The Mitochondrial Genome of the Globally Invasive Barnacle Megabalanus coccopoma Darwin 1854 (Crustacea: Balanomorpha): Rearrangement and Phylogenetic Consideration within Balanomorpha
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. PCR and Sequence Determination
2.3. Gene Identification
2.4. Genome Analysis and Phylogenetic Analysis
3. Results
3.1. General Characteristics
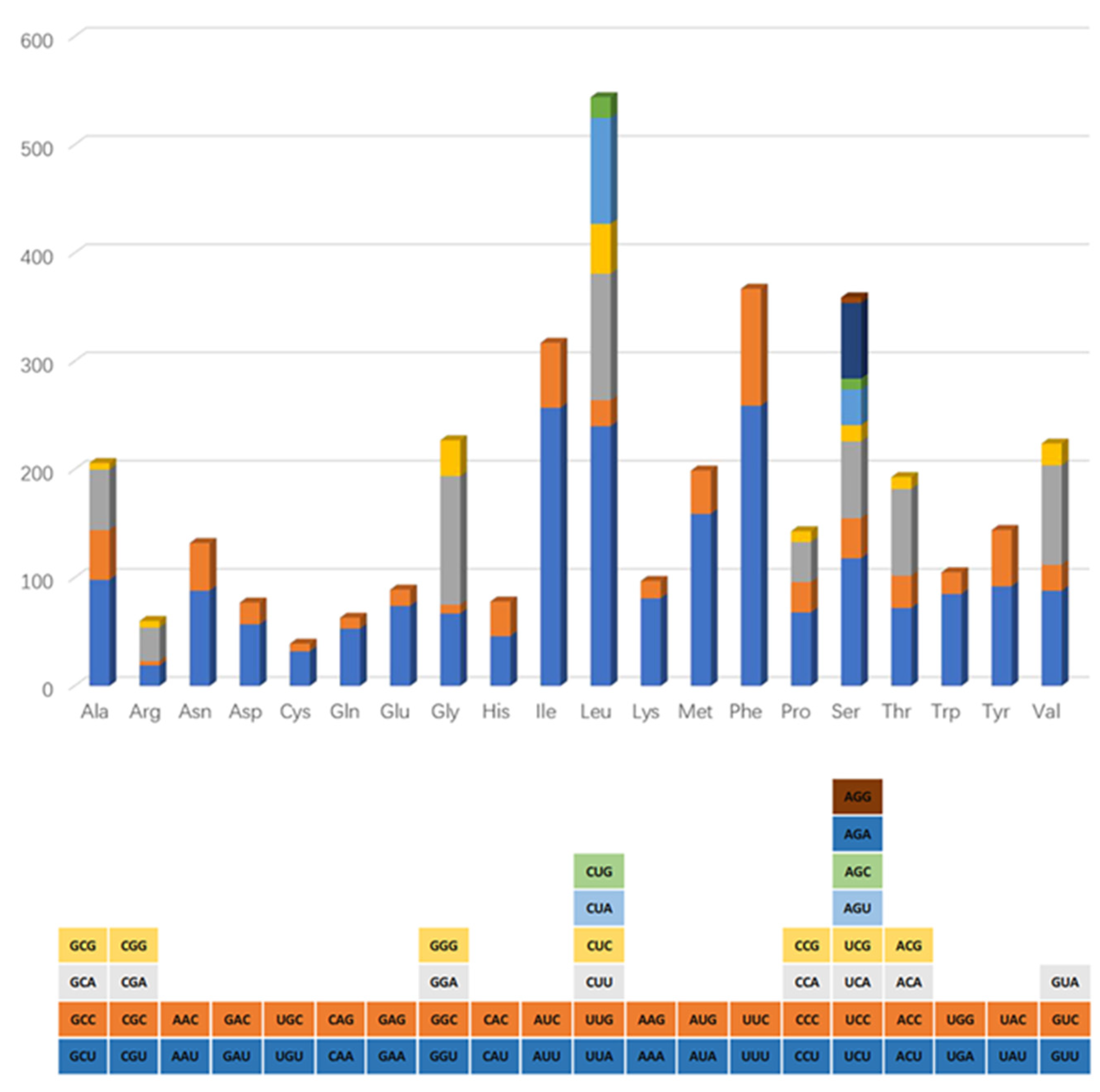
3.2. Codon Use and The Genetics of Protein Synthesis
3.3. Composition of The Base and Skew
3.4. Genes for Ribosomal and Transfer RNA
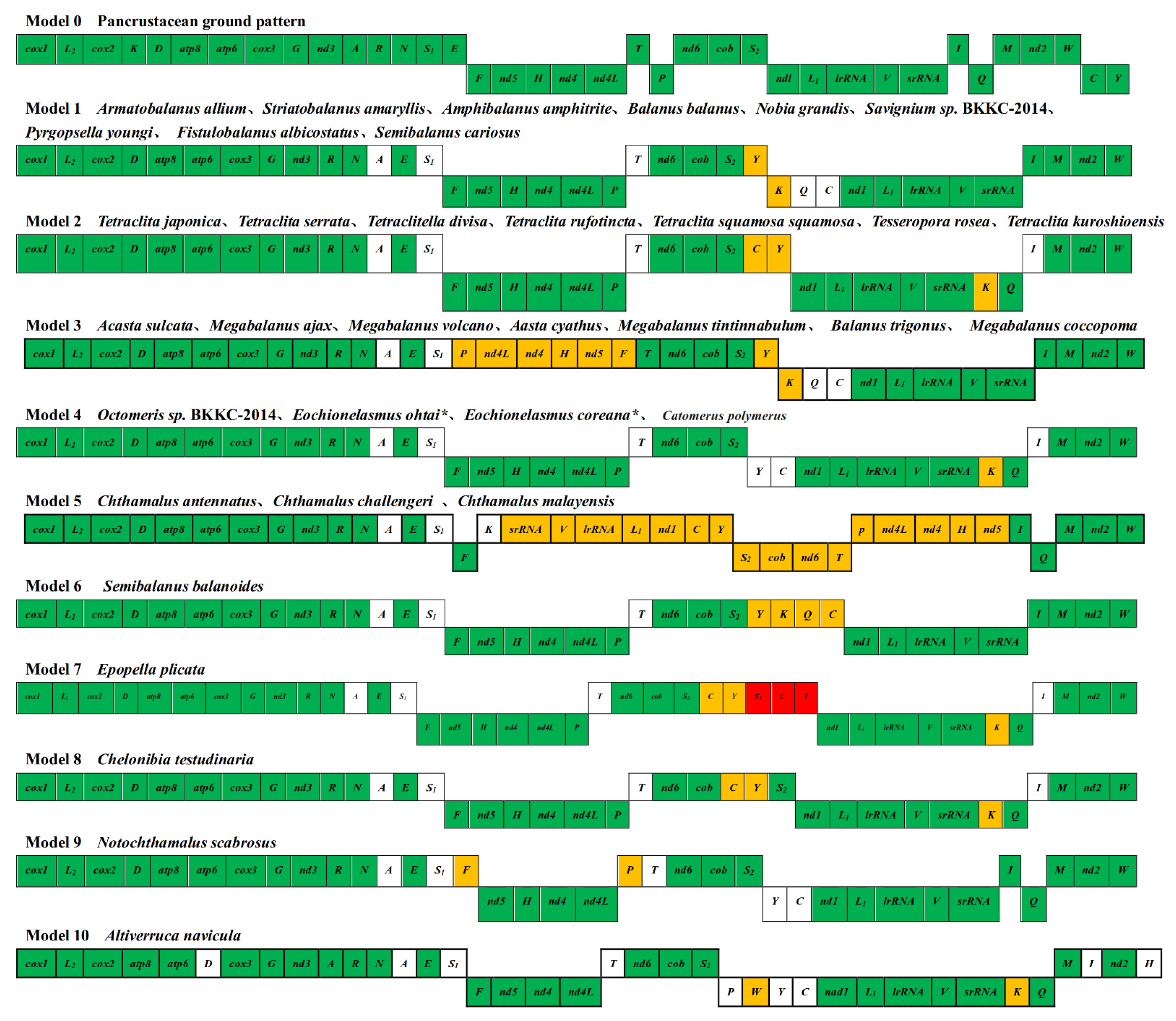
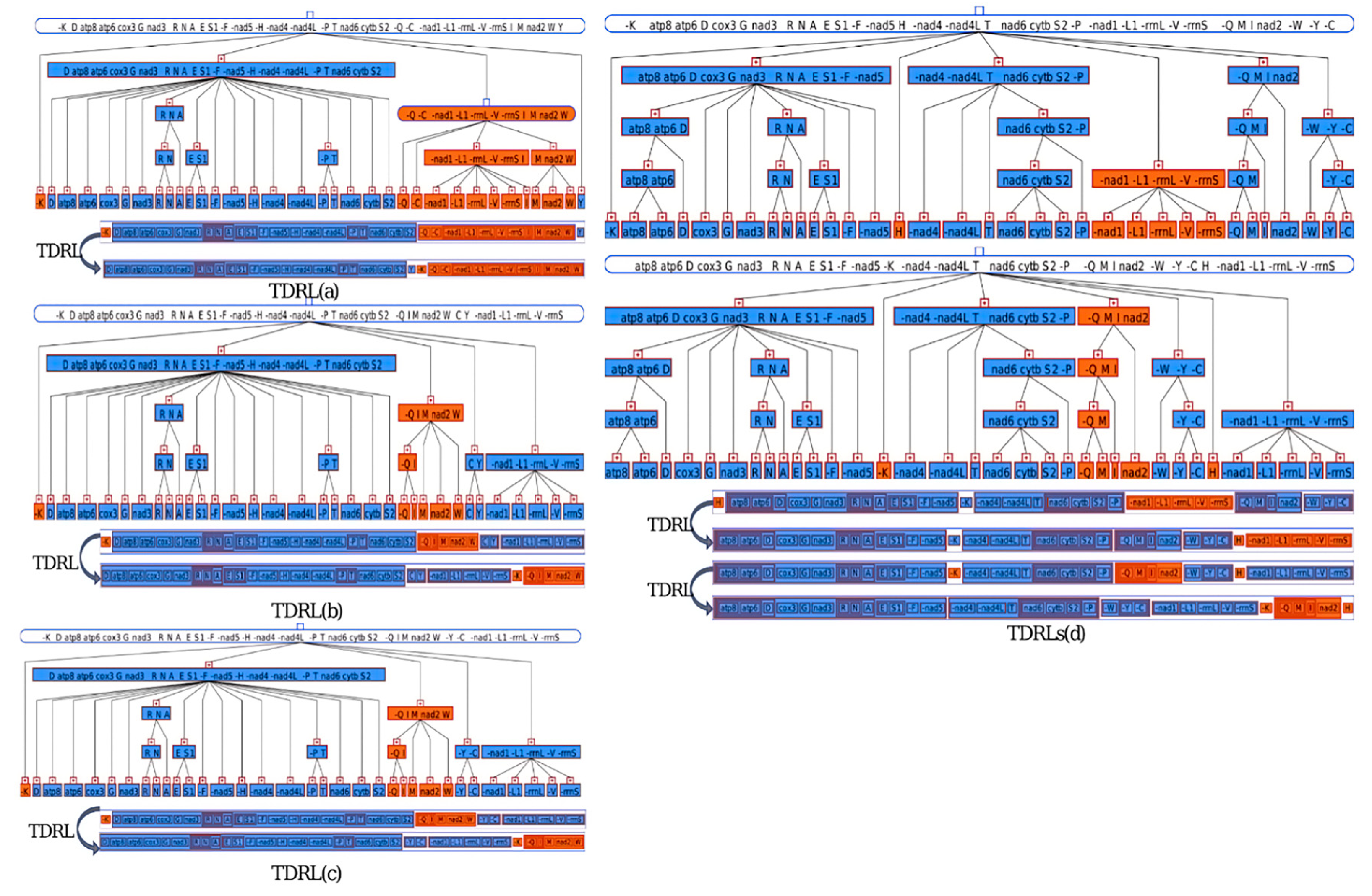
3.5. Gene Arrangement
3.6. Phylogeny Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chan, B.K.K.; Dreyer, N.; Gale, A.S.; Glenner, H.; Ewers-Saucedo, C.; Pérez-Losada, M.; Kolbasov, G.A.; Crandall, K.A.; Høeg, J.T. The evolutionary diversity of barnacles, with an updated classification of fossil and living forms. Zool. J. Linn. Soc. 2021, 193, 789–846. [Google Scholar] [CrossRef]
- Cao, W.; Yan, T.; Li, Z.; Li, J. Fouling acorn barnacles in China—A review. Chin. J. Oceanol. Limnol. 2013, 13, 699–711. [Google Scholar] [CrossRef]
- Tsang, L.M.; Achituv, Y.; Chu, K.H.; Chan, B.K.K. Zoogeography of intertidal communities in the West Indian Ocean as determined by ocean circulation systems: Patterns from the Tetraclita barnacles. PLoS ONE 2012, 7, e45120. [Google Scholar] [CrossRef] [PubMed]
- Abreu, N.M.N.; Marçal, I.; Duarte, A.B.; Pitombo, F.B.; Vilasboa, A.; Gusmão, J. Microsatellite markers for barnacle studies: Isolation and characterization of polymorphic microsatellite loci from the invasive barnacle Megabalanus coccopoma (Darwin, 1854) and its cross-amplification in the Southern Atlantic endemic species Megabalanus vesiculosus (Darwin, 1854). Biochem. Syst. Ecol. 2016, 66, 224–228. [Google Scholar] [CrossRef]
- Crickenberger, S. Predicting a range shift and range limits in an introduced tropical marine invertebrate using species distribution models. Hydrobiologia 2016, 763, 193–205. [Google Scholar] [CrossRef]
- Crickenberger, S.; Walther, K.; Moran, A.L. Lower thermal limits to larval development do not predict poleward range limits of the introduced tropical barnacle Megabalanus coccopoma. Invertebr. Biol. 2017, 136, 37–49. [Google Scholar] [CrossRef]
- Baek, Y.S.; Min, G.S.; Kim, S.; Choi, H.G. Complete mitochondrial genome of the Antarctic barnacle Lepas australis (Crustacea, Maxillopoda, Cirripedia). Mitochondrial DNA Part A 2016, 27, 1677–1678. [Google Scholar] [CrossRef]
- Moritz, C.; Dowling, T.E.; Brown, W.M. Evolution of animal mitochondrial dna: Relevance for population biology and systematics. Annu. Rev. Ecol. Syst. 1987, 18, 269–292. [Google Scholar] [CrossRef]
- Shen, X.; Tsang, L.M.; Chu, K.H.; Chan, B.K.K. A unique duplication of gene cluster (S2–C–Y) in (Crustacea) mitochondrial genome and phylogeny within Cirripedia. Mitochondrial DNA Part A 2015, 28, 285–287. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Ge, T.; Cai, Y.; Ji, N.; Shen, X. The mitochondrial genome of Chthamalus malayensis (Sessilia: Chthamalidae) and its molecular phylogeny within Cirripedia. Mitochondrial DNA Part B 2021, 6, 643–644. [Google Scholar] [CrossRef]
- Yang, Y.; Qi, L.; Lingfeng, K.; Hong, Y. The complete mitochondrial genome of the grand jackknife clam, Solen grandis (Bivalvia: Solenidae): A novel gene order and unusual non-coding region. Mol. Biol. Rep. 2012, 39, 1287–1292. [Google Scholar] [CrossRef]
- Xie, G.L.; Köhler, F.; Huang, X.C.; Wu, R.W.; Zhou, C.H.; Ouyang, S.; Wu, X.P. A novel gene arrangement among the Stylommatophora by the complete mitochondrial genome of the terrestrial slug Meghimatium bilineatum (Gastropoda, Arionoidea). Mol. Phylogenetics Evol. 2019, 135, 177–184. [Google Scholar] [CrossRef]
- Nunez, J.; Elyanow, R.G.; Ferranti, D.A.; Rand, D.M. Population Genomics and Biogeography of the Northern Acorn Barnacle (Semibalanus balanoides) Using Pooled Sequencing Approaches. In Population Genomics: Marine Organisms; Springer: Cham, Switzerland, 2018; pp. 139–168. [Google Scholar] [CrossRef]
- Lim, J.T.; Hwang, U.W. The complete mitochondrial genome of Pollicipes mitella (Crustacea, Maxillopoda, Cirripedia): Non-monophylies of maxillopoda and crustacea. Mol. Cells 2006, 22, 314–322. [Google Scholar] [CrossRef]
- Shen, X.; Kwok-Ho, T.; Chi-Chiu, C. The model barnacle Balanus balanus Linnaeus, 1758 (Crustacea: Maxillopoda: Sessilia) mitochondrial genome and gene rearrangements within the family Balanidae. Mitochondrial DNA Part A 2014, 27, 2112. [Google Scholar] [CrossRef]
- Shen, X.; Tsang, L.M.; Chu, K.H.; Achituv, Y. Mitochondrial genome of the intertidal acorn barnacle Tetraclita serrata Darwin, 1854 (Crustacea: Sessilia): Gene order comparison and phylogenetic consideration within Sessilia. Mar. Genom. 2015, 22, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Song, J.; Shen, X.; Cai, Y.; Chu, K.H.; Li, Y.; Tian, M. Mitochondrial genome of Chthamalus challengeri(Crustacea:Sessilia): Gene order comparison within Chthamalidae and phylogenetic consideration within Balanomorpha. Acta Oceanol. Sin. 2019, 38, 25–31. [Google Scholar] [CrossRef]
- Tsang, L.M.; Shen, X.; Chu, K.H.; Chan, B. Complete mitochondrial genome of the acorn barnacle Striatobalanus amaryllis (Crustacea: Maxillopoda): The first representative from Archaeobalanidae. Mitochondrial DNA Part A 2014, 26, 761–762. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Chen, P.; Tian, M.; Ji, N.; Shen, X. The first mitochondrial genome of Fistulobalanus albicostatus (Crustacea: Maxillopoda: Sessilia) and phylogenetic consideration within the superfamily Balanoidea. Mitochondrial DNA Part B 2020, 5, 2779–2781. [Google Scholar] [CrossRef]
- Lee, W.K.; Chan, B.; Ju, S.J.; Kim, D.; Kim, S.J. The mitochondrial genome of hydrothermal vent barnacle Eochionelasmus coreana (Cirripedia: Thoracica) from the Indian Ocean. Mitochondrial DNA Part B 2021, 6, 710–712. [Google Scholar] [CrossRef]
- Chan, B.K.K.; Aguilar, L.; Hou, B.K.; Kang, H.M.; Kim, S.J. Complete mitochondrial genome of the catophragmid barnacle Catomerus polymerus (Cirripedia, Thoracica, Balanomorpha, Catophragmidae). Mitochondrial DNA Part B 2018, 3, 1286–1287. [Google Scholar] [CrossRef]
- Nunez, J.; Rong, S.; Damian-Serrano, A.; Burley, J.T.; Rand, D.M. Ecological load and balancing selection in circumboreal barnacles. Cold Spring Harb. Lab. 2020, 38, 676–685. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Ren, X. Fauna Sinica Invertebrata Vol. 42 Crustacea Cirripedia Thoracica; Science Press: Beijing, China, 2007; ISBN 9787030188113. [Google Scholar]
- Drummond, A. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Swindell, S.R.; Plasterer, T.N. SEQMAN. Contig assembly. Methods Mol. Biol. 1997, 70, 75. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovli, I.; Zou, H.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Marc, L.; Oliver, D.; Sabine, K.; Ralph, B. OrganellarGenomeDRAW--a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef]
- Bernt, M.; Merkle, D.; Kai, R.; Fritzsch, G.; Perseke, M.; Bernhard, D.; Schlegel, M.; Stadler, P.F.; Middendorf, M. CREx: Inferring genomic rearrangements based on common intervals. Bioinformatics 2007, 23, 2957–2958. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.; Haeseler, A.V.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Yan, J.; Zhou, J.; Li, P.; Sun, H.; Zhou, K. Nearly complete mitochondrial genome of Polyascus gregaria and the phylogenetic relationships among maxillopodans. Mol. Biol. Rep. 2012, 39, 7413–7419. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Cao, W.; Wang, C.; Lin, S.; Zhou, Y. Complete mitochondrial genome of Tetraclita squamosa squamosa (Sessilia: Tetraclitidae) from China and phylogeny within Cirripedia. Mitochondrial DNA Part B 2020, 5, 2121–2123. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.O.; Chan, B.K.K.; Hou, B.K.; Ju, S.J.; Kim, S.J. Complete mitochondrial genome of the deep-water epibiotic stalked barnacle, Glyptelasma annandalei (Cirripedia, Lepadiformes, Poecilasmatidae). Mitochondrial DNA Part B 2018, 4, 99–100. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Saccone, C.; Giorgi, C.D.; Gissi, C.; Pesole, G.; Reyes, A. Evolutionary genomics in Metazoa: The mitochondrial DNA as a model system. Gene 1999, 238, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Tsang, L.M.; Shen, X.; Cheang, C.C.; Chu, K.H.; Chan, B.K.K. Gene rearrangement and sequence analysis of mitogenomes suggest polyphyly of Archaeobalanid and Balanid barnacles (Cirripedia: Balanomorpha). Zool. Scr. 2017, 46, 729–739. [Google Scholar] [CrossRef]
- Shen, X.; Chu, H.K.; Chan, B.K.K.; Tasng, L.M. The complete mitochondrial genome of the fire coral-inhabiting barnacle Megabalanus ajax (Sessilia: Balanidae): Gene rearrangements and atypical gene content. Mitochondrial DNA Part A 2014, 27, 1173–1174. [Google Scholar] [CrossRef]
- Feng, M.P.; Lin, S.Q.; Wang, C.S.; Dong, S.; Dong, Z.Y.; Xin, B.Y.; Da, X.K. The first mitochondrial genome of Megabalanus tintinnabulum (Sessilia: Balanidae) from China: Phylogeny within Cirripedia based on mitochondrial genes. Mitochondrial DNA Part B 2019, 4, 4016–4018. [Google Scholar] [CrossRef]
- Moritz, C.; Brown, W.M. Tandem duplications in animal mitochondrial DNAs: Variation in incidence and gene content among lizards. Proc. Natl. Acad. Sci. USA 1987, 84, 7183–7187. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Chan, B.K.K.; Tsang, L.M. The complete mitochondrial genome of common fouling barnacle Amphibalanus amphitrite (Darwin, 1854) (Sessilia: Balanidae) reveals gene rearrangements compared to pancrustacean ground pattern. Mitochondrial DNA Part A 2014, 26, 773–774. [Google Scholar] [CrossRef] [PubMed]
- Ji, N.; Ge, T.; Mao, S.; Zhang, M.; Shen, X. The first mitochondrial genome of Tetraclita kuroshioensis (Crustacea: Sessilia) from China: Insight into the phylogeny within Cirripedia. Mitochondrial DNA Part B 2021, 6, 988–989. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.F.; Shen, X.; Zhou, L.; Chu, K.H.; Chan, B.K.K. Mitochondrial genome of Tesseropora rosea: Molecular evidence for non-monophyly of the genus Tetraclita. Mitochondrial DNA Part B 2018, 3, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Shen, X.; Chu, K.H.; Chan, B.K.K. Mitochondrial genome of the acorn barnacle Tetraclita rufotincta Pilsbry, 1916: Highly conserved gene order in Tetraclitidae. Mitochondrial DNA Part B 2017, 2017, 936–937. [Google Scholar] [CrossRef]
- Wares, J.P. Mitochondrial evolution across lineages of the vampire barnacle Notochthamalus scabrosus. Mitochondrial DNA Part A 2015, 26, 7–10. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Høeg, J.T.; Simon-Blecher, N.; Achituv, Y.; Jones, D.; Crandall, K.A. Molecular phylogeny, systematics and morphological evolution of the acorn barnacles (Thoracica: Sessilia: Balanomorpha). Mol. Phylogenetics Evol. 2014, 81, 147–158. [Google Scholar] [CrossRef]
- Kim, S.J.; Lee, W.K.; Kim, R.O.; Ju, S.J. Complete mitochondrial genome of the hydrothermal vent barnacle Eochionelasmus ohtai (Cirripedia, Thoracica). Mitochondrial DNA Part B 2018, 3, 46–47. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Chen, P.P.; Song, J.; He, F.; Shen, X. The first mitochondrial genome of Capitulum mitella (Crustacea: Cirripedia) from China: Revealed the phylogenetic relationship within Thoracica. Mitochondrial DNA Part B 2020, 5, 2573–2575. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Brown, W.M.; Boore, J.L. Phylogenetic position of the Pentastomida and (pan)crustacean relationships. Proc. Biol. Sci. 2004, 271, 537–544. [Google Scholar] [CrossRef]
- Lee, W.K.; Kang, H.M.; Chan, B.K.K.; Ju, S.J.; Kim, S.J. Complete mitochondrial genome of the hydrothermal vent stalked barnacle Vulcanolepas fijiensis (Cirripedia, Scalpelliforms, Eolepadidae). Mitochondrial DNA Part B 2019, 4, 2725–2726. [Google Scholar] [CrossRef]
- Sha, Z.; Ren, X. A new species of the genus Arcoscalpellum (Cirripedia, Thoracica, Scalpellidae) from deep waters in the South China Sea. Chin. J. Oceanol. Limnol. 2015, 33, 732–734. [Google Scholar] [CrossRef]
- Kim, S.J.; Lee, W.K.; Hou, B.K.; Chan, B.K.K.; Ju, S.J. Complete mitochondrial genome of the deep-sea asymmetrical barnacle Altiverruca navicula (Cirripedia, Thoracica, Verrucumorpha). Mitochondrial Dna Part B 2017, 2, 934–935. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Lu, G.; Bork, P.; Hu, S.; Lercher, M.J. Energy efficiency trade-offs drive nucleotide usage in transcribed regions. Nat. Commun. 2016, 7, 11334. [Google Scholar] [CrossRef]
- Shen, X.; Chan, B.K.K.; Tasng, L.M. The mitochondrial genome of Nobia grandis Sowerby, 1839 (Cirripedia: Sessilia): The first report from the coral-inhabiting barnacles family Pyrgomatidae. Mitochondrial DNA Part A 2014, 27, 339–341. [Google Scholar] [CrossRef]
- Newman, W.A.; Ross, A. Revision of the balanomorph barnacles, including a catalog of the species. San Diego Soc. Nat. Hist. Mem. 1976, 9, 1–108. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Harp, M.; Høeg, J.T.; Achituv, Y.; Jones, D.; Watanabe, H.; Crandall, K.A. The tempo and mode of barnacle evolution. Mol. Phylogenetics Evol. 2008, 46, 328–346. [Google Scholar] [CrossRef]
- Chan, B.K.K.; Corbari, L.; Rodriguez, M.P.A.; Tsang, L.M. Molecular phylogeny of the lower acorn barnacle families (Bathylasmatidae, Chionelasmatidae, Pachylasmatidae and Waikalasmatidae) (Cirripedia: Balanomorpha) with evidence for revisions in family classification. Zool. J. Linn. Soc. 2017, 180, 542–555. [Google Scholar] [CrossRef]



| Primer Name | Sequence (5′–3′) |
|---|---|
| dgLCO | GGTCAACAAATCATAAAGAYATYGG |
| dgHCO | TAAACTTCAGGGTGACCAAARAAYCA |
| H3-F1 | ATGGCTCGTACCAAGCAGACVGC |
| H3-R1 | ATATCCTTRGGCATRATRGTGAC |
| Gene | Stand | Position | Nucleotide | Codons | Anti-Codon | Intergenic Sequence * | ||
|---|---|---|---|---|---|---|---|---|
| Start | Stop | Start | Stop | |||||
| cox1 | H | 10 | 1545 | 1,536 | CGA | TAA | 2 | |
| L2 | H | 1548 | 1615 | 68 | taa | 5 | ||
| cox2 | H | 1621 | 2304 | 684 | ATG | TAA | 0 | |
| D | H | 2305 | 2368 | 64 | gtc | 0 | ||
| atp8 | H | 2369 | 2527 | 159 | ATT | TAA | −7 | |
| atp6 | H | 2521 | 3186 | 666 | ATG | TAA | −1 | |
| cox3 | H | 3186 | 3972 | 787 | ATG | T- | 0 | |
| G | H | 3973 | 4036 | 64 | tcc | 0 | ||
| nad3 | H | 4037 | 4388 | 352 | ATT | T- | 0 | |
| R | H | 4389 | 4451 | 63 | tcg | 0 | ||
| N | H | 4452 | 4515 | 64 | gtt | 0 | ||
| A | H | 4516 | 4581 | 66 | tgc | 1 | ||
| E | H | 4583 | 4648 | 66 | ttc | 0 | ||
| S1 | H | 4649 | 4707 | 59 | gct | 75 | ||
| P | H | 4783 | 4846 | 64 | tgg | 0 | ||
| nad4L | H | 4847 | 5140 | 294 | GTG | TAA | −7 | |
| nad4 | H | 5134 | 6463 | 1330 | ATG | T- | 0 | |
| H | H | 6464 | 6528 | 65 | gtg | 0 | ||
| nad5 | H | 6529 | 8230 | 1702 | ATT | T- | 0 | |
| F | H | 8231 | 8294 | 64 | gaa | 5 | ||
| T | H | 8300 | 8366 | 67 | tgt | 0 | ||
| nad6 | H | 8367 | 8855 | 489 | ATG | TAA | −1 | |
| cob | H | 8855 | 9994 | 1140 | ATG | TA- | −2 | |
| S2 | H | 9993 | 10,062 | 70 | tga | 24 | ||
| Y | H | 10,087 | 10,150 | 64 | gta | 24 | ||
| K | L | 10,175 | 10,240 | 66 | ttt | 9 | ||
| Q | L | 10,250 | 10,317 | 68 | ttg | 1 | ||
| C | L | 10,319 | 10,380 | 62 | gca | −2 | ||
| nad1 | L | 10,379 | 11,305 | 927 | ATA | TAA | −3 | |
| L1 | L | 11,303 | 11,370 | 68 | tag | 0 | ||
| rrnL | L | 11,371 | 12,689 | 1319 | 0 | |||
| V | L | 12,690 | 12,737 | 48 | tac | 1 | ||
| rrnS | L | 12,739 | 13,486 | 748 | 415 | |||
| I | H | 13,902 | 13,969 | 68 | gat | 0 | ||
| M | H | 13,970 | 14,035 | 66 | cat | 0 | ||
| nad2 | H | 14,036 | 15,034 | 999 | ATG | TAA | −2 | |
| W | H | 15,033 | 15,098 | 66 | tca | 9 | ||
| Gene | Nucleotide Proportion | A + T (%) | AT Skew | GC Skew | |||
|---|---|---|---|---|---|---|---|
| A (%) | C (%) | G (%) | T (%) | ||||
| atp6 | 30.3 | 18.6 | 11.0 | 40.1 | 70.4 | −0.139 | −0.257 |
| atp8 | 34.6 | 20.1 | 8.2 | 37.1 | 71.7 | −0.035 | −0.420 |
| cob | 27.0 | 20.8 | 14.5 | 37.7 | 64.7 | −0.165 | −0.178 |
| cox1 | 27.5 | 18.6 | 16.9 | 36.9 | 64.5 | −0.146 | −0.048 |
| cox2 | 30.6 | 19.7 | 14.0 | 35.7 | 66.2 | −0.077 | −0.169 |
| cox3 | 26.3 | 20.8 | 15.8 | 37.1 | 63.4 | −0.170 | −0.137 |
| nd1 | 25.2 | 11.0 | 17.8 | 46.0 | 71.2 | −0.292 | 0.236 |
| nd2 | 28.2 | 17.8 | 13.7 | 40.2 | 68.5 | −0.175 | −0.130 |
| nd3 | 28.7 | 18.2 | 12.5 | 40.6 | 69.3 | −0.172 | −0.186 |
| nd4 | 27.4 | 15.6 | 15.3 | 41.6 | 69.0 | −0.206 | −0.010 |
| nd4L | 29.4 | 14.3 | 15.5 | 40.8 | 70.2 | −0.162 | 0.040 |
| nd5 | 28.6 | 16.9 | 15.7 | 38.8 | 67.5 | −0.151 | −0.037 |
| nd6 | 31.7 | 22.1 | 7.8 | 38.5 | 70.1 | −0.097 | −0.478 |
| srRNA | 34.4 | 12.8 | 21.5 | 31.3 | 65.6 | 0.047 | 0.254 |
| lrRNA | 36.4 | 10.2 | 16.9 | 36.6 | 73.0 | −0.003 | 0.247 |
| All PCGs | 28.2 | 17.8 | 14.8 | 39.3 | 67.5 | −0.164 | −0.092 |
| All genes | 31.3 | 18.1 | 13.8 | 36.9 | 68.2 | −0.082 | −0.136 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Cai, Y.; Ji, N.; Chan, B.K.K.; Shen, X. The Mitochondrial Genome of the Globally Invasive Barnacle Megabalanus coccopoma Darwin 1854 (Crustacea: Balanomorpha): Rearrangement and Phylogenetic Consideration within Balanomorpha. Diversity 2023, 15, 117. https://doi.org/10.3390/d15010117
Zhang M, Cai Y, Ji N, Chan BKK, Shen X. The Mitochondrial Genome of the Globally Invasive Barnacle Megabalanus coccopoma Darwin 1854 (Crustacea: Balanomorpha): Rearrangement and Phylogenetic Consideration within Balanomorpha. Diversity. 2023; 15(1):117. https://doi.org/10.3390/d15010117
Chicago/Turabian StyleZhang, Mengjuan, Yuefeng Cai, Nanjing Ji, Benny Kwok Kan Chan, and Xin Shen. 2023. "The Mitochondrial Genome of the Globally Invasive Barnacle Megabalanus coccopoma Darwin 1854 (Crustacea: Balanomorpha): Rearrangement and Phylogenetic Consideration within Balanomorpha" Diversity 15, no. 1: 117. https://doi.org/10.3390/d15010117
APA StyleZhang, M., Cai, Y., Ji, N., Chan, B. K. K., & Shen, X. (2023). The Mitochondrial Genome of the Globally Invasive Barnacle Megabalanus coccopoma Darwin 1854 (Crustacea: Balanomorpha): Rearrangement and Phylogenetic Consideration within Balanomorpha. Diversity, 15(1), 117. https://doi.org/10.3390/d15010117







