Abstract
Bioprospecting of novel antibiotics has been the conventional norm of research fostered by researchers worldwide to combat drug resistance. With the exhaustion of incessant leads, the search for new chemical entities moves into uncharted territories such as the deep sea. The deep sea is a furthermost ecosystem with much untapped biodiversity thriving under extreme conditions. Accordingly, it also encompasses a vast pool of ancient natural products. Actinobacteria are frequently regarded as the bacteria of research interest due to their inherent antibiotic-producing capabilities. These interesting groups of bacteria occupy diverse ecological habitats including a multitude of different deep-sea habitats. In this review, we provide a recent update on the novel species and compounds of actinomycetes from the deep-sea environments within a period of 2016–2022. Within this period, a total of 24 new species of actinomycetes were discovered and characterized as well as 101 new compounds of various biological activities. The microbial communities of various deep-sea ecosystems are the emerging frontiers of bioprospecting.
1. Introduction
The prevalence and pervasiveness of drug-resistant pathogens create havoc in the health sector by surpassing the exigent issue of microbial infections with increased mortality and morbidity [1,2]. Thus, a dire need of drug discovery and development of novel antibiotics with improved modes of action is required. Natural products are the preeminent storehouse and foundation for drug production [3]. Its putative contribution along with its analogous metabolites (synthetic or combinatorial libraries) toward people’s health has intensified over the years [4]. Bioprospecting of terrestrial biomes have been largely exhausted; thus, a paradigm shift of focus was progressively redirected towards the heterogeneity of marine habitats for natural products [5]. Marine natural products can yield a much greater estimate of potency than its terrestrial counterparts [6,7]. The marine environment is an attractive field of research which continues to receive great attention, as more natural products are being uncovered over the years [8,9,10,11]. The de-replication strategy of recurrent known compounds has led to the shortage of novel compounds. Strategies taken to address this issue involve screening extreme biomes such as the deep-sea on the premise that extreme abiotic conditions give rise to complex chemistry [12]. Therefore, deep-sea peregrination is diverted from geological and ecological surveys to bioprospecting for natural bioactive compounds [13].
Actinomycetes are an intermediary between fungi and bacteria and exist as both symbiont and pathogens. They are diverse, wealthy, and complex groups of bacteria naturally gifted with antibiotic mechanisms [14,15]. Actinomycetes constitute prolific sources of viable metabolites for pharmaceutical utilization. Consequently, a remarkable number of antibiotics are derived from actinomycetes, exclusively from the genus Streptomyces, which contributes towards the betterment of global health by treating malignant diseases [16,17]. Actinomycetes excel at using available organic sources as nutrition which contributes to organic matter turnovers and replenishment of nutrients [15]. They are widely recognized as economically friendly and environmentally sound engineers of practical applications such as bioremediation [18] and antibiotics [19]. Actinomycetes abundantly dwell in terrestrial ecosystems, mainly soils [20], but evidence indicates they are common in marine habitats including deep sea [21,22]. Studies have intensely reviewed the deep-sea actinomycete literature for their cultivation techniques and new compounds and species for the past decades [21,23]. In the sequel, we herein reviewed the recent progress on deep-sea actinomycetes and their natural products from the literature during the years 2016–2022. We reported new species and new compounds of actinomycetes belonging to the phylum Actinomycetota (Actinobacteria) (http://www.bacterio.net/-classifphyla.html#actinobacteria, accessed on 11 November 2022) for the benefit of ongoing compatible research and giving momentum to the growing frontier of deep-sea bioprospecting.
2. The Overview of the Deep-Sea Biosphere and Novelty
2.1. The Deep-Sea Habitats and Biodiversity: A Realm of Uncharted Biowealth
The marine realm is the largest inhabitable space for living biomass, since it encompasses 71% of the earth’s surface with deep-sea area being geographically vast [24,25,26]. The deep sea is home to unprecedented and tremendous marine biodiversity, rivalling that of coral reefs and rainforest [27,28]. In tandem with this, it has a water depth of 200 m and beyond and yet remains poorly explored [29]. The deep-sea biodiversity holds significant economic values and contributes to key ecological processes of the ocean [30,31]. Deep sea is an unique and extreme environment, primarily due to the documented characteristics: (1) elevated hydrostatic pressure; (2) low levels of oxygen capacity or anoxia; (3) temperature drops to abyssal values; and (4) decline of light intensity or photons [24,32]. These challenging conditions hone its inhabitant’s biology through mutation of gene expressions and primary and secondary metabolite pathways, which enables them to thrive and survive [33].
The deep sea comprises a collection of heterogeneous habitats including abyssal plain, hydrothermal vents, continental margins, cold water corals, seamounts, hadal zone, and oxygen minimum zones that nest numerous life forms [34]. Although the deep-sea area is geographically greater, the census of deep-sea prokaryote is meager [25]. The uncharted territories of deep sea have made them potential reservoirs of specialized metabolites and curiosity destinations for bioprospecting [35]. Limitations in research capabilities and resources during the past have hindered and set constrains on sourcing marine organisms at depths unreachable by scuba [36]. However, the advent and introduction of sophisticated ocean technologies such as remotely operated vehicles (ROVs) and submersible technology have unlocked new opportunities for bioprospecting programs; thus, deep-sea bioprospecting have become more accessible and feasible with continuous upgrades of these technologies [37,38]. Of all the known marine natural products, 2% circa is derived from the deep-sea environment [27]. Deep-sea-derived natural products have various biological activities such as antiviral, antibacterial, antifungal, anti-inflammatory, insecticidal, and antitumor [27,28,35,39,40,41]. More than 70% of deep-sea natural products are biologically active [42]. Despite the ubiquity of microbial richness, most indigenous marine microbes remain uncultured due to limited knowledge about their physiology and environmental interactions [43,44]. Thus, endeavors to devise specific strategies in overcoming the technological as well as the culturing barrier are relevant despite its chronological challenges [21,34,40,45]. The percentage of undiscovered species from the deep ocean is higher, relatively awaiting description [46]. In this context, access to deep sea brings with it the potential to discover valuable and beneficial compounds. Microorganisms undebatably dominate the Earth’s land and ocean masses [47]. The deep-sea microbes engage in complex and undeciphered interspecies interactions which boost their fitness [48].
2.2. The Deep-Sea Actinobacteria: A Biogenic Repository of Specialized Metabolites
Actinomycetes are highly evolved and well-suited to thrive in deep-sea extremities. They are multifarious and sophistically equipped with an impressive array of unique molecular and chemical scaffolds deemed to be exquisite and complex [49]. The particularity and complexity of deep-sea habitats forces actinomycetes to exhibit a remarkable adaptation strategy due to their genomic evolution, which brings about unique physiology and metabolism [50]. Higher diversity of microorganisms including actinomycete genera occurs at greater depth (>1000 m) [51,52]. Generally, the distribution and composition of marine actinobacteria is dependent on the physicochemical parameters including pressure, pH, temperature, salinity, and total organic carbon across sparse locations [53]. In this view, the biogeography and diversity of deep-sea actinobacteria remains a persistent mystery [54]. Marine actinobacteria, including those thriving in deep-sea habitats occupy a multitude of habitats, either as a symbiont in invertebrates such corals, sponge, and other marine organisms or as ecosystem engineer responsible for the turnover of complex molecules, contributing to biogeochemical cycles [53,55], across various deep-sea ecosystems such as the water columns (warm/cold water currents), the ocean bed or sedimentary mats, and hydrothermal vents. However, the trace of deep-sea actinobacteria is abundant in deep-sea sediments [54].
The deep-sea actinomycetes are capable of producing bioactive compounds. In recent years, an inventory of studies revealed that deep-sea actinomycetes occupy a spectrum of interesting metabolites [21,56]. Natural compounds recovered from terrestrial and marine environments are specific to each environment with specific margins of similarity. Studies have suggested that although deep-sea are part of the marine environment, it may well represent a separate and unique ecosystem that harbors actinomycetes with enhanced bioactivity. [5]. To highlight, a study by [57] analyzed the antimicrobial activity of tunicamycins from the deep-sea-derived Streptomyces xinghaiensis SCSIO S15077. Interesting results showed that tunicamycins were reported for the first time to be effective against Bacillus thuringiensis, Candida albicans CMCC (F) 98001, and Candida albicans ATCC 96901. Another study by [58] highlighted the anticancer effects of cyclic dipeptides and phenolic compounds from the deep-sea derived Streptomyces xiamenensis MCCC 1A01570 due to their ability to influence the transcription activation function of retinoid X receptor-α (RXRα). Additionally, deep-sea sediment-derived Streptomyces sp. YB104 is considered to produce relatively large quantities of inthomycin B (a bioactive compound with antimicrobial, herbicidal, and anticancer properties) production compared to other species in industrial settings [59]. A wealth of natural products derived from deep-sea actinomycetes are proven to be effective and therefore be considered for further pharmaceutical development.
2.3. Genomic Insights of Deep-Sea Actinobacteria: Unveiling the Hidden Biosynthetic Gene Clusters
Metagenomic research with DNA extraction of microorganisms have revolutionized the bioprospecting strategy [60]. Studies have substantially revealed that microbes genetically encode their natural products in biosynthetic gene clusters (BGCs). A BGC can be defined as a physically clustered group of two or more genes in a particular genome that together encode a biosynthetic pathway for the production of a specialized metabolite (including its chemical variants) [61]. Different structural classes of BGCs exist, including non-ribosomal peptide synthetases (NRPS), polyketide synthases (PKS), terpenes, and bacteriocins. NRPS and PKS are popular targets for natural product discovery, as they are known to synthesize a diversity of antibiotics and immunosuppressants with enormous pharmaceutical potential [62,63,64,65]. The number of BGCs is species-dependent, but individual BGCs has a family of cognate compounds that share a core with a variety in substituents [66]. Each strain of actinomycetes genome on average contains 30–40 secondary metabolism biosynthetic gene clusters, and yet most of which are cryptic [67]. For instance, Streptomyces genus is the largest natural producer of bioactive compounds [17], and its genome contains 25–70 BGCs, most of which are cryptic and are not expressed under normal laboratory conditions [68,69]. Doroghazi et al. [68] did an extensive comparative study by analyzing the genomes of six actinomycetes genera, namely Mycobacterium, Corynebacterium, Rhodococcus, Arthrobacter, Frankia, and Streptomyces, in detail to determine the extent to which natural product gene clusters are conserved within each genus. They showed that within the immense gene clusters diversity, there are patterns showing that some genera have higher prevalence of NRPS or PKS natural products compared to other genera. For example, some groups found conservation of the spore pigment and desferrioxamine class of siderophores in Streptomyces, along with mycolic acid, mycobactin and phthiocerol in Mycobacterium. Further, Hifnawy et al. [70] discuss in their review that the genus Micromonospora is a model system for natural product research. Using biosynthetic gene clusters and gene cluster families from 87 Micromonospora genomes, they showed that this genus contains 2387 BGCs that could be grouped into 1033 BGC-families. The majority of BGC-families belong to the type 1 polyketide synthases (T1PKS) and the non-ribosomal peptide synthetases (NRPS) [70]. This highlights the immense potential that the actinobacteria phylum possesses.
Nonetheless, there are available molecular tools designated for the genetic engineering of actinomyces in order to harvest their valuable compounds [71]. Apart from that, the functions of genome mining have complemented the classical chemistry-driven screening [72]. To illustrate, we analyzed the literature of the following deep-sea actinomycete genomes. A study by [73] indicated that Streptomyces koyangensis SCSIO 5802 produces two active metabolites. However, the analysis of its complete genome revealed its potential to produce 21 categories of natural products. Another study [74] unveiled 37 putative BGCs of deep-sea-derived Streptomyces olivaceus SCSIO T05. Attempts to activate the cryptic BGCs resulted in the discovery of a known compound, lobophorin CR4 (antibacterial and antitumor properties). The genomic data of Janibacter limosus, a deep-sea actinobacterium, revealed a gene cluster for degrading phenol and its derivatives [75]. Two deep-sea Streptomyces isolates (MA3_2.13 and S07_1.15) have 32 and 24 BGCs, respectively. About 30 percent of the characterized BGCs have gene homologies with known clusters of compounds previously described [76]. Genome mining of the deep-sea-derived Streptomyces atratus SCSIO ZH16 leads to the activation of a cyclodepsipeptide gene cluster and the recovery of a potent compound known as atratumycin, which has antibacterial activity [77]. Branchybacterium ginsengisoli B129SM11, an actinobacterium isolated from a deep-sea sponge, is capable of producing enzyme that degrades polyethylene terephthalate (PET). The functions of genome sequencing coupled with genome mining identifies an encoded putative PET hydrolase gene that produces a polyesterase-type enzyme tagged as BgP [78]. Additionally, the complete sequenced genome of Mycetocola spongiae MSC19T has 2887 coding sequences which harbor genes for heavy metal resistance, natural product synthesis and multidrug resistance [79]. The sequencing of deep-sea actinomycete whole genomes magnifies their biosynthetic potential for producing pharmaceutical metabolites.
2.4. Novel Species and Compounds of Deep-Sea Actinomycetes
An impressive yield of novel species and compounds from deep-sea actinomycetes was found between 2016 and 2022. This review has identified 24 new actinomycete species belonging to nine different actinomycete families (Table 1 and Figure 1). The latter are Pseudonocardiaceae (4 species), Brevibacteriaceae (2 species), Rubrobacteraceae (3 species), Micrococcaceae (4 species), Micromonosporaceae (3 species), Streptomycetaceae (1 species), Microbacteriaceae (4 species), Lamiaceae (1 species), and Miltoncostaeaceae (2 species). Notably, most of the new species were derived from deep-sea sediments. A few of the novel species of actinobacteria described were further evaluated for their diverse innate potentials. To illustrate, Micromonospora provocatoris harbors n-acetylgutaminyl glutamine amide and deferoxamine B [80]. Kocuria oceani has the ability to reduce iron (III) in both aqueous and solid forms [81]. Micromonospora ferruginea has the capacity to produce natural products such as kosinostatin and isoquinocycline B, which exhibit both antibiotic and antitumor properties [82]. The novel compounds discovered belong to various chemical classes, and most of them yield bioactive properties with different modes of action such as cytotoxic activity, anti-allergic activity, antifungal activity, antibacterial activity, and antiviral activity (Table 2). The structures of the reported novel compounds are shown in Table 2. The majority of the discovery of novel actinomycete species and compounds occurred as of the years 2021 and 2017, respectively. A total of 101 new compounds were classified, which belong to the actinomycete genera Streptomyces, Dermacoccus, Pseudonocardia, Williamsia, Microbacterium, Amycolatopsis, Nesterenkonia, Micromonospora, Agrococcus, Actinomadura, Rhodococcus, Nonomuraea, and Saccharopolyspora.
From the 101 compounds discovered, 40 of them have no visibility of biological activity against the numerous cancer cell lines and pathogenic test strains. For instance, 3R-OH-1,6-diene-cyclohexylacetic acid, linear tetradepsipeptide, N1-N5-di-p-(EE)-coumaroyl-N10-acetylsepermidine, and furan fatty acid are compounds produced by Agrococcus sp. SCSIO 52902 from a deep-sea sediment that display no cytotoxic activity against human tumor cell lines (A-549, HL-60, and HCT-116) and no antibacterial activity against Gram-positive and Gram-negative bacteria [83]. However, 39 compounds reported have a vivid trace of potency either against specific human cancer cell lines, pathogenic fungal strain and both Gram-positive and Gram-negative bacteria. For example, paulomycin G is a compound produced by Micromonospora matsunotoense M-412 that displays a strong cytotoxic activity against tumor cell lines of pancreatic adenocarcinoma, breast adenocarcinoma and hepatocellular carcinoma [84]. Interestingly, Actinomadura sp. KD439 is reported to harbor potent compounds such as kumemicinones A-G, which are cytotoxic against P388 murine leukemia cells [85]. Selective compounds such as streptothiazolidine A, streptodiketopiperazine A and B, and (S)-1-(3-ethylphenyl)-1,2-ethanediol belonging to Streptomyces sp. SY1965 were reported to have antifungal effect towards Candida albicans [86]. Further, nocardiopsistins A−C derived from Nocardiopsis sp. HB-J378 have a cytotoxic activity against methicillin-resistant Staphylococcus aureus [87]. Apart from that, a collection of novel compounds, i.e., amycolachromones A−F (Amycolatopsis sp. WP1) [88] and georgenione A (Georgenia sp. 40DY180) [89], display weak inhibitory activity against the ABH2 enzyme and anti-tyrosinase activity against tyrosinase enzyme, respectively. Moreover, 14 of the total novel compounds were reported with no bioactive analyses. These compounds are produced by various deep-sea actinomycetes such as Streptomyces sp. SCSIO ZS0520 (actinopyrones E-G, seco-salinomycins A-E and minipyrone) [90,91], Micromonospora echinospora SCSIO 04089 (nenestatin B) [92], Dermacoccus abyssi MT1.1 (dermacozine M) [93], Streptomyces olivaceus SCSIO T05 (olimycin A and B) [94], and Williamsia sp. MCCC 1A11233 (3-benzyl-3α,4β-dihydroxypentan-2-one) [95]. Additionally, albisporachelin is a compound isolated from Amycolatopsis albisporachelin WP1T, which functions as a chelating agent for coordinating iron update [96]. From the inventory of species from which the new compounds are discovered, the genus Streptomyces is frequent. Overall, new species and compounds of actinomycetes are being discovered on a constant basis, as studies multiply in that environment.

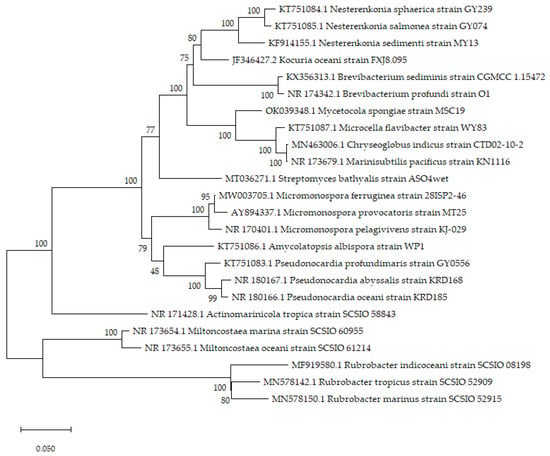


Table 1.
New species of actinobacteria from deep-sea niches reported between 2016 and 2022. Superscript T designates type strains.
Table 1.
New species of actinobacteria from deep-sea niches reported between 2016 and 2022. Superscript T designates type strains.
| Novel Species | Family | Source, Depth, and Geographic Location | Ref. |
|---|---|---|---|
| Amycolatopsis albispora (KCTC 39642T = MCCC 1A10745T) | Pseudonocardiaceae | Sediment at 2945 m The Indian Ocean | [97] |
| Brevibacterium sediminis (CGMCC 1.15472T = DSM 102229T) | Brevibacteriaceae | Sediment at 3690 m The Carlsberg Ridge | [98] |
| Kocuria oceani (CGMCC 4.6946T = DSM 24949T) | Micrococcaceae | Hydrothermal plume at 2800 m The Southwest Indian Ridge | [99] |
| Pseudonocardia profundimaris (MCCC 1A10574T = KCTC 39641T) | Pseudonocardiaceae | Sediment at 7118 m The Western Pacific Ocean | [100] |
| Rubrobacter indicoceani DSM 105148T = CGMCC 1.16398T) | Rubrobacteraceae | Sediment at 4602 m The Indian Ocean | [101] |
| Rubrobacter tropicus (KCTC 49412T = CGMCC 1.13853T) & Rubrobacter marinus (KCTC 49411T = CGMCC 1.13852T) | Rubrobacteraceae | Sediment at 3448 m The South China Sea | [102] |
| Nesterenkonia salmonea (KCTC 39639T = MCCC 1A11256T) & Nesterenkonia sphaerica (KCTC 39640T = MCCC 1A10688T) | Micrococcaceae | Sediment at 3223 and 2859 m The Southern Atlantic Ocean | [103] |
| Micromonospora pelagivivens (NBRC 113519T = TBRC 9233T) | Micromonosporaceae | Sediment at 226 m Kagoshima, Japan | [104] |
| Actinomarinicola tropica (KCTC 49408T = CGMCC 1.17503T) | Lamiaceae | Sediment at 460 m The South China Sea | [105] |
| Brevibacterium profundi (JCM 33845T = MCCC 1A16744T) | Brevibacteriaceae | Sediment at 7068 m The Western Pacific Ocean | [106] |
| Micromonospora provocatoris (NCIMB 15245T = TISTR 2834T) | Micromonosporaceae | Sediment at 10,898 m The Mariana Trench | [80] |
| Marinisubtillis pacificus (CGMCC 1.17143T = KCTC 49299T) | Microbacteriaceae | Seawater at 400 m The Tropical Western Pacific Ocean | [107] |
| Microcella flavibacter (KCTC 39637T = MCCC 1A07099T) | Microbacteriaceae | Sediment at 3039 m The Indian Ocean | [108] |
| Chryseoglobus indicus (JCM 33842T = MCCC 1A16619T) | Microbacteriaceae | Deep-sea water (depth not stated) The Indian Ocean | [109] |
| Pseudonocardia abyssalis (DSM 111918T = NCIMB 15270T) & Pseudonocardia oceani (DSM 111919T = NCIMB 15269T) | Pseudonocardiaceae | Sediment at 4539 and 4060 m The Southern Ocean | [110] |
| Streptomyces bathyalis (DSM 106605T = NCCB 100657T) | Streptomycetaceae | Sponge at 1000–4000 m The North Atlantic Ocean | [111] |
| Nesterenkonia sedimenti (LMG 28111T = MCCC 1A09979T = JCM 19767T = CGMCC 1.12784T) | Micrococcaceae | Sediment (depth not stated) The Western Pacific Ocean | [112] |
| Micromonospora ferruginea (NCTC 14469T = DSMZ 111791T) | Micromonosporaceae | Sponge at 971 m The Atlantic Ocean | [82] |
| Miltoncostaea marina (DSM 110281T = CGMCC 1.18757T) & Miltoncostaea oceani (KCTC 49527T = CGMCC 1.18758T) | Miltoncostaeaceae | Sediment at 460 and 323 m The South China Sea | [113] |
| Mycetocola spongiae (MCCC 1K06265T = KCTC 49701T) | Microbacteriaceae | Sponge at 2681 m The Mariana Trench | [114] |

Figure 1.
Maximum-likelihood tree of the novel species of actinobacteria isolated from deep-sea samples (constructed using MEGA11 software, State College, PA, USA, available from www.megasoftware.net.) [115]. The phylogenetic tree was constructed using the 16s rRNA gene sequence of 24 novel actinobacteria with GenBank/EMBL/DDBJ with accession numbers of KT751084, KT751085, KF914155, JF346427, KX356313, NR_174342, OK039348, KT751087, MN463006, NR_173679, MT036271, MW003705, AY894337, NR_170401, KT751086, KT751083, NR_180167, NR_180166, NR_171428, NR_173654, NR_173655, MF919580, MN578142, and MN578150 (the number of nucleotide pairs varied across species). The evolutionary distances are computed using the Tamura 3-parameter model. Bootstrap percentage values are given for 100 replicates. Bar: 0.05 substitution per nucleotide position.

Table 2.
New compounds of actinobacteria from deep-sea niches reported between 2016 and 2022.
Table 2.
New compounds of actinobacteria from deep-sea niches reported between 2016 and 2022.
| Name and Structure | Chemical Group | Region, Depth, Organism, and Bioactivity | Ref. |
|---|---|---|---|
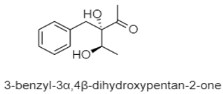 | Phenolics | The Southwestern Indian Ocean; 1654 m; Williamsia sp. MCCC 1A11233; no bioactive test. | [95] |
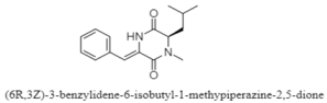 | Diketopiperazine | The South China Sea; 3536 m; Streptomyces sp. SCSIO 04496; no cytotoxic activity (concentration: 100 µM) against five tumor cell lines including SF-268, MCF-7, NCI-H460, HepG-2, and LX-2 cells. | [116] |
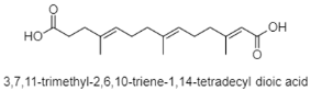 | Dioic acid | The South China Sea; 3536 m; Streptomyces somaliensis SCSIO ZH66; no cytotoxic activity against the hepatic carcinoma cell line (Huh 7.5). | [117] |
 | Bisindole pyrroles spiroindimicins | The Tautra Ridge in the Trondheim fjord, Norway; 450 m; Streptomyces sp. MP131-18; no antibacterial activity against Escherichia coli, Bacillus subtilis, and Pseudomonas putida. Spiroindimicin E displays weak cytotoxic activity against the growth of T24 bladder carcinoma cells. | [118] |
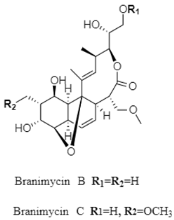 | Macrolide | The Cantabrian Sea; 3000 m; Pseudonocardia carboxydivorans M-227; antibacterial activity against the panel of Gram-positive bacteria (Corynebacterium urealyticum, Clostridium perfringens, and Micrococcus luteus) and Gram-negative bacterium (Neisseria meningitidis). | [119] |
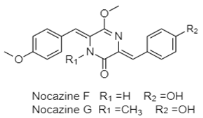 | Diketopiperazine | Deep-sea sediment (region and depth not specify); Nocardiopsis sp. YIM M13066; displaying cytotoxic activity against human cancer cell lines (H1299, HeLa, HL7702, MCF-7, PC3, and U251). | [120] |
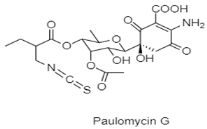 | Glycosylated paulomycins | Submarine Aviles Canyon; 1800 m; Micromonospora matsumotoense M-412; displaying strong cytotoxic activity against various human tumor cell lines (pancreatic adenocarcinoma (MiaPaca_2), breast adenocarcinoma (MCF-7), and hepatocellular carcinoma (HepG2)). | [84] |
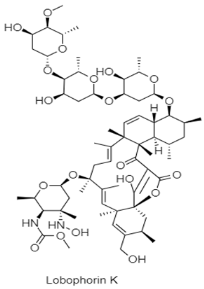 | Spirotetronate | Submarine Aviles Canyon; 1800 m; Streptomyces sp. M-207; displaying cytotoxic activity against human tumor cell lines (pancreatic adenocarcinoma (MiaPaca_2), and breast adenocarcinoma (MCF-7)); Moderate and selective antibacterial activity against Staphylococcus aureus. | [121] |
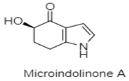 | Novel indole | The Southwestern Indian Ocean; 1603 m; Microbacterium sp. MCCC 1A11207; no significant cytotoxic activity against RBL-2H3 cells and no anti-allergic activity against RBL-2H3 cells. | [122] |
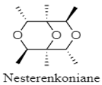 | Novel cyclic ether | The Eastern Pacific Ocean; 5302 m; Nesterenkonia flava MCCC 1K00610; moderate anti-allergic activity against RBL-2H3 cells. | [123] |
Nocapyrone S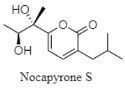 | α-pyrone | The Arctic Ocean; 2042 m; Nocardiopsis dassonvillei subsp. dassonvillei DMS 43111 (T); no cytotoxic activity against K562, MCF-7, SGC7901, A375, Hela, and HepG2 cell lines. | [124] |
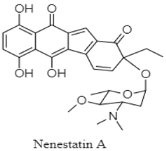 | Benzofluorene-containing angucyclines | The Northern South China Sea; 3025 m; Micromonospora echinospora SCSIO 04089; no antibacterial activity against seven bacterial strains (Escherichia coli, Staphylococcus aureus, Micrococcus luteus, Enterococcus faecalis, Acinetobacter baumannii, methicillin-resistant S. aureus, and Vibrio alginolyticus). | [125] |
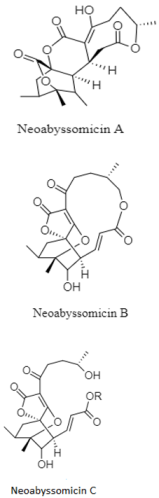 | Polycyclic macrolactones | The South China Sea; 3536 m; Streptomyces koyangensis SCSIO 5802; no cytotoxic against HIV-1 virus and no antibacterial activity against the panel of Gram positive (Bacillus thuringiensis, Micrococcus luteus, Enterococcus faecalis, & Staphylococcus aureus) and clinical isolates of methicillin-resistant S. aureus: MRSA-862, MRSA-669, MRSA-991, and MRSA-A1. | [126] |
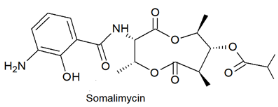 | Antimycin-type depsipeptide | The South China Sea; 3536 m; Streptomyces somaliensis SCSIO ZH66; weak cytotoxic activity against human umbilical vein endothelial cells. No antibacterial activity against Staphylococcus aureus, Escherichia coli, Enterococcus faecalis, Enterococcus faecium, and Salmonella typhimurium. | [127] |
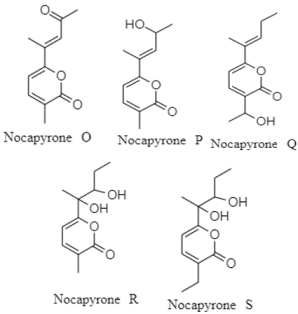 | α-pyrones | Deep-sea sediment (region and depth not specify); Nocardiopsis sp. YIM M13066; no cytotoxic activity against H1299, HeLa, HL7702, MCF-7, PC3 and U251 cell lines. | [128] |
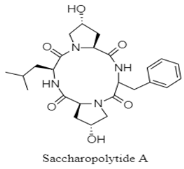 | Cyclic tetrapeptide | The Atlantic Ocean; 2875 m; Saccharopolyspora cebuensis MCCC 1A09850; weak anti-allergic and anti-proliferate activity against Hela and H1299 tumor cell lines. | [129] |
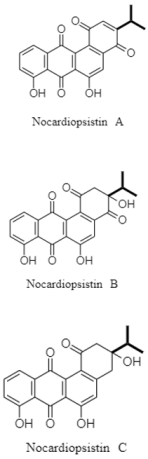 | Angucyclines | Deep-sea sponge (region and depth not specify); Nocardiopsis sp. HB-J378; Antimicrobial activity against methicillin-resistant Staphylococcus aureus. | [87] |
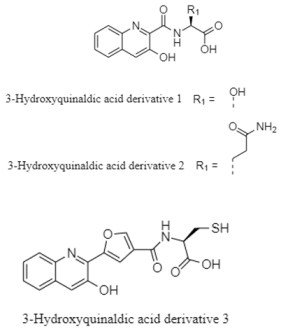 | 3-hydroxyquinaldic acid | The Cantabrian Sea; 2000 m; Streptomyces cyaneofuscatus M-157; no antibacterial activity against Gram positive (MRSA) and Gram negative (Escherichia coli and Acinetobacter baumannii); weak cytotoxic activity against human tumor cell line HepG2. | [130] |
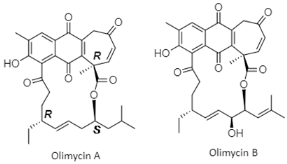 | Naphthoquinone macrolides | The Indian Ocean; 4617 m; Streptomyces olivaceus SCSIO T05; No bioactive test. | [94] |
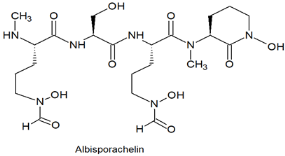 | Hydroxamatet-ype siderophore | The Indian Ocean; 2945 m; Amycolatopsis albisporachelin WP1T; chelating agent for coordinating iron uptake. | [96] |
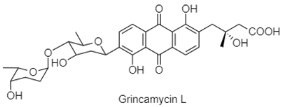 | Angucycline | The Indian Ocean; 4495 m; Streptomyces lusitanus OUCT16-27; antibacterial activity against multi-drug resistant strains of Enterococcus faecium, Enterococcus faecalis, and Staphylococcus aureus. | [131] |
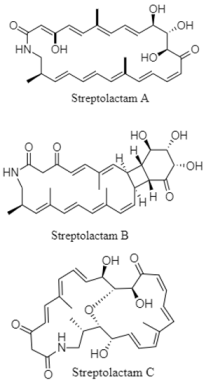 | Macrolactams | The South Mid-Atlantic Ridge; 2782 m; Streptomyces sp. OUCMDZ-3159; no cytotoxic activity against MCF-7, A549, K549, and HL-60 cell lines; no antimicrobial activity against pathogenic bacteria. Streptolactam A and C display antifungal activity against Candida albicans. | [132] |
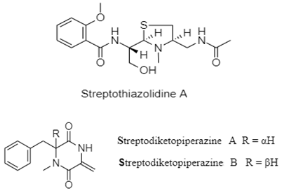 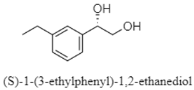 | Salicylamide, diketopiperazine, and phenylethanediol | The Mariana Trench; 11,000 m; Streptomyces sp. SY1965; antifungal activity against Candida albicans. | [86] |
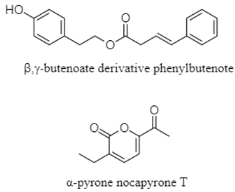 | β,γ-butanoate and α-pyrone | The Mariana Trench; 4448 m; Nocardiopsis sp. HDN 17-237; no antioxidant and antibacterial activity against Escherichia coli, Proteus vulgaris, Pseudomonas aeruginosa, Vibrio parahemolyticus, Bacillus subtilis, and Mycobacterium phlei. | [133] |
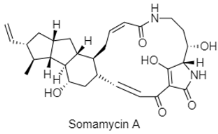 | Polycyclic tetramate macrolactams | The South China Sea; 3536 m; Streptomyces somaliensis SCSIO ZH66; antifungal activity against Fusarium oxysporum and moderate cytotoxic activity against human cell lines of HCT116 and K562. | [134] |
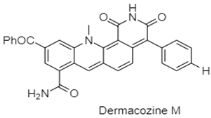 | New phenazine | The Mariana Trench; 10898 m; Dermacoccus abyssi MT1.1; no bioactive test. | [93] |
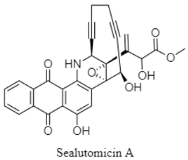 | Enediyne | The Japan Trench; 329 m; Nonomuraea sp. MM565M-173N2; strong antibacterial activity against carbapenem-resistant Enterobacteriaceae. | [135] |
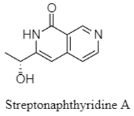 | Naphthyridine | The Mariana Trench; 11,000 m; Streptomyces sp. SY2111; antiproliferative activity against human glioma U87MG and U251 cells. | [136] |
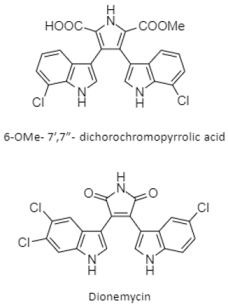 | bis-indole alkaloids | The South China Sea; 1765 m; Streptomyces sp. SCSIO 11791; dionemycin shows antibacterial activity against methicillin-resistant Staphylococcus aureus and cytotoxic activity against human cell lines (NCI-H460, MDA-MB-231, HCT-116, and HepG2) and noncancerous MCF10A. | [137] |
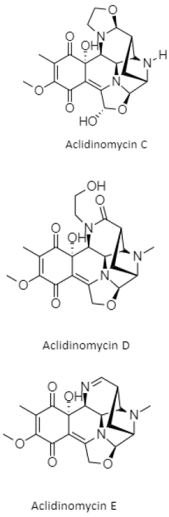 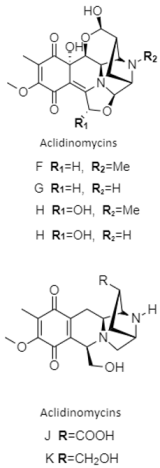 | Tetrahydroisoquinolines | The South China Sea; 3536 m; Streptomyces niveus SCSIO 3406; aclidinomycins D, E, G, J, and K display antibacterial activity against Bacillus subtilis, Micrococcus luteus, Staphylococcus aureus and a panel of MRSA (MRSA 991, MRSA 1862, and MRSA SH1). | [138] |
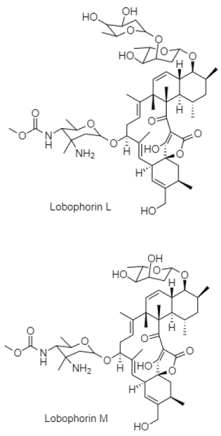 | Spirotetronate | The South China Sea; 3565 m; Streptomyces sp. 4506; antibacterial activity against Micrococcus luteus, Bacillus thuringiensis, Staphylococcus aureus, MRSA, Acinetobacter baumannii, Escherichia coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae. | [139] |
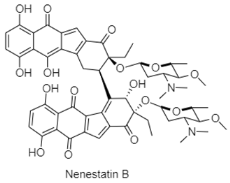 | Benzofluorene-containing angucyclines | The Northern South China Sea; 3025 m; Micromonospora echinospora SCSIO 04089; no bioactive test. | [92] |
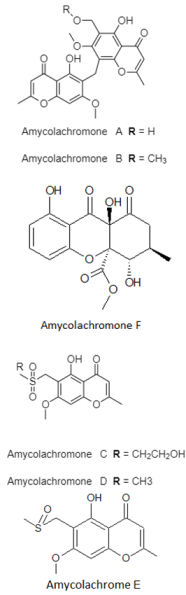 | Chromone derivatives | The Southwest Indian Ocean; 2945 m; Amycolatopsis sp. WP1; weak inhibitory activity against the ABH2 enzyme. | [88] |
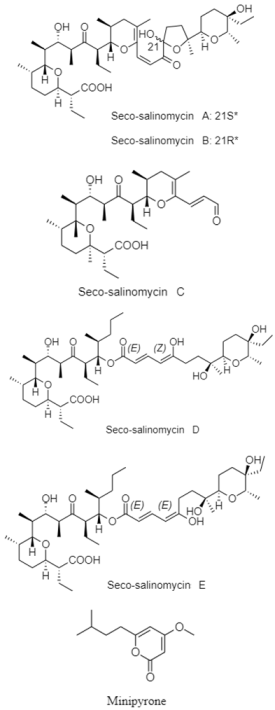 | Salinomycin and α-pyrone | The Okinawa Trough; 1039 m; Streptomyces sp. SCSIO ZS0520; no bioactive test. | [90] |
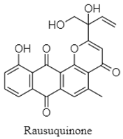 | Pluramycin-class polyketide | Deep water sample; Rhodococcus sp. RD015140; antimicrobial activity against Gram-positive bacteria. | [140] |
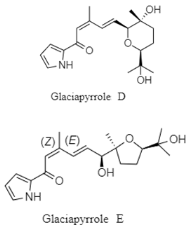 | Pyrrolosesquiterpenes | The East Sea of Korea; 2163 m; Streptomyces sp. GGS53; antiviral activity against influenza A virus. | [141] |
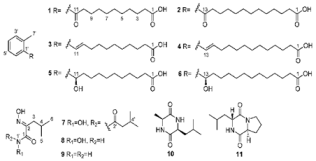 | Aromatic acids and leucine derivatives | The Indian Ocean; 3386 m; Streptomyces chumphonensis SCSIO 5079; no antibacterial and cytotoxic activity against bacterial pathogenic strains and human cancer cell lines. | [142] |
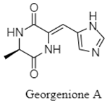 | Piperazinedione | The Pacific Ocean; 5591 m; Georgenia sp. 40DY180; anti-tyrosinase activity against mushroom tyrosinase enzyme. | [89] |
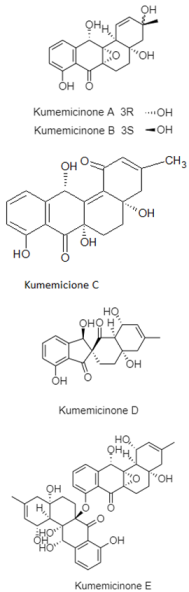 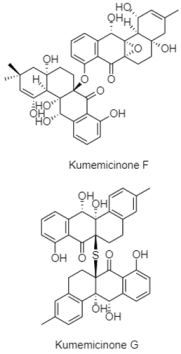 | Angucyclinones | Kumejima Island, Okinawa, Japan; 612 m; Actinomadura sp. KD439; cytotoxic activity against P388 murine leukemia cells. | [85] |
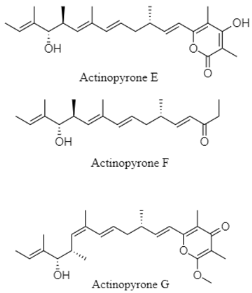 | Pyrone polyketide | The Okinawa Trough; 1039 m; Streptomyces sp. SCSIO ZS0520; no bioactive test. | [91] |
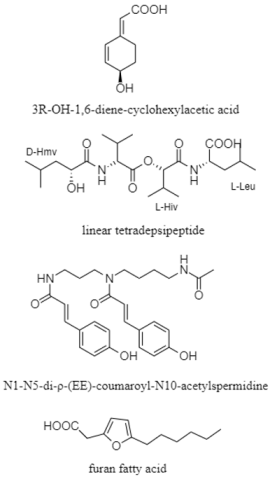 | Cyclohexylacetic acid, depsipeptide, spermidine, and fatty acid | The South China Sea; 2061 m; Agrococcus sp. SCSIO 52902; no cytotoxic activity against human tumor cell lines (A-549, HL-60, and HCT-116) and no antibacterial activity against Bacillus subtilis, Bacillus thuringiensis, Staphylococcus aureus, and Escherichia coli. | [83] |
3. Limitations and Challenges of Acquiring Natural Products from Actinobacteria
The success of recovering actinomycetes and their natural metabolites from diverse ecological niches is possible due to the improvement and advancement of research techniques, tools, and genomic data over the years. Despite that, access to the optimum biosynthetic potential of actinobacteria remains an ongoing objective among researchers worldwide. To date, a higher percent of microbes remain uncultivated including actinobacteria [143]. Actinobacteria species respond differently to stressors and have a varied sources of energy preferences [144]. Thus, understanding the characteristics of a particular species for cultivation is very crucial. Currently, there are modern cultivation techniques and their upgrades which function with its own challengers and limitations [145]. The advent of the genomic era has unveiled the hidden biosynthetic potential of actinobacteria, but progress made to activate and capture the natural products in the physical form often faces limitations [146]. Natural products from actinobacteria are mostly recovered through the process of diverse extraction techniques. Extraction is a fundamental step that transfers groups of compounds from a matrix into a different phase. The consecutive steps of natural product extractions are intercorrelated and affect the overall performance of the analysis. In this case, inconsistency and errors are inevitable, if one of the steps is not properly followed. Therefore, extraction of natural products should be comprehended in order to avoid compromising the compounds original profiles and quantities [147]. Moreover, extreme environments offer another major obstacle in terms of access and also replication of its natural conditions in a laboratory setting. Actinobacteria are versatile, but their cultivation in the laboratory is significantly dependent on the conditions under which they are accustomed to survive and reproduce [144]. Bioprospecting for actinobacteria natural products is profitable, but there is a lack of advanced multidisciplinary studies due to funding issues. In this regard, it limits the progress of scrutinizing the compounds for potential drug development and also hinders bio-accessibility to underexplored locations.
4. Conclusions
Deep-sea habitats represent a vast pool of essential compounds. Actinomycetes versatility to the harsh environmental conditions of the deep sea has enabled a diverse ingenuity and complexity of actinomycete-derived natural metabolites. This review presents 24 new species of actinobacteria as well as 101 new compounds obtained from actinomycetes isolated from various deep-sea niches. In addition, it outlines the tremendous potential of deep-sea actinobacteria for future prospects. A growing body of knowledge has been generated from deep-sea environments. Deep-sea microbiology is the emerging frontier of research.
More research should target deep-sea environments for actinomycete-derived natural metabolites. The Pacific Ocean is the largest and deepest among the Earth’s five oceanic divisions and represents an unlimited source of potential new bioactive compounds from actinobacteria.
Author Contributions
Conceptualization, G.S. and A.P.; writing—original draft preparation, G.S. and L.D.; writing, G.S., L.D., and A.P.; supervision, A.P.; review and editing, A.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank their respective departments for their support and cooperation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Akova, M. Epidemiology of antimicrobial resistance in bloodstream infections. Virulence 2016, 7, 252–266. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- Singh, I.P.; Ahmad, F.; Chatterjee, D.; Bajpai, R.; Sengar, N. Natural products: Drug Discovery and Development. In Drug Fiscovery and Development: From Targets and Molecules to Medicines; Poduri, R., Ed.; Springer: Singapore, 2021; pp. 11–65. [Google Scholar] [CrossRef]
- Trindade, M.; Van Zyl, L.J.; Navarro-Fernández, J.; Abd Elrazak, A. Targeted metagenomics as a tool to tap into marine natural product diversity for the discovery and production of drug candidates. Front. Microbiol. 2015, 6, 890. [Google Scholar] [CrossRef]
- Voser, T.M.; Campbell, M.D.; Carroll, A.R. How different are marine microbial natural products compared to their terrestrial counterparts? Nat. Prod. Rep. 2022, 39, 7–19. [Google Scholar] [CrossRef]
- Haefner, B. Drugs from the deep: Marine natural products as drug candidates. Drug Discov. Today 2003, 8, 536–544. [Google Scholar] [CrossRef]
- Sigwart, J.D.; Blasiak, R.; Jaspars, M.; Jouffray, J.B.; Tasdemir, D. Unlocking the potential of marine biodiscovery. Nat. Prod. Rep. 2021, 38, 1235–1242. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2022, 39, 1122–1171. [Google Scholar] [CrossRef]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2021, 38, 362–413. [Google Scholar] [CrossRef]
- Stonik, V.A.; Makarieva, T.N.; Shubina, L.K. Antibiotics from marine bacteria. Biochemistry 2020, 85, 1362–1373. [Google Scholar] [CrossRef]
- Subramani, R.; Narayanasamy, M. Screening of marine actinomycetes isolated from the Bay of Bengal, India for antimicrobial activity and industrial enzymes. World J. Microbiol. Biotechnol. 2009, 25, 2103–2111. [Google Scholar] [CrossRef]
- Leary, D.; Vierros, M.; Hamon, G.; Arico, S.; Monagle, C. Marine genetic resources: A review of scientific and commercial interest. Mar. Policy 2009, 33, 183–194. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef]
- Anandan, R.; Dharumadurai, D.; Manogaran, G.P. An Introduction to Actinobacteria. In Actinobacteria-Basics and Biotechnological Applications; Dhanasekaran, D., Jiang, Y., Eds.; IntechOpen: London, UK, 2016; pp. 3–37. [Google Scholar] [CrossRef]
- Subramani, R.; Sipkema, D. Marine rare actinomycetes: A promising source of structurally diverse and unique novel natural products. Mar. Drugs 2019, 17, 249. [Google Scholar] [CrossRef]
- Donald, L.; Pipite, A.; Subramani, R.; Owen, J.; Keyzers, R.A.; Taufa, T. Streptomyces: Still the biggest producer of new natural secondary metabolites, a current perspective. Microbiol. Res. 2022, 13, 418–465. [Google Scholar] [CrossRef]
- Verma, S.; Kuila, A. Bioremediation of heavy metals by microbial process. Environ. Technol. Innov. 2019, 14, 100369. [Google Scholar] [CrossRef]
- Mast, Y.; Stegmann, E. Actinomycetes: The antibiotics producers. Antibiotics 2019, 8, 105. [Google Scholar] [CrossRef]
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 2017, 111, 458–467. [Google Scholar] [CrossRef]
- Kamjam, M.; Sivalingam, P.; Deng, Z.; Hong, K. Deep-sea actinomycetes and their secondary metabolites. Front. Microbiol. 2017, 8, 760. [Google Scholar] [CrossRef]
- Subramani, R.; Aalbersberg, W. Marine actinomycetes: An ongoing source of novel bioactive metabolites. Microbiol. Res. 2012, 167, 571–580. [Google Scholar] [CrossRef]
- Sun, C.; Mudassir, S.; Zhang, Z.; Feng, Y.; Chang, Y.; Che, Q.; Gu, Q.; Zhu, T.; Zhang, G.; Li, D. Secondary metabolites from deep-sea derived microorganisms. Curr. Med. Chem. 2019, 27, 6244–6273. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Llodra, E.; Brandt, A.; Danovaro, R.; De Mol, B.; Escobar, E.; German, C.R.; Levin, L.A.; Martinez Arbizu, P.; Menot, L.; Buhl-Mortensen, P.; et al. Deep, diverse and definitely different: Unique attributes of the world’s largest ecosystem. Biogeosciences 2010, 7, 2851–2899. [Google Scholar] [CrossRef]
- Das, S.; Lyla, P.S.; Khan, S.A. Marine microbial diversity and ecology: Importance and future perspectives. Curr. Sci. 2006, 90, 1325–1335. [Google Scholar]
- Kaur, J.; Vishnu, A.L.; Khipla, N.; Kaur, J. Microbial Life in Cold Regions of the Deep-Sea. In Survival Strategies in Cold-Adapted Microorganisms; Goel, R., Soni, R., Suyal, D.C., Khan, M., Eds.; Springer: Singapore, 2022; pp. 63–86. [Google Scholar] [CrossRef]
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef]
- Skropeta, D. Deep-sea natural products. Nat. Prod. Rep. 2008, 25, 1131–1166. [Google Scholar] [CrossRef]
- Costello, M.J.; Chaudhary, C. Marine biodiversity, biogeography, deep-sea gradients, and conservation. Curr. Biol. 2017, 27, 2051. [Google Scholar] [CrossRef]
- Thurber, A.R.; Sweetman, A.K.; Narayanaswamy, B.E.; Jones, D.O.B.; Ingels, J.; Hansman, R.L. Ecosystem function and services provided by the deep-sea. Biogeosciences 2014, 11, 3941–3963. [Google Scholar] [CrossRef]
- Folkersen, M.V.; Fleming, C.M.; Hasan, S. The economic value of the deep-sea: A systematic review and meta-analysis. Mar. Policy 2018, 94, 71–80. [Google Scholar] [CrossRef]
- Thistle, D. The Deep-Sea Floor: An Overview. In Ecosystems of the Deep Oceans; Tyler, P.A., Ed.; Elsevier: Amsterdam, The Netherlands, 2003; p. 5. [Google Scholar]
- Jebbar, M.; Franzetti, B.; Girard, E.; Oger, P. Microbial diversity and adaptation to high hydrostatic pressure in deep-sea hydrothermal vents prokaryotes. Extremophiles 2015, 19, 721–740. [Google Scholar] [CrossRef]
- Paulus, E. Shedding light on deep-sea biodiversity—A highly vulnerable habitat in the face of anthropogenic change. Front. Mar. Sci. 2021, 8, 667048. [Google Scholar] [CrossRef]
- Wang, Y.N.; Meng, L.H.; Wang, B.G. Progress in research on bioactive secondary metabolites from deep-sea derived microorganisms. Mar. Drugs 2020, 18, 614. [Google Scholar] [CrossRef] [PubMed]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Danovaro, R.; Carugati, L.; Berzano, M.; Cahill, A.E.; Carvalho, S.; Chenuil, A.; Cinzia, C.; Sonia, C.; Romain, D.; Antonio, D. Implementing and innovating marine monitoring approaches for assessing marine environmental status. Front. Mar. Sci. 2016, 3, 213. [Google Scholar] [CrossRef]
- Feng, J.C.; Liang, J.; Cai, Y.; Zhang, S.; Xue, J.; Yang, Z. Deep-sea organisms research oriented by deep-sea technologies development. Sci. Bull. 2022, 67, 1802–1816. [Google Scholar] [CrossRef] [PubMed]
- Saide, A.; Lauritano, C.; Ianora, A. A treasure of bioactive compounds from the deep-sea. Biomedicines 2021, 9, 1556. [Google Scholar] [CrossRef]
- Tortorella, E.; Tedesco, P.; Fortunato, P.E.; January, G.G.; Fani, R.; Jaspars, M.; Donatella, D.P. Antibiotics from deep-sea microorganisms: Current discoveries and perspectives. Mar. Drugs 2018, 16, 355. [Google Scholar] [CrossRef]
- Song, C.; Yang, J.; Zhang, M.; Ding, G.; Jia, C.; Qin, J.; Guo, L. Marine natural products: The important resource of biological insecticide. Chem. Biodivers. 2021, 18, e2001020. [Google Scholar] [CrossRef]
- Pilkington, L. A chemometric analysis of deep-sea natural products. Molecules 2019, 24, 3942. [Google Scholar] [CrossRef]
- Stewart, E.J. Growing unculturable bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef]
- Bérdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, Y.; Zhang, X.H. Cultivation of microbes from the deep-sea environments. Top. Stud. Oceanogr. 2018, 155, 34–43. [Google Scholar] [CrossRef]
- Mora, C.; Tittensor, D.P.; Adl, S.; Simpson, A.G.B.; Worm, B. How many species are there on earth and in the ocean? PLoS Biol. 2011, 9, e1001127. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. Prokaryote diversity and taxonomy: Current status and future challenges. Philos. Trans. R. Soc. Lond. B 2004, 359, 623–638. [Google Scholar] [CrossRef]
- Nawaz, M.Z.; Subin Sasidharan, R.; Alghamdi, H.A.; Dang, H. Understanding interaction patterns within deep-sea microbial communities and their potential applications. Mar. Drugs 2022, 20, 108. [Google Scholar] [CrossRef]
- Hui, M.L.Y.; Tan, L.T.H.; Letchumanan, V.; He, Y.W.; Fang, C.M.; Chan, K.G.; Law, J.W.F.; Lee, L.H. The extremophilic actinobacteria: From microbes to medicine. Antibiotics 2021, 10, 682. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Zhang, Z.; Yang, T.; Chen, M.; Li, J.; Chen, F.; Yang, J.; Li, W.; Zhang, B. Comparative genomics analysis of Streptomyces species reveals their adaptation to the marine environment and their diversity at the genomic level. Front. Microbiol. 2016, 7, 998. [Google Scholar] [CrossRef] [PubMed]
- Kamjam, M.; Xie, Q.; Deng, Z.; Hong, K. Isolation and diversity of actinomycetes from sediments of different depths between 34 m and 3235 m in south china sea. Chiang Mai J. Sci. 2018, 45, 1595–1609. [Google Scholar]
- Fan, S.; Wang, M.; Ding, W.; Li, Y.X.; Zhang, Y.Z.; Zhang, W. Scientific and technological progress in the microbial exploration of the hadal zone. Mar. Life Sci. Technol. 2022, 4, 127–137. [Google Scholar] [CrossRef]
- Jagannathan, S.V.; Manemann, E.M.; Rowe, S.E.; Callender, M.C.; Soto, W. Marine actinomycetes, new sources of biotechnological products. Mar. Drugs 2021, 19, 365. [Google Scholar] [CrossRef]
- Chen, P.; Zhang, L.; Guo, X.; Dai, X.; Liu, L.; Xi, L.; Wang, J.; Song, L.; Wang, Y.; Zhu, W. Diversity, biogeography, and biodegradation potential of actinobacteria in the deep-sea sediments along the southwest indian ridge. Front. Microbiol. 2016, 7, 1340. [Google Scholar] [CrossRef]
- Siro, G.; Pipite, A.; Christi, K.; Srinivasan, S.; Subramani, R. Marine actinomycetes associated with stony corals: A potential hotspot for specialized metabolites. Microorganisms 2022, 10, 1349. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Han, L.; Li, C.; Cao, Q.; Zhu, D.; Barrett, N.H.; Harmody, D.; Chen, J.; Zhu, H. Bioprospecting deep-sea actinobacteria for novel anti-infective natural products. Front. Microbiol. 2018, 9, 787. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gui, C.; Shao, M.; Kumar, P.S.; Huang, H.; Ju, J. Antimicrobial tunicamycin derivatives from the deep-sea derived Streptomyces xinghaiensis SCSIO S15077. Nat. Prod. Res. 2020, 34, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.X.; Xie, C.L.; Zhou, M.; Xia, M.L.; Zhou, T.T.; Chen, H.F.; Yang, X.W.; Yang, Q. Chemical constituents from the deep-sea derived Streptomyces xiamenensis MCCC 1A01570 and their effects on RXRα transcriptional regulation. Nat. Prod. Res. 2020, 34, 1461–1464. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, G.; Wang, B.; Li, X.; Yue, S.; Chen, J.; Zhang, H.; Wang, H. Production and identification of inthomycin B produced by a deep-sea sediment-derived Streptomyces sp. YB104 Based on cultivation-dependent approach. Curr. Microbiol. 2018, 75, 942–951. [Google Scholar] [CrossRef]
- Palazzotto, E.; Weber, T. Omics and multi-omics approaches to study the biosynthesis of secondary metabolites in microorganisms. Curr. Opin. Microbiol. 2018, 45, 109–116. [Google Scholar] [CrossRef]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef]
- Schwecke, T.; Aparicio, J.F.; Molnar, I.; König, A.; Khaw, L.E.; Haydock, S.F.; Oliynyk, M.; Caffrey, P.; Cortés, J.; Lester, J.B. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc. Natl. Acad. Sci. USA 1995, 92, 7839–7843. [Google Scholar] [CrossRef]
- Tillett, D.; Dittmann, E.; Erhard, M.; von Döhren, H.; Börner, T.; Neilan, B.A. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: An integrated peptide–polyketide synthetase system. Chem. Biol. 2000, 7, 753–764. [Google Scholar] [CrossRef]
- Wang, H.; Fewer, D.P.; Holm, L.; Rouhiainen, L.; Sivonen, K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc. Natl. Acad. Sci. USA 2014, 111, 9259–9264. [Google Scholar] [CrossRef]
- Chen, R.; Wong, H.L.; Kindler, G.S.; MacLeod, F.I.; Benaud, N.; Ferrari, B.C.; Burns, B.P. Discovery of an abundance of biosynthetic gene clusters in shark bay microbial mats. Front. Microbiol. 2020, 11, 1950. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Seyedsayamdost, M.R. Synergy and target promiscuity drive structural divergence in bacterial alkylquinolone biosynthesis. Cell Chem. Biol. 2017, 24, 1437–1444.e3. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Charusanti, P.; Musiol-Kroll, E.M.; Jiang, X.; Tong, Y.; Kim, H.U.; Lee, S.W. Metabolic engineering of antibiotic factories: New tools for antibiotic production in actinomycetes. Trends Biotechnol. 2015, 33, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Doroghazi, J.R.; Metcalf, W.W. Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genom. 2013, 14, 611. [Google Scholar] [CrossRef]
- Van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; Van Wezel, G.P. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef]
- Hifnawy, M.S.; Fouda, M.M.; Sayed, A.M.; Mohammed, R.; Hassan, H.M.; AbouZid, S.F.; Rateb, M.E.; Keller, A.; Adamek, M.; Ziemert, N. The genus Micromonospora as a model microorganism for bioactive natural product discovery. RSC Adv. 2020, 10, 20939–20959. [Google Scholar] [CrossRef] [PubMed]
- Mitousis, L.; Thoma, Y.; Musiol-Kroll, E.M. An update on molecular tools for genetic engineering of actinomycetes—The source of important antibiotics and other valuable compounds. Antibiotics 2020, 9, 494. [Google Scholar] [CrossRef] [PubMed]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes–a review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef]
- Ding, W.; Tu, J.; Zhang, H.; Wei, X.; Ju, J.; Li, Q. Genome mining and metabolic profiling uncover polycyclic tetramate macrolactams from Streptomyces koyangensis SCSIO 5802. Mar. Drugs 2021, 19, 440. [Google Scholar] [CrossRef]
- Zhang, C.; Ding, W.; Qin, X.; Ju, J. Genome sequencing of Streptomyces olivaceus SCSIO T05 and activated production of lobophorin CR4 via metabolic engineering and genome mining. Mar. Drugs 2019, 17, 593. [Google Scholar] [CrossRef]
- Su, S.; Liao, L.; Yu, Y.; Zhang, J.; Chen, B. Genomic data mining of an antarctic deep-sea actinobacterium, Janibacter limosus P3-3-X1. Mar. Genom. 2019, 48, 100684. [Google Scholar] [CrossRef]
- Albuquerque, P.; Ribeiro, I.; Correia, S.; Mucha, A.P.; Tamagnini, P.; Braga-Henriques, A.; Carvalho, M.F.; Mendes, M.V. Complete genome sequence of two deep-sea Streptomyces isolates from madeira archipelago and evaluation of their biosynthetic potential. Mar. Drugs 2021, 19, 621. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yang, Z.; Zhang, C.; Liu, Z.; He, J.; Liu, Q.; Zhang, T.; Ju, J.; Ma, J. Genome mining of Streptomyces atratus SCSIO ZH16: Discovery of atratumycin and identification of its biosynthetic gene cluster. Org. Lett. 2019, 21, 1453–1457. [Google Scholar] [CrossRef] [PubMed]
- Carr, C.M.; de Oliveira, B.F.R.; Jackson, S.A.; Laport, M.S.; Clarke, D.J.; Dobson, A.D.W. Identification of BgP, a cutinase-like polyesterase from a deep-sea sponge-derived actinobacterium. Front. Microbiol. 2022, 13, 888343. [Google Scholar] [CrossRef]
- Chen, Y.; Pan, T.; Chai, G.; Li, Z. Complete genome of Mycetocola spongiae MSC19T isolated from deep-sea sponge Cacospongia mycofijiensis indicates the adaptation to deep-sea environment and sponge-microbe symbioses. Mar. Genom. 2022, 63, 100955. [Google Scholar] [CrossRef]
- Abdel-Mageed, W.M.; Al-Wahaibi, L.H.; Lehri, B.; Al-Saleem, M.S.M.; Goodfellow, M.; Kusuma, A.B.; Nouioui, I.; Soleh, H.; Pathom-Aree, W. Biotechnological and ecological potential of Micromonospora provocatoris sp. nov., a gifted strain isolated from the challenger deep of the mariana trench. Mar. Drugs 2021, 19, 243. [Google Scholar] [CrossRef]
- Zhang, L.; Zeng, Q.; Liu, X.; Chen, P.; Guo, X.; Ma, L.Z.; Dong, H.; Huiang, Y. Iron reduction by diverse actinobacteria under oxic and pH-neutral conditions and the formation of secondary minerals. Chem. Geol. 2019, 525, 390–399. [Google Scholar] [CrossRef]
- Back, C.R.; Stennett, H.L.; Williams, S.E.; Wang, L.; Ojeda Gomez, J.; Abdulle, O.M.; Duffy, T.; Neal, C.; Mantell, J.; Jepson, M.A. A new Micromonospora strain with antibiotic activity isolated from the microbiome of a mid-Atlantic deep-sea sponge. Mar. Drugs 2021, 19, 105. [Google Scholar] [CrossRef]
- Ding, W.; Li, Y.; Tian, X.; Chen, M.; Xiao, Z.; Chen, R.; Yin, Y.; Zhang, S. Investigation on metabolites in structural diversity from the deep-sea sediment-derived bacterium Agrococcus sp. SCSIO 52902 and their biosynthesis. Mar. Drugs 2022, 20, 431. [Google Scholar] [CrossRef]
- Sarmiento-Vizcaíno, A.; Braña, A.F.; Pérez-Victoria, I.; Martín, J.; De Pedro, N.; Cruz, M.D.; Diaz, C.; Vicente, F.; Acuña, J.L.; Reyes, F. Paulomycin G: A new natural product with cytotoxic activity against tumor cell lines produced by deep-sea sediment derived Micromonospora matsumotoense M-412 from the avilés canyon in the cantabrian sea. Mar. Drugs 2017, 15, 271. [Google Scholar] [CrossRef]
- Zhang, Z.; In, Y.; Fukaya, K.; Yang, T.; Harunari, E.; Urabe, D.; Imada, C.; Oku, N.; Igarashi, Y. Kumemicinones A–G, cytotoxic angucyclinones from a deep-sea-derived actinomycete of the genus Actinomadura. J. Nat. Prod. 2022, 85, 1098–1108. [Google Scholar] [CrossRef] [PubMed]
- Yi, W.; Qin, L.; Lian, X.Y.; Zhang, Z. New antifungal metabolites from the mariana trench sediment-associated actinomycete Streptomyces sp. SY1965. Mar. Drugs 2020, 18, 385. [Google Scholar] [CrossRef]
- Xu, D.; Nepal, K.K.; Chen, J.; Harmody, D.; Zhu, H.; McCarthy, P.J.; Wright, A.E.; Wang, G. Nocardiopsistins A-C: New angucyclines with anti-MRSA activity isolated from a marine sponge-derived Nocardiopsis sp. HB-J378. Synth. Syst. Biotechnol. 2018, 3, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, J.; Wang, S.; Bao, X.; Li, S.; Wei, B.; Zhang, H.; Whang, H. Amycolachromones A-F, isolated from a Streptomycin-resistant strain of the deep-sea marine actinomycete Amycolatopsis sp. WP1. Mar. Drugs 2022, 20, 162. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, S.; Xu, Y.; Zhang, H.; Wang, H. Anti-tyrosinase compounds from the deep-sea-derived actinomycete Georgenia sp. 40DY180. Chem. Biodivers. 2022, 19, e202200037. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, Y.; Li, Y.; Song, Y.; Ma, J.; Ju, J. Secondary metabolites and biosynthetic gene clusters analysis of deep-sea hydrothermal vent-derived Streptomyces sp. SCSIO ZS0520. Mar. Drugs 2022, 20, 393. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.; Huang, Y.; Yuan, J.; Wei, X.; Ju, J. Discovery, structure correction, and biosynthesis of actinopyrones, cytotoxic polyketides from the deep-sea hydrothermal-vent-derived Streptomyces sp. SCSIO ZS0520. J. Nat. Prod. 2022, 85, 625–633. [Google Scholar] [CrossRef]
- Jiang, X.; Fang, Z.; Zhang, Q.; Liu, W.; Zhang, L.; Zhang, W.; Yang, C.; Zhang, H.; Zhu, W.; Zhang, C. Discovery of a new asymmetric dimer nenestatin B and implications of a dimerizing enzyme in a deep-sea actinomycete. Org. Biomol. Chem. 2021, 19, 4243–4247. [Google Scholar] [CrossRef]
- Abdel-Mageed, W.M.; Juhasz, B.; Lehri, B.; Alqahtani, A.S.; Nouioui, I.; Pech-Puch, D.; Tabudravu, J.N.; Goodfellow, M.; Rodríguez, J.; Jaspars, M.; et al. Whole genome sequence of Dermacoccus abyssi MT1.1 isolated from the challenger deep of the mariana trench reveals phenazine biosynthesis locus and environmental adaptation factors. Mar. Drugs 2020, 18, 131. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, C.; Qin, X.; Wei, X.; Liu, Q.; Li, Q.; Ju, J. Genome mining of Streptomyces olivaceus SCSIO T05: Discovery of olimycins A and B and assignment of absolute configurations. Tetrahedron 2018, 74, 199–203. [Google Scholar] [CrossRef]
- Xie, C.L.; Niu, S.W.; Zhou, T.T.; Zhang, G.Y.; Yang, Q.; Yang, X.W. Chemical constituents and chemotaxonomic study on the marine actinomycete Williamsia sp. MCCC 1A11233. Biochem. Syst. Ecol. 2016, 67, 129–133. [Google Scholar] [CrossRef]
- Wu, Q.; Deering, R.W.; Zhang, G.; Wang, B.; Li, X.; Sun, J.; Chen, J.; Zhang, H.; Rowley, D.C.; Wang, H. Albisporachelin, a new hydroxamate type siderophore from the deep ocean sediment-derived actinomycete Amycolatopsis albispora WP1T. Mar. Drugs 2018, 16, 199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wang, L.; Li, J.; Zhou, Y. Amycolatopsis albispora sp. nov., isolated from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 2016, 66, 3860–3864. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zhang, L.; Wang, J.; Ruan, J.; Han, X.; Huang, Y. Brevibacterium sediminis sp. nov., isolated from deep-sea sediments from the Carlsberg and Southwest Indian Ridges. Int. J. Syst. Evol. Microbiol. 2016, 66, 5268–5274. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xi, L.; Ruan, J.; Huang, Y. Kocuria oceani sp. nov., isolated from a deep-sea hydrothermal plume. Int. J. Syst. Evol. Microbiol. 2017, 67, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wang, L.; Li, J.; Zhou, Y. Pseudonocardia profundimaris sp. nov., isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2017, 67, 1693–1697. [Google Scholar] [CrossRef]
- Chen, R.W.; Wang, K.X.; Wang, F.Z.; He, Y.Q.; Long, L.J.; Tian, X.P. Rubrobacter indicoceani sp. nov., a new marine actinobacterium isolated from Indian Ocean sediment. Int. J. Syst. Evol. Microbiol. 2018, 68, 3487–3493. [Google Scholar] [CrossRef]
- Chen, R.W.; Li, C.; He, Y.Q.; Cui, L.Q.; Long, L.J.; Tian, X.P. Rubrobacter tropicus sp. nov. and Rubrobacter marinus sp. nov., isolated from deep-sea sediment of the South China Sea. Int. J. Syst. Evol. Microbiol. 2020, 70, 5576–5585. [Google Scholar] [CrossRef]
- Zhang, G.; Wang, L.; Xie, F.; Pei, S.; Jiang, L. Nesterenkonia salmonea sp. nov. and Nesterenkonia sphaerica sp. nov., isolated from the southern atlantic ccean. Int. J. Syst. Evol. Microbiol. 2020, 70, 923–928. [Google Scholar] [CrossRef]
- Intra, B.; Panbangred, W.; Inahashi, Y.; Také, A.; Mori, M.; Ōmura, S.; Matsumoto, A. Micromonospora pelagivivens sp. nov., a new species of the genus Micromonospora isolated from deep-sea sediment in Japan. Int. J. Syst. Evol. Microbiol. 2020, 70, 3069–3075. [Google Scholar] [CrossRef]
- He, Y.Q.; Chen, R.W.; Li, C.; Shi, S.B.; Cui, L.Q.; Long, L.J.; Tian, X.P. Actinomarinicola tropica gen. nov. sp. nov., a new marine actinobacterium of the family Iamiaceae, isolated from South China Sea sediment environments. Int. J. Syst. Evol. Microbiol. 2020, 70, 3852–3858. [Google Scholar] [CrossRef] [PubMed]
- Pei, S.; Xie, F.; Niu, S.; Ma, L.; Zhang, R.; Zhang, G. Brevibacterium profundi sp. nov., isolated from deep-sea sediment of the Western Pacific Ocean. Int. J. Syst. Evol. Microbiol. 2020, 70, 5818–5823. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; He, W.X.; Zhang, D.C. Marinisubtilis pacificus gen. nov., sp. nov., a member of the family Microbacteriaceae isolated from a deep-sea seamount. Curr. Microbiol. 2021, 78, 2136–2142. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Pei, S.; Huang, X.; Wang, L.; Kou, J.; Zhang, G. Microcella flavibacter sp. nov., isolated from marine sediment, and reclassification of Chryseoglobus frigidaquae, Chryseoglobus indicus, and Yonghaparkia alkaliphila as Microcella frigidaquae comb. nov., Microcella indica nom. nov., and Microcella alkali. Antonie Leeuwenhoek 2021, 114, 2133–2145. [Google Scholar] [CrossRef] [PubMed]
- Pei, S.; Xie, F.; Wang, W.; Zhang, S.; Zhang, G. Chryseoglobus indicus sp. nov., isolated from deep-sea water. Int. J. Syst. Evol. Microbiol. 2021, 71, 4564. [Google Scholar] [CrossRef]
- Parra, J.; Soldatou, S.; Rooney, L.M.; Duncan, K.R. Pseudonocardia abyssalis sp. nov. and Pseudonocardia oceani sp. nov., two novel actinomycetes isolated from the deep Southern Ocean. Int. J. Syst. Evol. Microbiol. 2021, 71, 005032. [Google Scholar] [CrossRef]
- Risdian, C.; Landwehr, W.; Rohde, M.; Schumann, P.; Hahnke, R.L.; Spröer, C.; Bunk, B.; Kämpfer, P.; Schupp, P.J.; Wink, J. Streptomyces bathyalis sp. nov., an actinobacterium isolated from the sponge in a deep-sea. Antonie Van Leeuwenhoek 2021, 114, 425–435. [Google Scholar] [CrossRef]
- Xie, F.; Pei, S.; Zhang, Y.; Tian, Y.; Zhang, G. Nesterenkonia sedimenti sp. nov., isolated from marine sediment. Arch. Microbiol. 2021, 203, 6287–6293. [Google Scholar] [CrossRef]
- Li, C.; He, Y.Q.; Cui, L.Q.; Albuquerque, L.; Chen, R.W.; Long, L.J.; Tian, X.P. Miltoncostaea marina gen. nov. sp. nov., and Miltoncostaea oceani sp. nov., a novel deep branching phylogenetic lineage within the class Thermoleophilia isolated from marine environments, and proposal of Miltoncostaeaceae fam. nov. and Miltoncostaeales or. Syst. Appl. Microbiol. 2021, 44, 126216. [Google Scholar] [CrossRef]
- Chen, Y.; Sang, J.; Sun, W.; Song, Q.; Li, Z. Mycetocola spongiae sp. nov., isolated from deep-sea sponge Cacospongia mycofijiensis. Int. J. Syst. Evol. Microbiol. 2022, 72, 5291. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Tang, G.; Ju, J.; Lu, L.; Huang, H. A new diketopiperazine derivative from a deep-sea derived Streptomyces sp. SCSIO 04496. Nat. Prod. Res. 2016, 30, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Li, H.; Qiu, Y.; Hou, L.; Ju, J.; Li, W. A new dioic acid from a wbl gene mutant of deep-sea derived Streptomyces somaliensis SCSIO ZH66. Mar. Drugs 2016, 14, 184. [Google Scholar] [CrossRef] [PubMed]
- Paulus, C.; Rebets, Y.; Tokovenko, B.; Nadmid, S.; Terekhova, L.P.; Myronovskyi, M.; Zotchev, S.B.; Rückert, C.; Braig, S. New natural products identified by combined genomics-metabolomics profiling of marine Streptomyces sp. MP131-18. Sci. Rep. 2017, 7, 42382. [Google Scholar] [CrossRef]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Pérez-Victoria, I.; Otero, L.; Fernández, J.; Palacios, J.J.; Martín, J.; de la Cruz, M.; Díaz, C.; Vicente, F. Branimycins B and C antibiotics produced by the abyssal actinobacterium Pseudonocardia carboxydivorans M-227. J. Nat. Prod. 2017, 80, 569–573. [Google Scholar] [CrossRef]
- Sun, M.; Chen, X.; Li, W.; Lu, C.; Shen, Y. New diketopiperazine derivatives with cytotoxicity from Nocardiopsis sp. YIM M13066. J. Antibiot. 2017, 70, 795–797. [Google Scholar] [CrossRef]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Osset, M.; Pérez-Victoria, I.; Martín, J.; De Pedro, N.; De la Cruz, M.; Díaz, C.; Vicente, F.; Reyes, F.; et al. Lobophorin K: A new natural product with cytotoxic activity produced by Streptomyces sp. M-207 associated with the deep-sea coral Lophelia pertusa. Mar. Drugs 2017, 15, 144. [Google Scholar] [CrossRef]
- Niu, S.; Zhou, T.T.; Xie, C.L.; Zhang, G.Y.; Yang, X.W. Microindolinone A, a novel 4, 5, 6, 7-tetrahydroindole, from the deep-sea-derived actinomycete Microbacterium sp. MCCC 1A11207. Mar. Drugs 2017, 15, 230. [Google Scholar] [CrossRef]
- Xie, C.L.; Liu, Q.; Xia, J.M.; Gao, Y.; Yang, Q.; Shao, Z.Z.; Liu, G.; Yang, X.-W. Anti-allergic compounds from the deep-sea-derived actinomycete Nesterenkonia flava MCCC 1K00610. Mar. Drugs 2017, 15, 71. [Google Scholar] [CrossRef]
- Zou, G.; Liao, X.J.; Peng, Q.; Chen, G.D.; Wei, F.Y.; Xu, Z.X.; Zhao, B.X.; Xui, S.H. A new α-pyrone from the deep-sea actinomycete Nocardiopsis dassonvillei subsp. dassonvillei DSM 43111(T). J. Asian Nat. Prod. Res. 2017, 19, 1232–1238. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, Q.; Zhu, Y.; Nie, F.; Wu, Z.; Yang, C.; Zhang, L.; Tian, X.; Zhang, C. Isolation, structure elucidation and biosynthesis of benzo[b]fluorene nenestatin A from deep-sea derived Micromonospora echinospora SCSIO 04089. Tetrahedron 2017, 73, 3585–3590. [Google Scholar] [CrossRef]
- Song, Y.; Li, Q.; Qin, F.; Sun, C.; Liang, H.; Wei, X.; Wong, N.K.; Ye, L.; Zhang, Y.; Shao, M.; et al. Neoabyssomicins A–C, polycyclic macrolactones from the deep-sea derived Streptomyces koyangensis SCSIO 5802. Tetrahedron 2017, 73, 5366–5372. [Google Scholar] [CrossRef]
- Li, H.; Huang, H.; Hou, L.; Ju, J.; Li, W. Discovery of antimycin-type depsipeptides from a wbl gene mutant strain of deepsea-derived Streptomyces somaliensis SCSIO ZH66 and their effects on pro-inflammatory cytokine production. Front. Microbiol. 2017, 8, 678. [Google Scholar] [CrossRef]
- Zhang, X.M.; Sun, M.W.; Shi, H.; Lu, C.H. α-pyrone derivatives from a marine actinomycete Nocardiopsis sp. YIM M13066. Nat. Prod. Res. 2017, 31, 2245–2249. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.L.; Niu, S.; Xia, J.M.; Peng, K.; Zhang, G.Y.; Yang, X.W. Saccharopolytide A, a new cyclic tetrapeptide with rare 4-hydroxy-proline moieties from the deep-sea derived actinomycete Saccharopolyspora cebuensis MCCC 1A09850. Nat. Prod. Res. 2018, 32, 1627–1631. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-López, F.J.; Alcalde, E.; Sarmiento-Vizcaíno, A.; Díaz, C.; Cautain, B.; García, L.A.; Blanco, G.; Reyes, F. New 3-Hydroxyquinaldic acid derivatives from cultures of the marine derived actinomycete Streptomyces cyaneofuscatus M-157. Mar. Drugs 2018, 16, 371. [Google Scholar] [CrossRef]
- Yang, L.; Hou, L.; Li, H.; Li, W. Antibiotic angucycline derivatives from the deep-sea derived Streptomyces lusitanus. Nat. Prod. Res. 2019, 34, 3444–3450. [Google Scholar] [CrossRef]
- Wang, P.; Wang, D.; Zhang, R.; Wang, Y.; Kong, F.; Fu, P.; Zhu, W. Novel macrolactams from a deep-sea derived Streptomyces species. Mar. Drugs 2020, 19, 13. [Google Scholar] [CrossRef]
- Wang, J.X.; Sun, C.X.; Shah, M.; Zhang, G.J.; Gu, Q.Q.; Zhu, T.J.; Che, Q.; Li, D.H. New metabolites from a mariana trench-derived actinomycete Nocardiopsis sp. HDN 17-237. J. Asian Nat. Prod. Res. 2020, 22, 1031–1036. [Google Scholar] [CrossRef]
- Hou, L.; Liu, Z.; Yu, D.; Li, H.; Ju, J.; Li, W. Targeted isolation of new polycyclic tetramate macrolactams from the deepsea-derived Streptomyces somaliensis SCSIO ZH66. Bioorg. Chem. 2020, 101, 103954. [Google Scholar] [CrossRef]
- Igarashi, M.; Sawa, R.; Umekita, M.; Hatano, M.; Arisaka, R.; Hayashi, C.; Ishizaki, Y.; Suzuki, M.; Kato, K. Sealutomicins, new enediyne antibiotics from the deep-sea actinomycete Nonomuraea sp. MM565M-173N2. J. Antibiot. 2021, 74, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Yong, K.; Lian, X.Y.; Zhang, Z. Streptonaphthyridine A: A new naphthyridine analogue with antiproliferative activity against human glioma cells from mariana trench-associated actinomycete Streptomyces sp. SY2111. Nat. Prod. Res. 2021, 17, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yang, J.; Yu, J.; Li, J.; Yuan, J.; Wong, N.K.; Ju, J. Chlorinated bis-indole alkaloids from deep-sea derived Streptomyces sp. SCSIO 11791 with antibacterial and cytotoxic activities. J. Antibiot. 2020, 73, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Song, Y.; Tang, M.C.; Li, M.; Deng, J.; Wong, N.K.; Ju, J. Genome-directed discovery of tetrahydroisoquinolines from deep-sea derived Streptomyces niveus SCSIO 3406. J. Org. Chem. 2021, 86, 11107–11116. [Google Scholar] [CrossRef]
- Luo, M.; Tang, L.; Dong, Y.; Huang, H.; Deng, Z.; Sun, Y. Antibacterial natural products lobophorin L and M from the marine-derived Streptomyces sp. 4506. Nat. Prod. Res. 2021, 35, 5581–5587. [Google Scholar] [CrossRef]
- Harunari, E.; Bando, M.; Igarashi, Y. Rausuquinone, a non-glycosylated pluramycin-class antibiotic from Rhodococcus. J. Antibiot. 2022, 75, 86–91. [Google Scholar] [CrossRef]
- Ko, K.; Kim, S.H.; Park, S.; Han, H.S.; Lee, J.K.; Cha, J.W.; Hwang, S.; Choi, K.Y.; Song, Y.-J.; Nam, S.-J.; et al. Discovery and photoisomerization of new pyrrolosesquiterpenoids glaciapyrroles D. and, E.; from deep-sea sediment Streptomyces sp. Mar. Drugs 2022, 20, 281. [Google Scholar] [CrossRef]
- Su, Z.; Li, K.; Luo, X.; Zhu, Y.; Mai, S.Y.; Zhu, Q.; Yang, B.; Zhou, X.; Tao, H. Aromatic acids and leucine derivatives produced from the deep-sea actinomycetes Streptomyces chumphonensis SCSIO15079 with antihyperlipidemic Activities. Mar. Drugs 2022, 20, 259. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Rosselló-Móra, R.; Amann, R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017, 11, 2399–2406. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, Q.; Chen, X.; Jiang, C. Isolation and Cultivation Methods of Actinobacteria. In Actinobacteria-Basics and Biotechnological Applications; Dhanasekaran, D., Jiang, Y., Eds.; InTechOpen: London, UK, 2016; pp. 39–50. [Google Scholar]
- Girão, M.; Ribeiro, I.; Carvalho, M.D.F. Actinobacteria from Marine Environments: A Unique Source of Natural Products. In Natural Products from Actinomycetes: Diversity, Ecology and Drug Discovery; Rai, R.V., Bai, J.A., Eds.; Springer: Singapore, 2022; pp. 1–45. [Google Scholar] [CrossRef]
- Van Bergeijk, D.A.; Terlouw, B.R.; Medema, M.H.; van Wezel, G.P. Ecology and genomics of actinobacteria: New concepts for natural product discovery. Nat. Rev. Microbiol. 2020, 18, 546–558. [Google Scholar] [CrossRef]
- Palma, M.; Barbero, G.F.; Pineiro, Z.; Liazid, A.; Barroso, C.G.; Rostagno, M.A.; Prado, J.M.; Meireles, M.A.A. Extraction of Natural Products: Principles and Fundamental Aspects. In Natural Product Extraction: Principles and Applications; Rostagno, M., Prado, J., Eds.; Royal Society of Chemistry: London, UK, 2022; pp. 58–88. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).