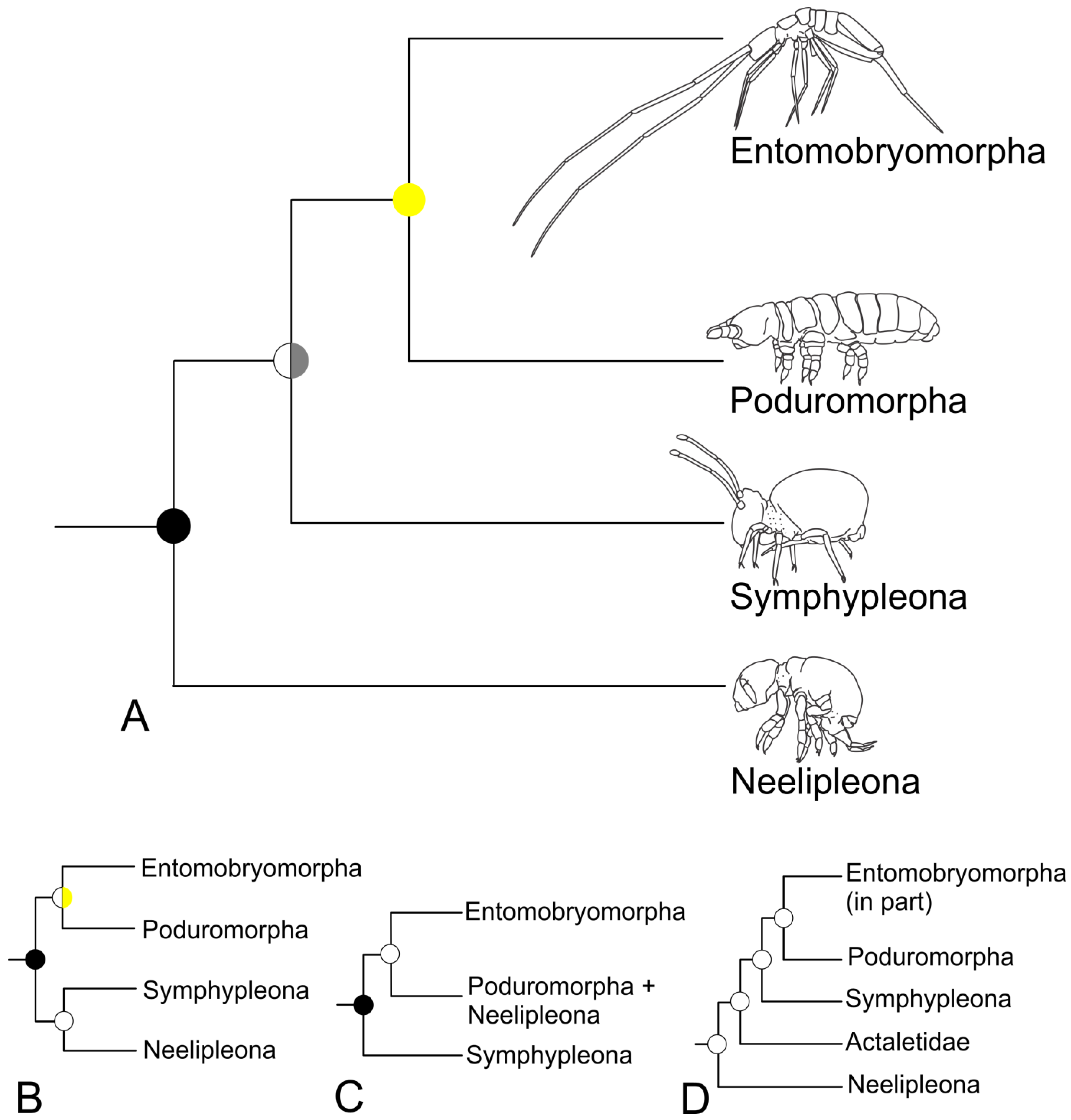
3.2. Phylogeny of the Orders
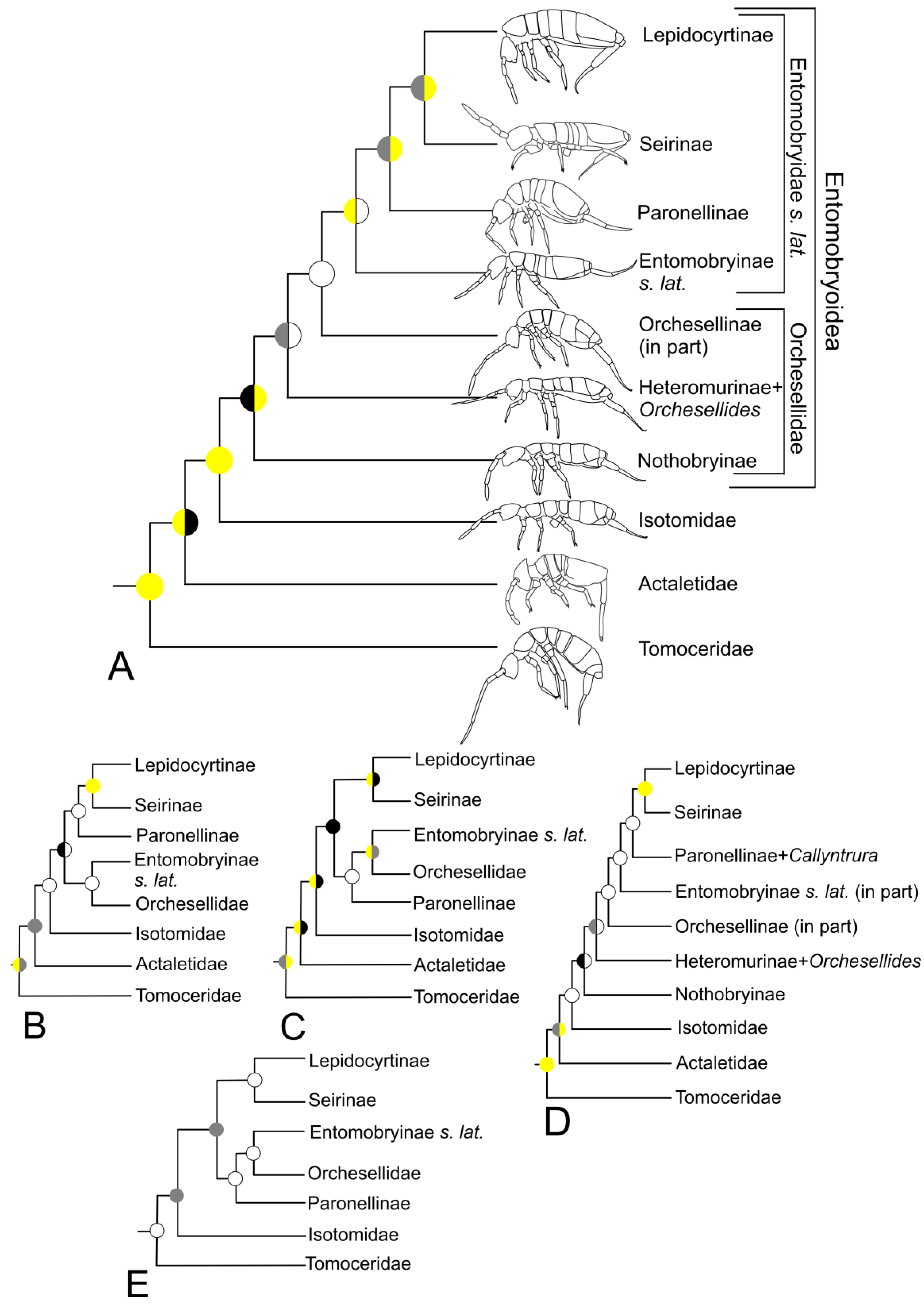
3.3. Entomobryomorpha
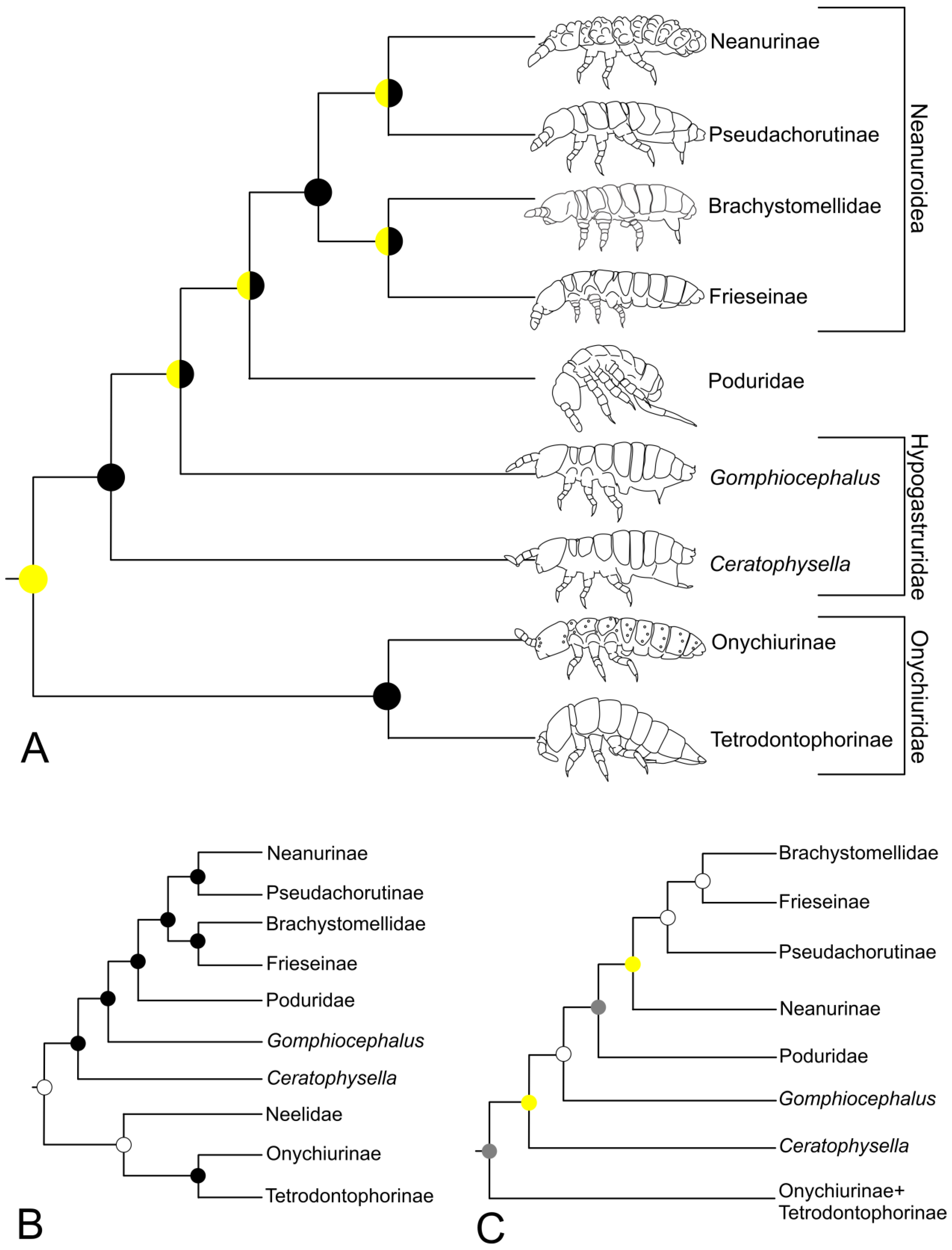
3.4. Poduromorpha
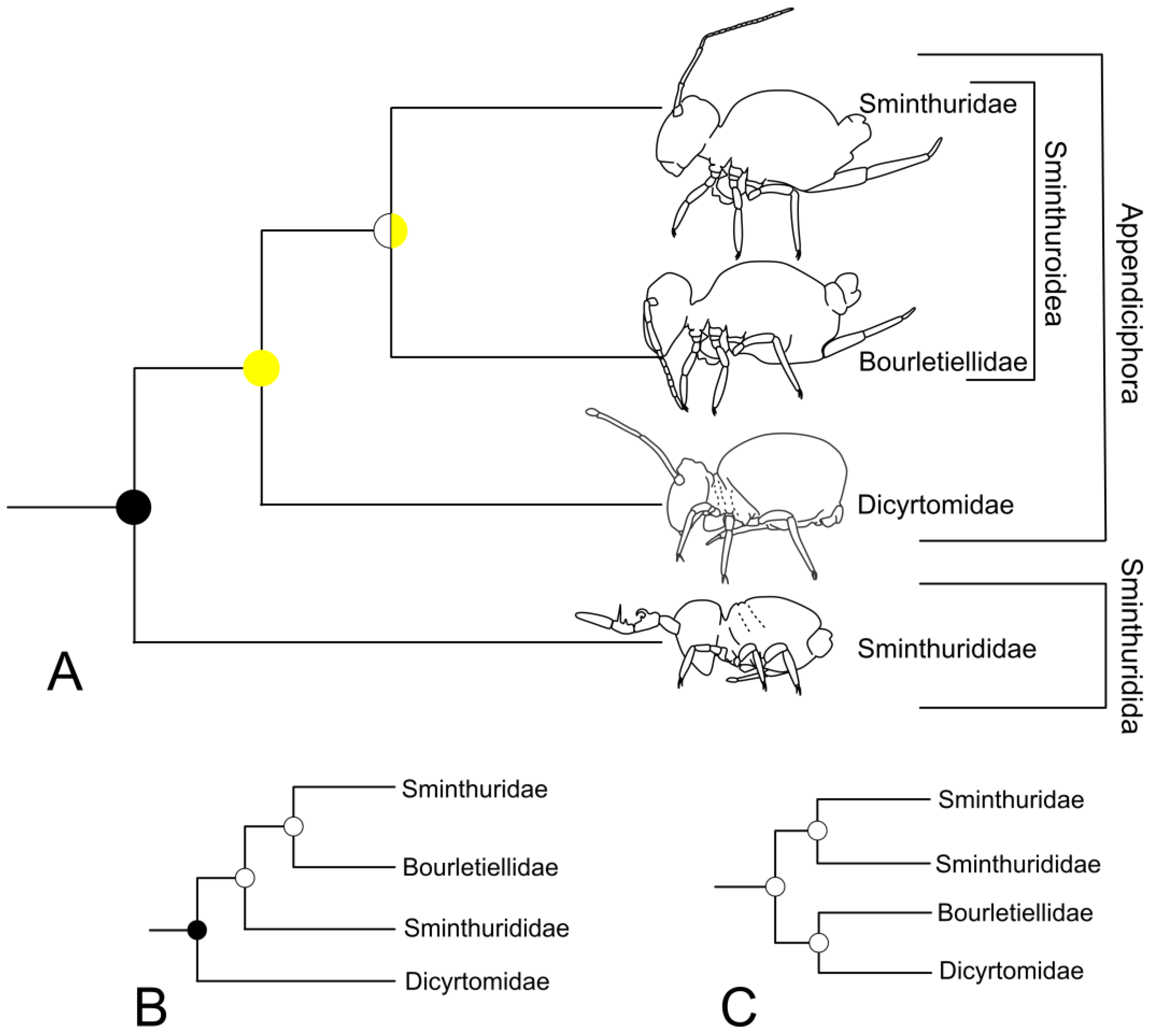
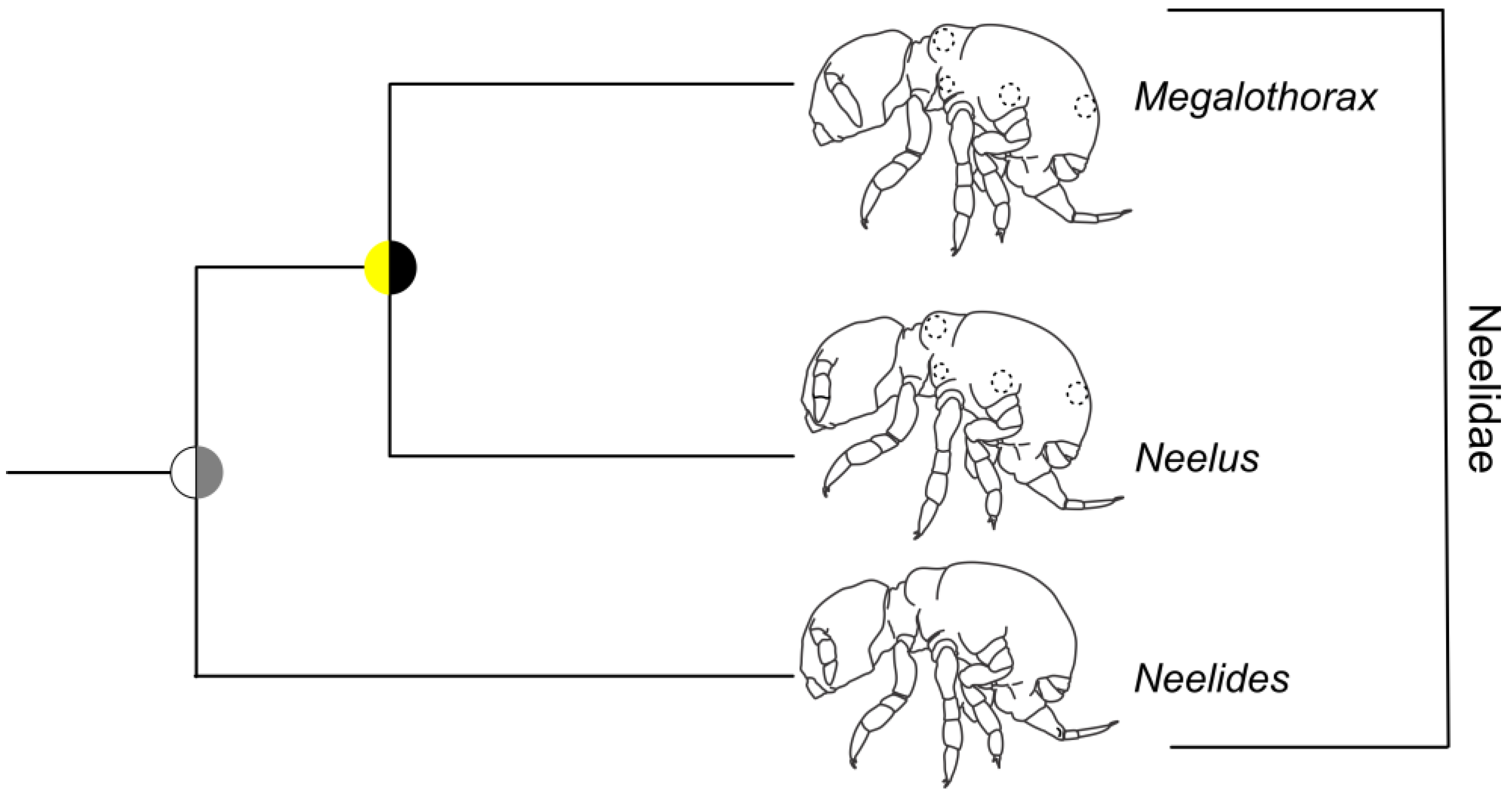
3.5. Symphypleona and Neelipleona
4. Discussion
4.1. Phylogeny of the Orders
4.2. Entomobryomorpha
4.3. Poduromorpha
4.4. Symphypleona and Neelipleona
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bellinger, P.F.; Christiansen, K.A.; Janssens, F. Checklist of the Collembola of the World. Available online: http://www.collembola.org (accessed on 16 November 2022).
- Potapov, A.; Bellini, B.C.; Chown, S.L.; Deharveng, L.; Janssens, F.; Kovác, L.; Kuznetsova, N.; Ponge, J.-F.; Potapov, M.; Querner, P.; et al. Towards a global synthesis of Collembola knowledge—Challenges and potential solutions. Soil Org. 2020, 92, 161–188. [Google Scholar] [CrossRef]
- Zhang, Z.-Q. Animal biodiversity: An outline of higher-level classification and survey of taxonomic richness. Zootaxa 2011, 3148, 3–6. [Google Scholar] [CrossRef]
- Cicconardi, F.; Fanciulli, P.P.; Emerson, B.C. Collembola, the biological species concept and the underestimation of global species richness. Mol. Ecol. 2013, 22, 5382–5396. [Google Scholar] [CrossRef] [PubMed]
- Börner, C. Neue Collembolenformen und zur Nomenclatur der Collembola Lubbock. Zool. Anz. 1901, 24, 696–712. [Google Scholar]
- Börner, C. Das System der Collembolen nebst Beschreibung neuer Collembolen des Hamburger Naturhistorischen Museums. Mitt. Nat. Mus. Hambg. 1906, 23, 147–188. [Google Scholar]
- Paclt, J. Biologie der primär flügellosen Insekten. Jena 1956, 6, 1–258. [Google Scholar]
- Yosii, R. Phylogenetische bedeutung der chaetotaxie bei den Collembolen. Contrib. Biol. Lab. Kyoto Univ. 1961, 12, 1–37. [Google Scholar]
- Salmon, J.T. An Index to the Collembola. R. Soc. N. Z. Bull. 1964, 7, 1–144. [Google Scholar]
- Richards, W.R. Generic classification, evolution, and biogeography of the Sminthuridae of the world (Collembola). Mem. Entomol. Soc. Can. 1968, 53, 3–54. [Google Scholar] [CrossRef]
- Uchida, H. Tentative key to the Japanese genera of Collembola, in relation to the world genera of this order I. Sci. Rep. Hirosaki Univ. 1971, 18, 63–76. [Google Scholar]
- Cassagnau, P. La phylogénie des Collemboles à la lumière des structures endocrines rétrocérébrales. I Symp. Inter. Zoolfilogenia 1971, 1, 333–349. [Google Scholar]
- Massoud, Z. Essai de synthèse sur la phylogénie des Collemboles. Rev. Ecol. Biol. Sol. 1976, 13, 241–252. [Google Scholar]
- Szeptycki, A. Chaetotaxy of the Entomobryidae and Its Phylogenetical Significance. Morpho-Systematic Studies on Collembola; Polska Akademia Nauk: Kraków, Poland, 1979; Volume IV, pp. 1–219. [Google Scholar]
- Betsch, J.M. Éléments pour une monographie des Collemboles Symplyplêones (Hexapodes, Aptérygotes). Mém. Mus. Natl. Hist. Nat. Sér. A Zool. 1980, 116, 1–227. [Google Scholar]
- Moen, P.; Ellis, W.N. Morphology and Taxonomic Position of Podura aquatica (Collembola). Entomol. Gen. 1984, 9, 193–204. [Google Scholar] [CrossRef]
- Bretfeld, G. Phylogenetic systematics of the higher taxa of Symphypleona Börner, 1901 (Insecta, Entognatha, Collembola). Proc. 2nd Intern. Sem. Apterygota 1986, 1, 302–311. [Google Scholar]
- Soto-Adames, F.N.; Barra, J.A.; Christiansen, K.; Jordana, R. Suprageneric classification of Collembola Entomobryomorpha. Ann. Entomol. Soc. Am. 2008, 101, 501–513. [Google Scholar] [CrossRef]
- Cassagnau, P. Des hexapodes vieux de 400 millions d’années: Les collemboles. 1.—Biologie et Évolution. Ann. Biol. 1990, 29, 1–69. [Google Scholar]
- Deharveng, L. Recent advances in Collembola systematics. Pedobiologia 2004, 48, 415–433. [Google Scholar] [CrossRef]
- Fjellberg, A. Redescription of Mackenziella psocoides Hammer, 1953 and discussion of its systematic position (Collembola, Mackenziellidae). Proc. 2nd Intern. Sem. Apterygota 1989, 1, 93–105. [Google Scholar]
- Fanciulli, P.P.; Melegari, D.; Carapelli, A.; Frati, F.; Dallai, R. Population structure, gene flow and evolutionary relationships in four species of the genera Tomocerus and Pogonognathellus (Collembola, Tomoceridae). Biol. J. Linn. Soc. 2000, 70, 221–238. [Google Scholar] [CrossRef][Green Version]
- Fanciulli, P.P.; Summa, D.; Dallai, R.; Frati, F. High levels of genetic variability and population differentiation in Gressittacantha terranova (Collembola, Hexapoda) from Victoria Land, Antarctica. Antarct. Sci. 2001, 13, 246–254. [Google Scholar] [CrossRef]
- Timmermans, M.J.T.N.; Ellers, J.; Mariën, J.; Verhoef, S.C.; Ferwerda, E.B.; Van Straalen, N.M. Genetic structure in Orchesella cincta (Collembola): Strong subdivision of European populations inferred from mtDNA and AFLP markers. Mol. Ecol. 2005, 14, 2017–2024. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.I.; Frati, F.; McGaughran, A.; Spinsanti, G.; Hogg, I.D. Phylogeographic structure suggests multiple glacial refugia in northern Victoria Land for the endemic Antarctic springtail Desoria klovstadi (Collembola, Isotomidae). Zool. Scr. 2007, 36, 201–212. [Google Scholar] [CrossRef]
- Torricelli, G.; Frati, F.; Convey, P.; Telford, M.; Carapelli, A. Population structure of Friesea grisea (Collembola, Neanuridae) in the Antarctic Peninsula and Victoria Land: Evidence for local genetic differentiation of pre-Pleistocene origin. Antarct. Sci. 2010, 22, 757–765. [Google Scholar] [CrossRef]
- Schneider, C.; Cruaud, C.; D’Haese, C.A. Unexpected diversity in Neelipleona revealed by molecular phylogeny approach (Hexapoda, Collembola). Soil Org. 2011, 83, 383–398. [Google Scholar]
- Schneider, C.; Zon, S.D.; D’Haese, C.A. Megalothorax laevis (Neelipleona, Neelidae): Account of a neglected springtail widely distributed in the intertropical zone. Int. J. Trop. Insect Sci. 2018, 38, 168–191. [Google Scholar] [CrossRef]
- Katz, A.D.; Giordano, R.; Soto-Adames, F.N. Operational criteria for cryptic species delimitation when evidence is limited, as exemplified by North American Entomobrya (Collembola: Entomobryidae). Zool. J. Linn. Soc. 2015, 173, 818–840. [Google Scholar] [CrossRef]
- Zhang, F.; Jantarit, S.; Nilsai, A.; Stevens, M.I.; Ding, Y.; Satasook, C. Species delimitation in the morphologically conserved Coecobrya (Collembola: Entomobryidae): A case study integrating morphology and molecular traits to advance current taxonomy. Zool. Scr. 2018, 47, 342–356. [Google Scholar] [CrossRef]
- Yu, D.; Qin, C.; Ding, Y.; Hu, F.; Zhang, F.; Liu, M. Revealing species diversity of Tomocerus ocreatus complex (Collembola: Tomoceridae): Integrative species delimitation and evaluation of taxonomic characters. Arthropod Syst. Phylogeny 2018, 76, 147–172. [Google Scholar]
- Mateos, E.; Winkler, D.; Riutort, M.; Álvarez-Presas, M. New morphological and molecular data reveal an important underestimation of species diversity and indicate evolutionary patterns in European Lepidocyrtus (Collembola: Entomobryidae). Invertebr. Syst. 2021, 35, 471–492. [Google Scholar] [CrossRef]
- Dukes, C.D.; Janssens, F.; Recuero, E.; Caterino, M.S. Specific and Intraspecific Diversity of Symphypleona and Neelipleona (Hexapoda: Collembola) in Southern High Appalachia (USA). Diversity 2022, 14, 847. [Google Scholar] [CrossRef]
- Soto-Adames, F.N. Molecular phylogeny of the Puerto Rican Lepidocyrtus and Pseudosinella (Hexapoda: Collembola), a validation of Yoshii’s “color pattern species”. Mol. Phylogenet. Evol. 2002, 25, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; D’Haese, C.A. Morphological and molecular insights on Megalothorax: The largest Neelipleona genus revisited (Collembola). Invertebr. Syst. 2013, 27, 317–364. [Google Scholar] [CrossRef]
- Mateos, E.; Escuer, P.; Busmachiu, G.; Riutort, M.; Álvarez-Presas, M. Untangling Lepidocyrtus (Collembola, Entomobryidae): New molecular data shed light on the relationships of the European groups. Invertebr. Syst. 2018, 32, 639–651. [Google Scholar] [CrossRef]
- Winkler, D.; Mateos, E.; Traser, G.; Lakatos, F.; Tóth, V. New Insight into the Systematics of European Lepidocyrtus (Collembola: Entomobryidae) Using Molecular and Morphological Data. Insects 2020, 11, 302. [Google Scholar] [CrossRef]
- Zhang, F.; Pan, Z.; Wu, J.; Ding, Y.; Yu, D.; Wang, B. Dental scales could occur in all scaled subfamilies of Entomobryidae (Collembola): New definition of Entomobryinae with description of a new genus and three new species. Invertebr. Syst. 2016, 30, 598–615. [Google Scholar] [CrossRef]
- Zhang, F.; Ma, Y.; Greenslade, P. New Australian Paronellidae (Collembola) reveal anomalies in existing tribal diagnoses. Invertebr. Syst. 2017, 31, 375–393. [Google Scholar] [CrossRef]
- Nunes, R.C.; Godeiro, N.N.; Pacheco, G.; Liu, S.; Gilbert, M.T.P.; Alvarez-Valin, F.; Zhang, F.; Bellini, B.C. The discovery of Neotropical Lepidosira (Collembola, Entomobryidae) and its systematic position. Zool. Scr. 2019, 48, 783–800. [Google Scholar] [CrossRef]
- Godeiro, N.N.; Pacheco, G.; Liu, S.; Cipola, N.G.; Berbel-Filho, W.M.; Zhang, F.; Gilbert, M.T.; Bellini, B.C. Phylogeny of Neotropical Seirinae (Collembola, Entomobryidae) based on mitochondrial genomes. Zool. Scr. 2020, 49, 329–339. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, Z.; Dong, R.R.; Deharveng, L.; Stevens, M.I.; Huang, Y.H.; Zhu, C.D. Molecular phylogeny reveals independent origins of body scales in Entomobryidae (Hexapoda: Collembola). Mol. Phylogenet. Evol. 2014, 70, 231–239. [Google Scholar] [CrossRef]
- Zhang, F.; Sun, D.; Yu, D.; Wang, B. Molecular phylogeny supports S-chaetae as a key character better than jumping organs and body scales in classification of Entomobryoidea (Collembola). Sci. Rep. 2015, 5, 12471. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Bellini, B.C.; Soto-Adames, F.N. New insights into the systematics of Entomobryoidea (Collembola: Entomobryomorpha): First instar chaetotaxy, homology and classification. Zool. Syst. 2019, 44, 249–278. [Google Scholar] [CrossRef]
- Zhang, F.; Deharveng, L. Systematic revision of Entomobryidae (Collembola) by integrating molecular and new morphological evidence. Zool. Scr. 2015, 44, 298–311. [Google Scholar] [CrossRef]
- Yu, D.; Zhang, F.; Stevens, M.I.; Yan, Q.; Liu, M.; Hu, F. New insight into the systematics of Tomoceridae (Hexapoda, Collembola) by integrating molecular and morphological evidence. Zool. Scr. 2016, 45, 286–299. [Google Scholar] [CrossRef]
- Yu, D.; Ding, Y.; Tihelka, E.; Cai, C.; Hu, F.; Liu, M.; Zhang, F. Phylogenomics of Elongate-Bodied Springtails Reveals Independent Transitions from Aboveground to Belowground Habitats in Deep Time. Syst. Biol. 2022, 71, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- D’Haese, C.A. Were the first springtails semi-aquatic? A phylogenetic approach by means of 28S rDNA and optimization alignment. Proc. R. Soc. Lond. 2002, 269, 1143–1151. [Google Scholar] [CrossRef]
- Luan, Y.-X.; Mallatt, J.M.; Xie, R.D.; Yang, Y.M.; Yin, W.Y. The phylogenetic positions of three basal-hexapod groups (Protura, Diplura, and Collembola) based on ribosomal RNA gene sequences. Mol. Biol. Evol. 2005, 22, 1579–1592. [Google Scholar] [CrossRef]
- Gao, Y.; Bu, Y.; Luan, Y.-X. Phylogenetic Relationships of Basal Hexapods Reconstructed from Nearly Complete 18S and 28S rRNA Gene Sequences. Zool. Sci. 2008, 25, 1139–1145. [Google Scholar] [CrossRef]
- Xiong, Y.; Gao, Y.; Yin, W.Y.; Luan, Y.X. Molecular phylogeny of Collembola inferred from ribosomal RNA genes. Mol. Phylogenet. Evol. 2008, 49, 728–735. [Google Scholar] [CrossRef]
- Carapelli, A.; Convey, P.; Nardi, F.; Frati, F. The mitochondrial genome of the antarctic springtail Folsomotoma octooculata (Hexapoda; Collembola), and an update on the phylogeny of collembolan lineages based on mitogenomic data. Entomologia 2014, 2, 46–55. [Google Scholar] [CrossRef]
- Sun, X.; Yu, D.; Xie, Z.; Dong, J.; Ding, Y.; Yao, H.; Greenslade, P. Phylomitogenomic analyses on collembolan higher taxa with enhanced taxon sampling and discussion on method selection. PLoS ONE 2020, 15, e0230827. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Ding, Y.; Orr, M.C.; Zhang, F. Streamlining universal single-copy orthologue and ultraconserved element design: A case study in Collembola. Mol. Ecol. Resour. 2020, 20, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Leo, C.; Carapelli, A.; Cicconardi, F.; Frati, F.; Nardi, F. Mitochondrial genome diversity in Collembola: Phylogeny, dating and gene order. Diversity 2019, 11, 169. [Google Scholar] [CrossRef]
- Cucini, C.; Fanciulli, P.P.; Frati, F.; Convey, P.; Nardi, F.; Carapelli, A. Re-evaluating the internal phylogenetic relationships of Collembola by means of Mitogenome Data. Genes 2021, 12, 44. [Google Scholar] [CrossRef]
- Ma, Y.; Huang, C.-W.; Luan, Y.-X.; Chen, W.J. The mitochondrial genome of a minute springtail species Megalothorax incertus (Collembola: Neelipleona: Neelidae). Mitochondrial DNA B Resour. 2021, 6, 2430–2432. [Google Scholar] [CrossRef]
- Song, F.; Li, H.; Jiang, P.; Zhou, X.G.; Liu, J.P.; Sun, C.H.; Vogler, A.P.; Cai, W.Z. Capturing the phylogeny of Holometabola with mitochondrial genome data and Bayesian site-heterogeneous mixture models. Genome Biol. Evol. 2016, 8, 1411–1426. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, J.J.; Li, N.; Ma, G.Y.; Li, W.H. The first mitochondrial genome from Scopuridae (Insecta: Plecoptera) reveals structural features and phylogenetic implications. Int. J. Biol. Macromol. 2019, 122, 893–902. [Google Scholar] [CrossRef]
- Godeiro, N.N.; Bellini, B.C.; Ding, N.; Xu, C.; Ding, Y.; Zhang, F. A mitogenomic phylogeny of the Entomobryoidea (Collembola): A comparative perspective. Zool. Scr. 2021, 50, 658–666. [Google Scholar] [CrossRef]
- Godeiro, N.N.; Zhang, F.; Cipola, N.G. First partial mitogenome of a new Seira Lubbock species (Collembola, Entomobryidae, Seirinae) from Cambodia reveals a possible separate lineage from the Neotropical Seirinae. Zootaxa 2020, 4890, 451–472. [Google Scholar] [CrossRef]
- Godeiro, N.N.; Zhang, F. First record of Seira dowlingi (Wray, 1953) (Collembola, Entomobryidae, Seirinae) from China and mitogenome comparison with the New World specimens. Zootaxa 2021, 5020, 191–196. [Google Scholar] [CrossRef]
- Godeiro, N.N.; Palacios-Vargas, J.G.; Gao, Y.; Bu, Y. Complete mitochondrial genome of the Mexican marine littoral hygrophilous Spinactaletes boneti (Collembola: Actaletidae) and its phylogenetic placement. Mitochondrial DNA B Resour. 2022, 7, 755–757. [Google Scholar] [CrossRef] [PubMed]
- Nardi, F.; Cucini, C.; Leo, C.; Frati, F.; Fanciulli, P.P.; Carapelli, A. The complete mitochondrial genome of the springtail Allacma fusca, the internal phylogenetic relationships and gene order of Symphypleona. Mitochondrial DNA B Resour. 2020, 5, 3103–3105. [Google Scholar] [CrossRef] [PubMed]
- Beutel, R.G.; Yavorskaya, M.I.; Mashimo, Y.; Fukui, M.; Meusemann, K. The Phylogeny of Hexapoda (Arthropoda) and the Evolution of Megadiversity. Proc. Arthropod. Embryol. Soc. Jpn. 2017, 51, 1–15. [Google Scholar]
- Sasaki, G.; Ishiwata, K.; Machida, R.; Miyata, T.; Su, Z.H. Molecular phylogenetic analyses support the monophyly of Hexapoda and suggest the paraphyly of Entognatha. BMC Evol. Biol. 2013, 13, 236. [Google Scholar] [CrossRef] [PubMed]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Smirnov, V.; Warnow, T. MAGUS: Multiple sequence alignment using graph clustering. Bioinformatics 2021, 37, 1666–1672. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Criscuolo, A.; Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010, 10, 2–21. [Google Scholar] [CrossRef]
- Steenwyk, J.L.; Buida, T.J.; Labella, A.L.; Li, Y.; Shen, X.X.; Rokas, A. PhyKIT: A broadly applicable UNIX shell toolkit for processing and analyzing phylogenomic data. Bioinformatics 2021, 37, 2325–2331. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.; Wong, T.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Susko, E.; Minh, B.; Roger, A.J. Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Syst. Biol. 2018, 67, 216–235. [Google Scholar] [CrossRef] [PubMed]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J. PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef]
- Hoang, D.T.; Vinh, L.S.; Flouri, T.; Stamatakis, A.; von Haeseler, A.; Minh, B.Q. MPBoot: Fast phylogenetic maximum parsimony tree inference and bootstrap approximation. BMC Evol. Biol. 2018, 18, 11. [Google Scholar] [CrossRef]
- Rambaut, A.; Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. FigTree V1.3.1. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 16 November 2022).
- Soto-Adames, F.N. Revision de la familia Actaletidae Borner, 1902 (Insects: Collembola). Caribb. J. Sci. 1988, 24, 161–196. [Google Scholar]
- Bellini, B.C. Colêmbolos: Uma riqueza microscópica no Semiárido. In Conhecendo os artrópodes do Semiárido, 1st ed.; Bravo, F., Calor, A.R., Eds.; Métis Produção Editorial: São Paulo, Brazil, 2016; Volume 1, pp. 43–55. [Google Scholar]
- Sánchez-García, A.; Engel, M.S. Long-term stasis in a diverse fauna of Early Cretaceous springtails (Collembola: Symphypleona). J. Syst. Palaeontol. 2016, 15, 513–537. [Google Scholar] [CrossRef]
- Bretfeld, G. Synopses on Palaeartic Collembola, Volume 2. Symphypleona. Abh. Ber. Naturkundemus. Gorlitz 1999, 71, 1–318. [Google Scholar]
- D’Haese, C.A. Homology and morphology in Poduromorpha (Hexapoda, Collembola). Eur. J. Entomol. 2003, 101, 385–407. [Google Scholar] [CrossRef]
- Hopkin, S.P. Biology of the Springtails (Insecta: Collembola), 1st ed.; Oxford University Press: New York, NY, USA, 1997; pp. 1–330. [Google Scholar]
- Greenslade, P.; Stevens, M.I.; Torricelli, G.; D’Haese, C.A. An ancient Antarctic endemic genus restored: Morphological and molecular support for Gomphiocephalus hodgsoni (Collembola: Hypogastruridae). Syst. Entomol. 2011, 36, 223–240. [Google Scholar] [CrossRef]
- D’Haese, C.A. Morphological appraisal of Collembola phylogeny with special emphasis on Poduromorpha and a test of the aquatic origin hypothesis. Zool. Scr. 2003, 32, 563–586. [Google Scholar] [CrossRef]
- Zhang, F.; Deharveng, L. First instar tibiotarsal chaetotaxy supports the Entomobryidae and Symphypleona (Collembola) forming a cluster in a phylogenetic tree. Zootaxa 2015, 3955, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Bonet, F. Monografia de la familia Neelidae (Collembola). Rev. Soc. Mex. Hist. Nat. 1947, 8, 131–192. [Google Scholar]
- Betsch, J.M.; Waller, A. Chaetotaxic nomenclature of the head, thorax and abdomen in Symphypleona (Insecta, Collembola). Acta Zool. Fenn. 1994, 195, 5–12. [Google Scholar]
- Schneider, C. Morphological review of the order Neelipleona (Collembola) through the redescription of the type species of Acanthoneelidus, Neelides and Neelus. Zootaxa 2017, 4308, 1–94. [Google Scholar] [CrossRef]
- Betsch, J.M. An ontogenetically focused chaetotaxial scheme in Symphypleona (collembolan): The 6th abdominal segment. Pedobiologia 1997, 41, 13–18. [Google Scholar]
- Papáč, V.; Palacios-Vargas, J.G. A new genus of Neelidae (Collembola) from Mexican caves. ZooKeys 2016, 569, 37–51. [Google Scholar] [CrossRef][Green Version]
- Yosii, R. Studies on the Collembolan Genus Hypogastrura II. Neartic Forms collected by Prof. F. Bonet. Contrib. Biol. Lab. Kyoto Univ. 1962, 13, 1–25. [Google Scholar]
- Nayrolles, P. Chetotaxie tibiotarsale des collemboles symphyplèones. Trav. Lab. Ecobiol. Arthropodes Edaphiques 1988, 5, 1–19. [Google Scholar]
- Bretfeld, G.; Griegel, A. Description of a new Neelidae genus and species and of new specimens of Sminthurides annulicornis Axelson 1905 from Poland. Senckenberg. Biol. 1999, 79, 211–223. [Google Scholar]
- Havird, J.C.; Sloan, D.B. The roles of mutation, selection, and expression in determining relative rates of evolution in mitochondrial versus nuclear genomes. Mol. Biol. Evol. 2016, 33, 3042–3053. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Cipola, N.G.; Pan, Z.-X.; Ding, Y. New insight into the systematics of Heteromurini (Collembola: Entomobryidae: Heteromurinae) with special reference to Alloscopus and Sinodicranocentrus gen.n. Arth. Syst. Phylogeny 2020, 78, 1–16. [Google Scholar] [CrossRef]
- Ding, Y.-H.; Yu, D.-Y.; Guo, W.B.; Li, J.-N.; Zhang, F. Molecular phylogeny of Entomobrya (Collembola: Entomobryidae) from China: Color pattern groups and multiple origins. Insect Sci. 2018, 26, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Soto-Adames, F.N. Postembryonic development of the dorsal chaetotaxy in Seira dowlingi (Collembola, Entomobryidae); with an analysis of the diagnostic and phylogenetic significance of primary chaetotaxy in Seira. Zootaxa 2008, 1683, 1–31. [Google Scholar] [CrossRef]
- Potapov, M. Isotomidae. In Synopses on Palearctic Collembola, 1st ed.; Dunger, W., Ed.; Abdhandlungen und Berichte des Naturkundemuseums Görlitz: Görlitz, Germany, 2001; Volume 3, pp. 1–603. [Google Scholar]
- Nunes, R.C.; Bellini, B.C. A new species of Nothobrya Arlé, 1961 (Collembola: Entomobryidae) from Brazil and notes on key characters for Nothobryinae taxonomy, with an identification key to the species of the subfamily. Zootaxa 2019, 4615, 375–391. [Google Scholar] [CrossRef] [PubMed]
- Nunes, R.C.; Santos-Costas, R.C.; Bellini, B.C. The first Neotropical Capbrya Barra, 1999 (Collembola: Orchesellidae: Nothobryinae) and the reinterpretation of Nothobryinae systematics. Zool. Anz. 2020, 288, 24e42. [Google Scholar] [CrossRef]
- Massoud, Z. Monographie des Neanuridae, Collemboles Poduromorphes apiéces buccales modifiées. In Biologie de l’Amerique Australe, 1st ed.; Delamare Deboutteville, C., Rapoport, E.H., Eds.; Éditions du CNRS: Paris, France, 1967; Volume 3, pp. 7–399. [Google Scholar]
- Börner, C. Die Familien der Collembolen. Zool. Anz. 1913, 61, 315–322. [Google Scholar]
- Hammer, M. Investigations on the Microfauna of Northern Canada, Part II, Collembola. Acta Arct. 1953, 6, 1–108. [Google Scholar]
- Huther, W. Die systematische Stellung von Mackenziella psocoides Hammer (Collembola). Zool. Anz. 1964, 173, 119–126. [Google Scholar]
- Bretfeld, G. Sturmius epiphytus n. gen. n. spec. from Colombia, a taxon of the Symphypleona (Insecta, Colembola) with an unexpected character combination. Description and position in non-Linnean and Linnean classifications of the Symphypleona. Zool. Syst. Evol. Res. 1994, 32, 264–281. [Google Scholar] [CrossRef]
- Souza, P.G.C.; Medeiros, G.S.; Ferreira, R.L.; Souza-Silva, M.; Bellini, B.C. A Highly Troglomorphic New Genus of Sminthuridae (Collembola, Symphypleona) from the Brazilian Semiarid Region. Insects 2022, 13, 650. [Google Scholar] [CrossRef] [PubMed]
- Bernard, C.E.; Wynne, J.J. Disparrhopalites naasaveqw n. sp. from caves at Wupatki National Monument, Arizona, synonymy of Dietersminthurus Palacios-Vargas, Cuéllar & Vázquez, 1998 with Disparrhopalites Stach, 1956 and composition of Songhaicinae (Collembola: Sminthuridae). Zootaxa 2017, 4319, 77–90. [Google Scholar] [CrossRef]
| Type of Analysis | Partition Scheme | Models | Tree Code | Figure |
|---|---|---|---|---|
| Maximum Likelihood | Unpartitioned | mtZOA + F + R10 | ML_1 * | Figure 1 and Figure S6 |
| Maximum Likelihood | Partitioned | EX-EHO | ML_2 | Figure S1 |
| Maximum Likelihood | Unpartitioned | mtART + C60 + FO + R | ML_3 | Figure S2 |
| Maximum Likelihood | Partitioned | Table S2 | ML_4 | Figure S3 |
| Bayesian Inference | - | CAT + GTR | BI | Figure S4 |
| Maximum Parsimony | - | - | MP | Figure S5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellini, B.C.; Zhang, F.; de Souza, P.G.C.; dos Santos-Costa, R.C.; Medeiros, G.d.S.; Godeiro, N.N. The Evolution of Collembola Higher Taxa (Arthropoda, Hexapoda) Based on Mitogenome Data. Diversity 2023, 15, 7. https://doi.org/10.3390/d15010007
Bellini BC, Zhang F, de Souza PGC, dos Santos-Costa RC, Medeiros GdS, Godeiro NN. The Evolution of Collembola Higher Taxa (Arthropoda, Hexapoda) Based on Mitogenome Data. Diversity. 2023; 15(1):7. https://doi.org/10.3390/d15010007
Chicago/Turabian StyleBellini, Bruno Cavalcante, Feng Zhang, Paolla Gabryelle Cavalcante de Souza, Renata Clicia dos Santos-Costa, Gleyce da Silva Medeiros, and Nerivânia Nunes Godeiro. 2023. "The Evolution of Collembola Higher Taxa (Arthropoda, Hexapoda) Based on Mitogenome Data" Diversity 15, no. 1: 7. https://doi.org/10.3390/d15010007
APA StyleBellini, B. C., Zhang, F., de Souza, P. G. C., dos Santos-Costa, R. C., Medeiros, G. d. S., & Godeiro, N. N. (2023). The Evolution of Collembola Higher Taxa (Arthropoda, Hexapoda) Based on Mitogenome Data. Diversity, 15(1), 7. https://doi.org/10.3390/d15010007









