Bacteria Release from Microplastics into New Aquatic Environments
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Processing of Microplastics
2.2. Determination of Bacterial Abundance on Microplastics
2.3. Microcosm Experiment
2.4. Bacterial Diversity on the Plastic Bag (BA) Microcosms
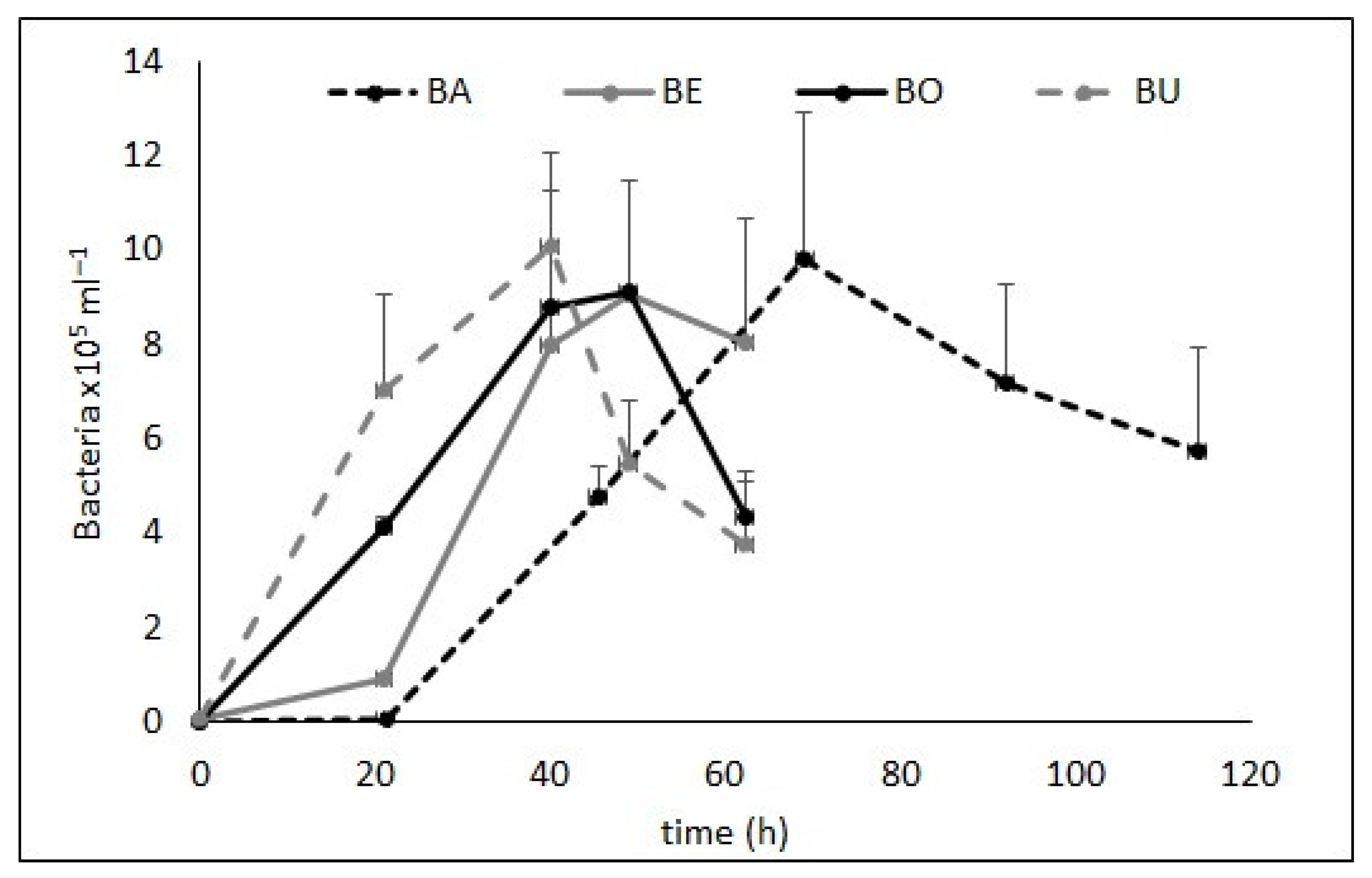
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Barnes, D.; Galgani, F.; Thompson, R.; Barlaz, M. Accumulation and fragmentation of plastic debris in global environments. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.G.; Moore, C.J.; Van Franeker, J.A.; Moloney, C.L. Monitoring the abundance of plastic debris in the marine environment. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Li, Υ.; Zhang, H.; Tang, C. A review of possible pathways of marine microplastics transport in the ocean. Anthr. Coasts 2019, 3, 6–13. [Google Scholar] [CrossRef]
- Browne, M.A.; Galloway, T.; Thompson, R. Microplastic—An Emerging Contaminant of Potential Concern? Integr. Environ. Assess. Manag. 2007, 3, 559–561. [Google Scholar] [CrossRef]
- Farrell, P.; Nelson, K. Trophic level transfer of microplastic: Mytilus edulis (L.) to Carcinus maenas (L.). Environ. Pollut. 2013, 177, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Horton, A.; Walton, A.; Spurgeon, D.J.; Lahive, E.; Svendsen, C. Microplastics in freshwater and terrestrial environments: Evaluating the current understanding to identify the knowledge gaps and future research priorities. Sci. Total Environ. 2017, 586, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Zettler, E.; Mincer, T.; Amaral-Zettler, L. Life in the “Plastisphere”: Microbial Communities on Plastic Marine Debris. Environ. Sci. Technol. 2013, 47, 7137–7146. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; Zettler, E.R.; Slikas, B.; Boyd, G.D. The biogeography of the plastisphere: Implications for policy. Front Ecol. Environ. 2015, 13, 541–546. [Google Scholar] [CrossRef]
- Carson, H.S. The incidence of plastic ingestion by fishes: From the prey’s perspective. Mar. Pollut. Bull. 2013, 74, 170–174. [Google Scholar] [CrossRef]
- Da Costa, J.P.; Santos, P.S.M.; Duarte, A.C.; Rocha-Santos, T. (Nano)plastics in the environment—Sources, fates and effects. Sci. Total Environ. 2016, 566–567, 15–26. [Google Scholar] [CrossRef]
- Gregory, M.R. Environmental implications of plastic debris in marine settings—Entanglement, ingestion, smothering, hangers-on, hitch-hiking and alien invasions. Philos. Trans. R. Soc. Lond. B, Biol. Sci. 2009, 364, 2013–2025. [Google Scholar] [CrossRef]
- Caruso, G. Microplastics in Marine Environments: Possible Interactions with the Microbial Assemblage. J. Pollut. Eff. Cont. 2015, 3, e111. [Google Scholar] [CrossRef]
- Viršek, M.K.; Lovšinb, M.N.; Korena, S.; Kržanc, A.; Peterlina, M. Microplastics as a vector for the transport of the bacterial fish pathogen species Aeromonas salmonicida. Mar. Pollut. Bull. 2017, 125, 301–309. [Google Scholar] [CrossRef]
- Maso, M.; Garces, E.; Pages, F.; Camp, J. Drifting plastic debris as a potential vector for dispersing Harmful Algal Bloom (HAB) species. Sci. Mar. 2003, 67, 107–111. [Google Scholar] [CrossRef]
- Yokota, K.; Waterfield, H.; Hastings, C.; Davidson, E.; Kwietniewski, E.; Wells, B. Finding the missing piece of the aquatic plastic pollution puzzle: Interaction between primary producers and microplastics. Limnol. Oceanogr. Lett. 2017, 2, 91–104. [Google Scholar] [CrossRef]
- Odobel, C.; Dussud, C.; Philip, L.; Derippe, G.; Lauters, M.; Eyheraguibel, B.; Burgaud, G.; Ter Halle, A.; Meistertzheim, A.-L.; Bruzaud, S.; et al. Bacterial Abundance, Diversity and Activity During Long-Term Colonization of Non-biodegradable and Biodegradable Plastics in Seawater. Front. Microbiol. 2021, 12, 734782. [Google Scholar] [CrossRef]
- Benavente, J.M.; Arévalo Caballero, M.J.; Silvero, G.; López-Coca, I.; Gómez Escobar, V. Cellulose Acetate Recovery from Cigarette Butts. Multidiscip. Digit. Publ. Inst. Proc. 2018, 2, 1447. [Google Scholar] [CrossRef]
- Masura, J.; Baker, J.; Foster, G.; Arthur, C.; Herring, C.; Technical Editor. Laboratory Methods for the Analysis of Microplastics in the Marine Environment: Recommendations for Quantifying Synthetic Particles in Waters and Sediments; NOAA Technical Memorandum NOS-OR&R-48; 2015. [Google Scholar]
- Andresen, M.; Kristensen, E. The importance of bacteria and microalgae in the diet of the deposit-feeding polychaete Arenicola marina. Ophelia 2002, 56, 179–196. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Kreikemeyer, B.; Labrenz, M. Environmental Factors Support the Formation of Specific Bacterial Assemblages on Microplastics. Front. Microbiol. 2018, 8, 2709. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Fu, J.; Zhang, Y.; Lin, S.; Zhang, W.; Shu, G.; Lin, J.; Li, H.; Xu, F.; Tang, H.; Peng, G.; et al. Strategies for Interfering With Bacterial Early Stage Biofilms. Front. Microbiol. 2021, 12, 675843. [Google Scholar] [CrossRef] [PubMed]
- Crespo, B.G.; Pommier, T.; Fernández-Gómez, B.; Pedrós-Alió, C. Taxonomic composition of the particle-attached and free-living bacterial assemblages in the Northwest Mediterranean Sea analyzed by pyrosequencing of the 16S rRNA. MicrobiologyOpen 2013, 2, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Xie, G.; Jiang, X.; Shao, K.; Tang, X.; Gao, G. The Relationships Between the Free-Living and Particle-Attached Bacterial Communities in Response to Elevated Eutrophication. Front. Microbiol. 2020, 11, 423. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, E.A.; Fonvielle, J.A.; Cottingham, S.; Zhang, Y.; Dittmar, T.; Aldridge, D.C.; Tanentzap, A.J. Plastic pollution fosters more microbial growth in lakes than natural organic matter. Nat. Commun. 2022, 13, 4175. [Google Scholar] [CrossRef] [PubMed]
- Fei, C.; Ochsenkühn, M.A.; Shibl, A.A.; Isaac, A.; Wang, C.; Amin, S.A. Quorum sensing regulates ‘swim-or-stick’ lifestyle in the phycosphere. Environ. Microbiol. 2020, 22, 4761–4778. [Google Scholar] [CrossRef]
- Villalba, L.A.; Kasada, M.; Zoccarato, L.; Wollrab, S.; Grossart, H.P. Differing Escape Responses of the Marine Bacterium Marinobacter adhaerens in the Presence of Planktonic vs. Surface-Associated Protist Grazers. Int. J. Mol. Sci. 2022, 23, 10082. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; Zettler, E.R.; Mincer, T.J. Ecology of the plastisphere. Nat. Rev. Microbiol. 2020, 18, 139–151. [Google Scholar] [CrossRef]
- Mhete, M.; Eze, N.P.; Rahube, O.T.; Akinyemia, F.O. Soil properties influence bacterial abundance and diversity under different land-use regimes in semi-arid environments. Sci. Afr. 2020, 7, e00246. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, J.-M.; Zhou, Y.-L.; Almeida, A.; Finn, R.D.; Danchin, A.; He, L.-S. Phylogenomics of expanding uncultured environmental Tenericutes provides insights into their pathogenicity and evolutionary relationship with Bacilli. BMC Genom. 2020, 21, 408. [Google Scholar] [CrossRef]
- Sun, R.; Zhang, P.; Riggins, C.W.; Zabaloy, C.M.; Rodríguez-Zas, S.; Villamil, M.B. Long-Term N Fertilization Decreased Diversity and Altered the Composition of Soil Bacterial and Archaeal Communities. Agronomy 2019, 9, 574. [Google Scholar] [CrossRef]
- Rahman, A.N.; Parks, D.H.; Vanwonterghem, I.; Morrison, M.; Tyson, G.W.; Hugenholtz, P. A Phylogenomic Analysis of the Bacterial Phylum Fibrobacteres. Front. Microbiol. 2016, 6, 1469. [Google Scholar] [CrossRef]
- Dussud, C.; Hudec, C.; George, M.; Fabre, P.; Higgs, P.; Bruzaud, S. Colonization of Non-biodegradable and Biodegradable Plastics by Marine Microorganisms. Front. Microbiol. 2018, 9, 1571. [Google Scholar] [CrossRef] [PubMed]
- Krizova, L.; Maixnerova, M.; Sedo, O.; Nemec, A. Acinetobacter bohemicus sp. nov. widespread in natural soil and water ecosystems in the Czech Republic. Syst. Appl. Microbiol. 2014, 37, 467–473. [Google Scholar] [CrossRef]
- Almasaudi, S.B. Acinetobacter spp. as nosocomial pathogens: Epidemiology and resistance features. Saudi J. Biol. Sci. 2018, 25, 586–596. [Google Scholar] [CrossRef]
- Guan, Y.; Jia, J.; Wu, L.; Xue, X.; Zhang, G.; Wang, Z. Analysis of bacterial community characteristics, abundance of antibiotics and antibiotic resistance genes along a pollution gradient of Ba River in xi’an, China. Front. Microbiol. 2018, 9, 3191. [Google Scholar] [CrossRef]
- Kelly, J.J.; London, M.G.; McCormick, A.R.; Rojas, M.; Scott, J.W.; Hoellein, T.J. Wastewater treatment alters microbial colonization of microplastics. PLoS ONE 2021, 16, e0244443. [Google Scholar] [CrossRef]
- Borde, X.; Guieysse, B.; Delgado, O.; Muñoz, R.; Hatti-Kaul, R.; Nugier-Chauvin, C.; Patin, H.; Mattiasson, B. Synergistic relationships in algal–bacterial microcosms for the treatment of aromatic pollutants. Bioresour. Technol. 2003, 86, 293–300. [Google Scholar] [CrossRef]
- Kirchman, D.L. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 2002, 39, 91–100. [Google Scholar] [CrossRef]
- Hoke, A.K.; Reynoso, G.; Smith, M.R.; Gardner, M.I.; Lockwood, D.J.; Gilbert, N.E. Genomic signatures of Lake Erie bacteria suggest interaction in the Microcystis phycosphere. PLoS ONE 2021, 16, e0257017. [Google Scholar] [CrossRef]
- Krishnan, A.; Zhang, Y.Q.; Mou, X. Isolation and Characterization of Microcystin-Degrading Bacteria from Lake Erie. Bull. Env. Contam. Toxicol. 2018, 101, 617–623. [Google Scholar] [CrossRef]
- Shigematsu, T.; Yumihara, K.; Ueda, Y.; Numaguchi, M.; Morimura, S.; Kida, K. Delftia tsuruhatensis sp. nov. a terephthalate assimilating bacterium isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 2003, 53, 1479–1483. [Google Scholar] [CrossRef] [PubMed]
- Juarez Jimenez, B.; Reboleiro Rivas, P.; Gonzalez Lopez, J.; Pesciaroli, C.; Barghini, P.; Fenice, M. Immobilization of Delftia tsuruhatensis in macro-porous cellulose and biodegradation of phenolic compounds in repeated batch process. J. Biotechnol. 2012, 157, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Ranc, A.; Dubourg, G.; Fournier, P.E.; Raoult, D.; Fenollar, F. Delftia tsuruhatensis, an Emergent Opportunistic Healthcare-Associated Pathogen. Emerg. Infect. Dis. 2018, 24, 594–596. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Khurana, H.; Singh, D.N.; Negi, R.K. The genus Sphingopyxis: Systematics, ecology, and bioremediation potential—A review. J. Environ. Manag. 2021, 280, 111744. [Google Scholar] [CrossRef]
- Godoy, F.; Vancanneyt, M.; Martínez, M.; Steinbüchel, A.; Swings, J.; Rehm, B.H.M. Sphingopyxis chilensis sp. nov. a chlorophenol-degrading bacterium that accumulates polyhydroxyalkanoate, and transfer of Sphingomonas alaskensis to Sphingopyxis alaskensis comb. nov. Int. J. Syst. Evol. 2003, 53, 473–477. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, T. Brevundimonas spp: Emerging global opportunistic pathogens. Virulence 2018, 9, 480–493. [Google Scholar] [CrossRef]

| Group | OTU | Closest Relatives | Functions |
|---|---|---|---|
| Cytophaga–Flavobacteria cluster | OTU12 | Arcicella sp. | Degradation of high-molecular-weight organic matter (e.g., cellulose, chitin, and pectin) [39]. |
| OTU563 | Flavobacterium succinicans | ||
| OTU7338 | Flavobacterium resistens | ||
| OTU19 | uncultured Flavobacterium | ||
| OTU53 | Emticicia sp. | ||
| OTU463 | Chryseobacterium indoltheticum | ||
| OTU88 | Flavobacterium aquatile | ||
| Alphaproteobacteria | OTU50 | Brevundimonas sp. | Degradation of aromatic compounds and environmental pollutants (e.g., chlorophenol) [45]. Emerging as global opportunistic pathogens [46]. |
| OTU484 | Sphingopyxis chilensis | ||
| Betaproteobacteria | OTU16 | Acidovorax sp. | Degradation of xenobiotics and environmental pollutants (e.g., phenol), terephthalate assimilation [42,43], close associations with bloom-forming cyanobacteria (e.g., Microcystis) and degradation of algal-derived metabolites (e.g., microcystins and glycolate) [38]. Emergent opportunistic healthcare-associated pathogens with antibiotic resistance [44]. |
| OTU7238 | Acidovorax sp. | ||
| OTU43 | Delftia tsuruhatensis | ||
| OTU120 | Comamonas jiangduensis | ||
| OTU1516 | uncultured Acidovorax | ||
| Gammaproteobacteria | OTU1 | Acinetobacter johnsonii | Degradation of phenanthrene and other aromatic compounds and xenobiotics. Opportunistic pathogens with synergistic relationships with algae and pathogens with antibiotic resistance [35,38]. |
| OTU9 | Acinetobacter piperi | ||
| OTU26 | Pseudomonas migulae |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikolopoulou, I.; Piperagkas, O.; Moschos, S.; Karayanni, H. Bacteria Release from Microplastics into New Aquatic Environments. Diversity 2023, 15, 115. https://doi.org/10.3390/d15010115
Nikolopoulou I, Piperagkas O, Moschos S, Karayanni H. Bacteria Release from Microplastics into New Aquatic Environments. Diversity. 2023; 15(1):115. https://doi.org/10.3390/d15010115
Chicago/Turabian StyleNikolopoulou, Ioanna, Odysseas Piperagkas, Stefanos Moschos, and Hera Karayanni. 2023. "Bacteria Release from Microplastics into New Aquatic Environments" Diversity 15, no. 1: 115. https://doi.org/10.3390/d15010115
APA StyleNikolopoulou, I., Piperagkas, O., Moschos, S., & Karayanni, H. (2023). Bacteria Release from Microplastics into New Aquatic Environments. Diversity, 15(1), 115. https://doi.org/10.3390/d15010115








