Genetic Diversity and Population Structure Derived from Body Remains of the Endangered Flightless Longhorn Beetle Iberodorcadion fuliginator in Grassland Fragments in Central Europe
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Species
2.2. Study Sites and Sampling
2.3. DNA Extraction and Microsatellite Markers
2.4. Genetic Analyses
3. Results
3.1. Genetic Diversity and Population Structure at the Regional Scale
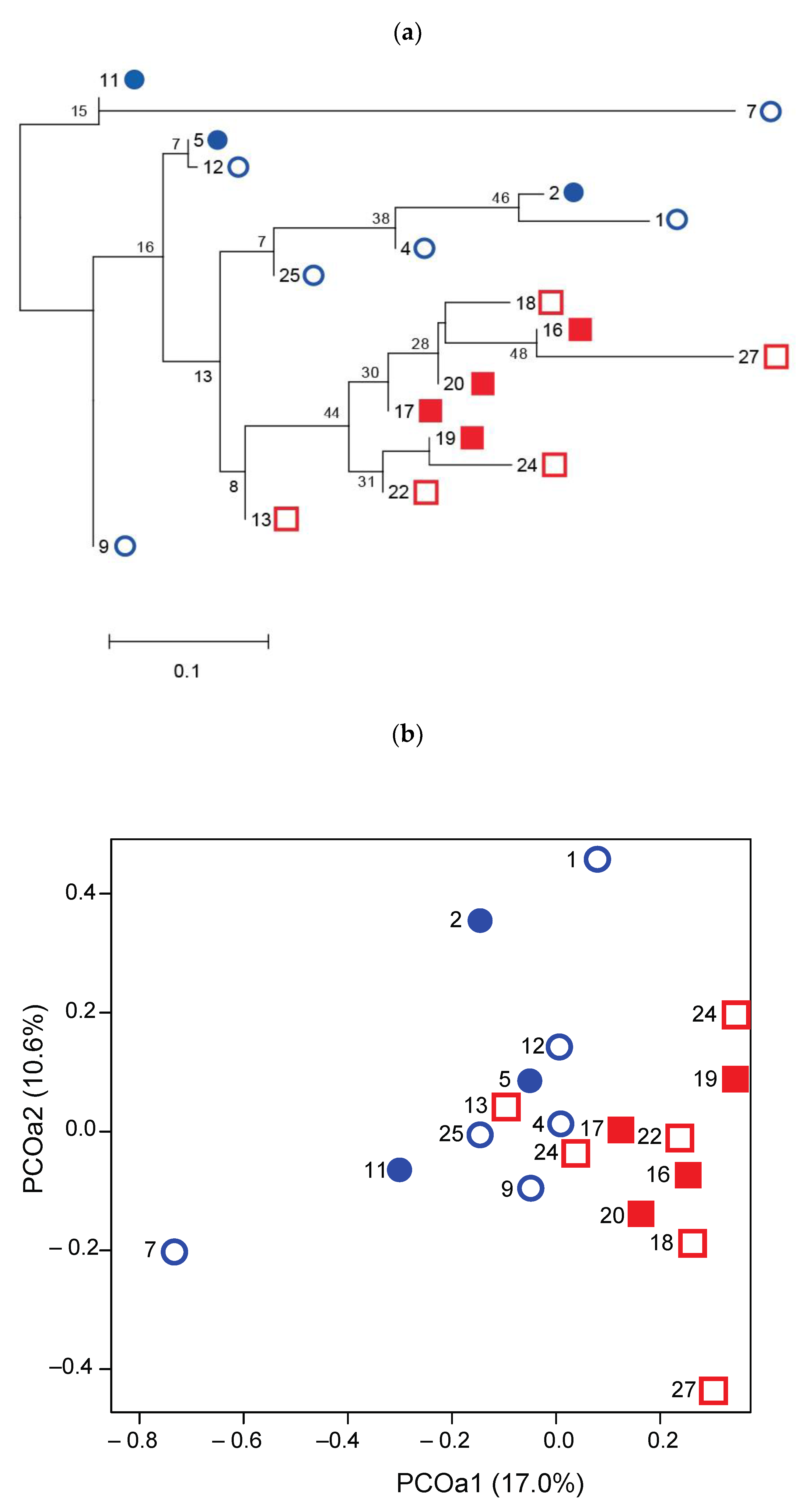
3.2. Spatial Genetic Structure
3.3. Genetic Diversity and Long-Term Population Dynamics
4. Discussion
4.1. Genetic Diversity and Inbreeding
4.2. Spatial Genetic Structure
4.3. Variation in Duration of Larval Development
4.4. Non-Invasive Approach
4.5. Implications for Conservation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baur, B.; Erhardt, A. Habitat fragmentation and habitat alteration: Principle threats to many animal and plant species. Gaia 1995, 4, 221–226. [Google Scholar] [CrossRef]
- Dirzo, R.; Young, H.S.; Galetti, M.; Ceballos, G.; Isaac, N.J.B.; Collen, B. Defaunation in the Anthropocene. Science 2014, 345, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.C.; Chen, X.-Y.; Corlett, R.T.; Didham, R.K.; Ding, P.; Holt, R.D.; Holyoak, M.; Hu, G.; Hughes, A.C.; Jiang, L.; et al. Habitat fragmentation and biodiversity conservation: Key findings and future challenges. Landsc. Ecol. 2016, 31, 219–227. [Google Scholar] [CrossRef]
- Saccheri, I.; Kuussaari, M.; Kankare, M.; Vikman, P.; Fortelius, W.; Hanski, I. Inbreeding and extinction in a butterfly metapopulation. Nature 1998, 392, 491–494. [Google Scholar] [CrossRef]
- Krauss, J.; Bommarco, R.; Guardiola, M.; Heikkinnen, M.; Lindborg, A.; Öckinger, E.; Pärtel, M.; Pino, J.; Pöyry, J.; Raatikainen, K.; et al. Habitat fragmentation causes immediate and time-delayed biodiversity loss at different trophic levels. Ecol. Lett. 2010, 13, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Ouborg, N.J.; Vergeer, P.; Mix, C. The rough edges of the conservation genetics paradigm for plants. J. Ecol. 2006, 94, 1233–1248. [Google Scholar] [CrossRef]
- Freeland, J.R.; Kirk, H.; Petersen, S.D. Molecular Ecology, 2nd ed.; Wiley-Blackwell: Oxford, UK, 2011; p. 464. [Google Scholar]
- Jump, A.S.; Marchant, R.; Peñuelas, J. Environmental change and the option value of genetic diversity. Trends Plant Sci. 2009, 14, 51–58. [Google Scholar] [CrossRef]
- Frankham, R. Challenges and opportunities of genetic approaches to biological conservation. Biol. Conserv. 2010, 143, 1919–1927. [Google Scholar] [CrossRef]
- Saccheri, I.J.; Brakefield, P.M.; Nichols, R.A. Severe inbreeding and rapid fitness rebound in the butterfly Bicyclus anynana (Satyridae). Evolution 1996, 50, 2000–2013. [Google Scholar] [CrossRef]
- Schlaepfer, D.R.; Braschler, B.; Rusterholz, H.-P.; Baur, B. Genetic effects of anthropogenic habitat fragmentation on remnant animal and plant populations: A meta-analysis. Ecosphere 2018, 9, e02488. [Google Scholar] [CrossRef]
- Samways, M.J. Insect Diversity Conservation; Cambridge University Press: New York, NY, USA, 2005; p. 342. [Google Scholar]
- Baur, B. Naturschutzbiologie; UTB 5416; Haupt Verlag: Bern, Switzerland, 2021; p. 440. [Google Scholar]
- Taberlet, P.; Waits, L.P.; Luikart, G. Noninvasive genetic sampling: Look before you leap. Trends Ecol. Evol. 1999, 14, 323–327. [Google Scholar] [CrossRef]
- Redlarski, A.J.; Klejdysz, T.; Kadej, M.; Meyza, K.; Vasilita, C.; Oleksa, A. Body remains left by bird predators as a reliable source for population genetic studies in the Great Capricorn beetle Cerambyx cerdo, a veteran oak specialist. Insects 2021, 12, 574. [Google Scholar] [CrossRef]
- Starks, P.T.; Peters, J.M. Semi-nondestructive genetic sampling from live eusocial wasps, Polistes dominula and Polistes fuscatus. Insectes Sociaux 2002, 49, 20–22. [Google Scholar] [CrossRef]
- Vila, M.; Auger-Rozenberg, M.A.; Goussard, F.; Lopez-Vaamonde, C. Effect of non-lethal sampling on life-history traits of the protected moth Graellsia isabellae (Lepidoptera: Saturniidae). Ecol. Entomol. 2009, 34, 356–362. [Google Scholar] [CrossRef]
- Marschalek, D.A.; Jesu, J.A.; Berres, M.E. Impact of non-lethal genetic sampling on the survival, longevity and behaviour of the Hermes copper (Lycaena hermes) butterfly. Insect Conserv. Divers. 2013, 6, 658–662. [Google Scholar] [CrossRef]
- Cox, K.; McKeown, N.; Vanden Broeck, A.; Van Breusegem, A.; Cammaerts, R.; Thomaes, A. Genetic structure of recently fragmented suburban populations of European stag beetle. Ecol. Evol. 2020, 10, 12290–12306. [Google Scholar] [CrossRef]
- Baur, B.; Coray, A.; Lenzin, H.; Schmera, D. Factors contributing to the decline of an endangered flightless longhorn beetle: A 20-year study. Insect Conserv. Divers. 2020, 13, 175–186. [Google Scholar] [CrossRef]
- Dennis, E.B.; Kéry, M.; Morgan, B.J.T.; Coray, A.; Schaub, M.; Baur, B. Integrated modelling of insect population dynamics at two temporal scales. Ecol. Model. 2021, 441, 109408. [Google Scholar] [CrossRef]
- Weibel, U. Zum Vorkommen und zur Phänologie des Berussten Erdbockes (Iberodorcadion fuliginator (Linnaeus 1758)) (Cerambycidae, Coleoptera) in der Region Schaffhausen. Mitt. Nat.forsch. Ges. Schaffhausen 2021, 49, 10–13. [Google Scholar]
- Horion, A. Faunistik der mitteleuropäischen Käfer. Band 12: Cerambycidae—Bockkäfer; Überlingen: Bodensee, Germany, 1974; p. 228. [Google Scholar]
- Villiers, A. Faune des Coléoptères de France. I. Cerambycidae; Lechevalier: Paris, France, 1978; p. 612. [Google Scholar]
- Vives, E. Revisión del género Iberodorcadion (Coleópteros, Cerambicidos); Consejo Superior de Investigaciones Cientificas, Instituto Español de Entomologia: Madrid, Spain, 1983; p. 171. [Google Scholar]
- Althoff, J.; Danilevsky, M.L. Seznam Kozlicev (Coleoptera, Cerambycoidea) Evrope (A check-list of longicorn beetles (Coleoptera, Cerambycoidea) of Europe); Slovensko Entomolosko Drustvo Stefana Michielija: Ljubljana, Slowenia, 1997; p. 64. [Google Scholar]
- Klausnitzer, B.; Sander, F. Die Bockkäfer Mitteleuropas. Die Neue Brehm Bücherei; A. Ziemsen: Wittenberg, Lutherstadt, Germany, 1978; p. 222. [Google Scholar]
- Coray, A.; Altermatt, F.; Birrer, S.; Buser, H.; Jäggi, C.; Reiss, T.; Schläpfer, M. Verbreitung, Habitat und Erscheinungsformen des Erdbockkäfers Dorcadion fuliginator (L.) (Coleoptera, Cerambycidae) in der Umgebung von Basel. Mitt. Entomol. Ges. Basel 2000, 50, 42–73. [Google Scholar]
- Baur, B.; Zschokke, S.; Coray, A.; Schläpfer, M.; Erhardt, A. Habitat characteristics of the endangered flightless beetle Dorcadion fuliginator (Coleoptera: Cerambycidae): Implications for conservation. Biol. Conserv. 2002, 105, 133–142. [Google Scholar] [CrossRef]
- Monnerat, C.; Barbalat, S.; Lachat, T.; Gonseth, Y. Rote Liste der Prachtkäfer, Bockkäfer, Rosenkäfer und Schröter. Gefährdete Arten der Schweiz; Umweltvollzug 1622; Bundesamt für Umwelt: Bern, Switzerland; Info Fauna CSCF: Neuenburg, Germany; Eidgenössische Forschungsanstalt WSL: Birmensdorf, Switzerland, 2016; p. 118. [Google Scholar]
- Niehuis, M. Bockkäfer—Rote Liste der Ausgestorbenen, Verschollenen und Gefährdeten Bockkäfer in Rheinland-Pfalz; Ministerium für Umwelt und Forsten: Mainz, Germany, 2000; p. 32. [Google Scholar]
- Schmidl, J.; Bussler, H. Rote Liste gefährdeter Bockkäfer (Coleoptera: Cerambycidae) Bayerns. Schriftenreihe Bayer. Landesamt Umweltschutz 2003, 166, 150–153. [Google Scholar]
- Pannel, J.R.; Charlesworth, B. Effects of metapopulation processes on measures of genetic diversity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 1851–1864. [Google Scholar] [CrossRef] [PubMed]
- Baur, B.; Coray, A.; Minoretti, N.; Zschokke, S. Dispersal of the endangered flightless beetle Dorcadion fuliginator (Coleoptera: Cerambycidae) in spatially realistic landscapes. Biol. Conserv. 2005, 124, 49–61. [Google Scholar] [CrossRef]
- Baur, B.; Burckhardt, D.; Coray, A.; Erhardt, A.; Heinertz, R.; Ritter, M.; Zemp, M. Der Erdbockkäfer, Dorcadion fuliginator (L., 1758) (Coleoptera: Cerambycidae), in Basel. Mitt. Entomol. Ges. Basel 1997, 47, 59–124. [Google Scholar]
- Baur, B. (University of Basel, Basel, Switzerland). Movements of Iberodorcadion fuliginator. 2011; Unpublished work. [Google Scholar]
- Rusterholz, H.P.; Ursenbacher, S.; Coray, A.; Weibel, U.; Baur, B. DNA quantity and quality in remnants of traffic-killed specimens of an endangered longhorn beetle: A comparison of different methods. J. Insect Sci. 2015, 15, 120. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT (Version 1.2): A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Chybicki, I.J.; Burczyk, J. Simultaneous estimation of null alleles and inbreeding coefficients. J. Hered. 2009, 100, 106–113. [Google Scholar] [CrossRef]
- Meirmans, P.G. GenoDive version 3.0: Easy-to-use software for the analysis of genetic data of diploids and polyploids. Mol. Ecol. Resour. 2020, 20, 1126–1131. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Mantel, N. Detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Rousset, F. Genepop’007: A complete reimplementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Takezaki, N.; Nei, M.; Tamura, K. POPTREE2: Software for constructing population trees from allele frequency data and computing other population statistics with Window interface. Mol. Biol. Evol. 2010, 27, 747–1624. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Goslee, S.C.; Urban, D.L. The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 2007, 22, 1–19. [Google Scholar] [CrossRef]
- Wright, S. Evolution in Mendelian populations. Genetics 1931, 16, 97–159. [Google Scholar] [CrossRef]
- Nei, M.; Maruyama, T.; Chakraborty, R. The bottleneck effect and genetic variability in populations. Evolution 1975, 29, 1–10. [Google Scholar]
- Allendorf, F.W. Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol. 1986, 5, 181–190. [Google Scholar] [CrossRef]
- Melosik, I.; Baraniak, E.; Przewozny, E.; Grzegorczyk, T.; Rzepka, M. Does gene flow balance the effect of habitat fragmentation in a population of the hermit beetle Osmoderma barnabita? Insect Conserv. Diver. 2020, 13, 360–373. [Google Scholar] [CrossRef]
- Robin, M.; Ferrari, G.; Akgül, G.; Münger, X.; von Seth, J.; Schuenemann, V.J.; Dalén, L.; Grossen, C. Ancient mitochondrial and modern whole genomes unravel massive genetic diversity loss during near extinction of Alpine ibex. Mol. Ecol. 2022, 31, 3548–3565. [Google Scholar] [CrossRef] [PubMed]
- Keller, I.; Largiadèr, C.R. Recent habitat fragmentation caused by major roads leads to reduction of gene flow and loss of genetic variability in ground beetles. Proc. R. Soc. Lond. B 2003, 270, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Keller, I.; Nentwig, W.; Largiadèr, C. Recent habitat fragmentation due to roads can lead to significant genetic differentiation in an abundant flightless ground beetle. Mol. Ecol. 2004, 13, 2983–2994. [Google Scholar] [CrossRef] [PubMed]
- Ursenbacher, S.; Alvarez, C.; Armbruster, G.F.J.; Baur, B. High population differentiation in the rock-dwelling land snail (Trochulus chaelatus) endemic to the Swiss Jura Mountains. Conserv. Genet. 2010, 11, 1265–1271. [Google Scholar] [CrossRef][Green Version]
- Hansen, A.K.; Justensen, M.J.; Olsen, M.T.; Solodovnikov, A. Genomic population structure and conservation of the red listed Carabus arcensis (Coleoptera: Carabidae) in island–mainland habitats of Northern Europe. Insect Conserv. Diver. 2018, 11, 255–266. [Google Scholar] [CrossRef]
- Brav-Cubitt, T.; Leschen, R.A.B.; Veale, A.J.; Buckley, T.R. Genetic diversity and differentiation in the leaf litter weevil Geochus politus across an urban-rural gradient. New Zealand J. Ecol. 2022, 46, 3459. [Google Scholar] [CrossRef]
- Holzmann, J.P.; Bohonak, A.J.; Kirkendall, L.R.; Gottlieb, D.; Harari, A.R.; Kelley, S.T. Inbreeding variability and population structure in the invasive haplodiploid palm-seed borer (Coccotrypes dactyliperda). J. Evol. Biol. 2009, 22, 1076–1087. [Google Scholar] [CrossRef]
- Keller, L.; Peer, K.; Bernasconi, C.; Taborsky, M.; Shuker, D.M. Inbreeding and selection on sex ratio in the bark beetle Xylosandrus germanus. BMC Evol. Biol. 2011, 11, 359. [Google Scholar] [CrossRef]
- Fisher, R.A. The Genetical Theory of Natural Selection; Oxford University Press: Oxford, UK, 1930; p. 272. [Google Scholar]
- Hanski, I. Metapopulation Ecology; Oxford University Press: Oxford, UK, 1999; p. 313. [Google Scholar]
- Frankham, R. Inbreeding and extinction: A threshold effect. Conserv. Biol. 1995, 9, 792–799. [Google Scholar] [CrossRef]
- Lozier, J.D.; Cameron, S.A. Comparative genetic analyses of historical and contemporary collections highlight contrasting demographic histories for the bumblebees Bombus pensylvanicus and B. impatiens in Illinois. Mol. Ecol. 2009, 18, 1875–1886. [Google Scholar] [CrossRef]
- Maebe, K.; Golsteyn, L.; Nunes-Silva, P.; Blochtein, B.; Smagghe, G. Temporal changes in genetic variability in three bumblebee species from Rio Grande do Sul, South Brazil. Apidologie 2018, 49, 415–429. [Google Scholar] [CrossRef]
- Life Science. Erdbock-Abklärungen (Schlussbericht); Unpublished Report; Life Science AG: Basel, Switzerland, 1996; p. 50. [Google Scholar]
- Buser, H.; Coray, A.; Reiss, T.; Schläpfer, M. Erdbock-Untersuchungen 1999. Unpublished Report; Basel, Switzerland, 1999; p. 14. [Google Scholar]
- Young, A.G.; Clarke, G.M. (Eds.) Genetics, Demography and Viability of Fragmented Populations; Cambridge University Press: Cambridge, UK, 2000; p. 438. [Google Scholar]
- Slatkin, M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution 1993, 47, 264–279. [Google Scholar] [CrossRef]
- Beebee, T.J.C. Population structure and its implications for conservation of the great silver beetle Hydrophilus piceus in Britain. Freshw. Biol. 2007, 52, 2101–2111. [Google Scholar] [CrossRef]
- Knutsen, H.; Rukke, B.A.; Jorde, P.E.; Ims, R.A. Genetic differentiation among populations of the beetle Bolitophagus reticulatus (Coleoptera: Tenebrionidae) in a fragmented and a continuous landscape. Heredity 2000, 84, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Desender, K.; Small, E.; Gaublomme, E.; Verdyck, P. Rural–urban gradients and the population genetic structure of woodland ground beetles. Conserv. Gen. 2005, 6, 51–62. [Google Scholar] [CrossRef]





| Population (Country) 1 | No. of Individuals Genotyped | State of Specimens 2 | Year(s) |
|---|---|---|---|
| 1 Basel, embankment of river Rhine (Switz) | 3 | c (1), f (1), i (1) | 2000 |
| 2 Allschwil (Switz) | 12 | c (4), f (2), i (6) | 1998–2017 |
| 4 St. Louis, E of airport (Fra) | 1 | i (1) | 1999 |
| 5 Blotzheim, E (Fra) | 17 | c (7), f (3), i (7) | 2012–2017 |
| 7 Blotzheim, NW of airport (Fra) | 1 | i (1) | 1998 |
| 9 Blotzheim, E (Fra) | 3 | c (2), f (1) | 2013 |
| 11 Blotzheim, Ruti SW (Fra) | 21 | c (12), f (5), i (4) | 1998–2017 |
| 12 Blotzheim, Rotfeld-Hattel (Fra) | 2 | c (2) | 2012 |
| 13 Sierentz, Hardt (Fra) | 4 | c (2), f (1), i (1) | 1998–1999 |
| 16 Istein, NW (Ger) | 63 | c (44), f (5), i (14) | 1998–2000 |
| 17 Istein, NE (Ger) | 10 | c (8), f (1), i (1) | 2000–2013 |
| 18 Huttingen, E (Ger) | 6 | c (4), f (1), i (1) | 2000 |
| 19 Huttingen, NE (Ger) | 12 | c (7), f (4), i (1) | 2000–2014 |
| 20 Huttingen, Tischlig (Ger) | 19 | c (8), f (5), i (6) | 1999–2017 |
| 21 Huttingen, Tischlig nature reserve (Ger) | 2 | i (2) | 1999–2000 |
| 22 Istein, N (Ger) | 2 | c (2) | 1999–2000 |
| 24 Efringen-Kirchen, N (Ger) | 1 | i (1) | 2001 |
| 25 Ötlingen, Tüllinger Berg (Ger) | 1 | i (1) | 1999 |
| 27 Istein, Isteiner Klotz (Ger) | 1 | c (1) | 2000 |
| 29 Thayngen, SH (Switz) | 14 | c (9), f (4), i (1) | 2010–2016 |
| 30 Altdorf, SH (Switz) | 34 | c (22), f (4), i (8) | 2005–2017 |
| 31 Taubergiessen (Ger) | 1 | c (1) | 2004 |
| 32 Kaiserstuhl (Ger) | 4 | c (3), f (1) | 1998 |
| 33 Westhalten (Fra) | 6 | c (1), f (2), i (3) | 1998 |
| 34 Bad Windsheim (Ger) | 3 | i (3) | 2012 |
| Population (Country) Long-Term Dynamics 1 | N | A | Ar | %P | I | PA | HO | HE | FIS |
|---|---|---|---|---|---|---|---|---|---|
| 2 Allschwil (Switz) s | 12 | 2.833 | 2.226 | 100.0 | 0.618 | 3 | 0.208 | 0.382 *** | 0.395 |
| 5 Blotzheim, E (Fra) d | 17 | 3.000 | 2.550 | 100.0 | 0.698 | 1 | 0.275 | 0.435 *** | 0.359 |
| 11 Blotzheim, Ruti SW (Fra) s | 21 | 2.667 | 2.386 | 100.0 | 0.752 | 0 | 0.340 | 0.507 *** | 0.231 |
| 16 Istein, NW (Ger) e | 63 | 2.667 | 1.995 | 100.0 | 0.497 | 2 | 0.241 | 0.310 *** | 0.144 |
| 17 Istein, NE (Ger) s | 10 | 2.000 | 2.000 | 100.0 | 0.523 | 0 | 0.360 | 0.368 | 0.021 |
| 19 Huttingen, NE (Ger) d | 12 | 1.833 | 1.832 | 83.3 | 0.481 | 0 | 0.194 | 0.345 *** | 0.336 |
| 20 Huttingen, Tischlig (Ger) d | 21 | 1.833 | 1.829 | 83.3 | 0.468 | 0 | 0.216 | 0.324 *** | 0.229 |
| 29 Thayngen, SH (Switz) s | 14 | 2.000 | 1.812 | 66.7 | 0.290 | 0 | 0.095 | 0.168 *** | 0.356 |
| 30 Altdorf, SH (Switz) s | 34 | 2.333 | 1.946 | 83.3 | 0.438 | 0 | 0.098 | 0.270 *** | 0.609 |
| Source of Variation | d.f. | Sum of Squares | Estimated Variance | Percentage of Variation | F | P |
|---|---|---|---|---|---|---|
| Between metapopulations | 1 | 39.25 | 0.241 | 17 | 0.165 | <0.001 |
| Among populations | 5 | 25.69 | 0.097 | 7 | 0.079 | <0.001 |
| Among individuals | 149 | 221.50 | 0.363 | 25 | 0.231 | <0.001 |
| Within individuals | 156 | 118.50 | 0.760 | 52 | 0.324 | <0.001 |
| Population | 2 | 5 | 11 | 16 | 17 | 19 | 20 |
|---|---|---|---|---|---|---|---|
| 2 | – | ** | *** | *** | *** | *** | *** |
| 5 | 0.104 | – | * | *** | *** | *** | *** |
| 11 | 0.171 | 0.035 | – | *** | ** | ** | *** |
| 16 | 0.391 | 0.235 | 0.200 | – | ** | ** | ** |
| 17 | 0.334 | 0.138 | 0.110 | 0.084 | – | ns | ns |
| 19 | 0.313 | 0.139 | 0.141 | 0.074 | 0.011 | – | ns |
| 20 | 0.404 | 0.205 | 0.137 | 0.060 | 0.020 | 0.052 | – |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rusterholz, H.-P.; Ursenbacher, S.; Weibel, U.; Coray, A.; Baur, B. Genetic Diversity and Population Structure Derived from Body Remains of the Endangered Flightless Longhorn Beetle Iberodorcadion fuliginator in Grassland Fragments in Central Europe. Diversity 2023, 15, 16. https://doi.org/10.3390/d15010016
Rusterholz H-P, Ursenbacher S, Weibel U, Coray A, Baur B. Genetic Diversity and Population Structure Derived from Body Remains of the Endangered Flightless Longhorn Beetle Iberodorcadion fuliginator in Grassland Fragments in Central Europe. Diversity. 2023; 15(1):16. https://doi.org/10.3390/d15010016
Chicago/Turabian StyleRusterholz, Hans-Peter, Sylvain Ursenbacher, Urs Weibel, Armin Coray, and Bruno Baur. 2023. "Genetic Diversity and Population Structure Derived from Body Remains of the Endangered Flightless Longhorn Beetle Iberodorcadion fuliginator in Grassland Fragments in Central Europe" Diversity 15, no. 1: 16. https://doi.org/10.3390/d15010016
APA StyleRusterholz, H.-P., Ursenbacher, S., Weibel, U., Coray, A., & Baur, B. (2023). Genetic Diversity and Population Structure Derived from Body Remains of the Endangered Flightless Longhorn Beetle Iberodorcadion fuliginator in Grassland Fragments in Central Europe. Diversity, 15(1), 16. https://doi.org/10.3390/d15010016





