Abstract
The genera Monostroma and Gayralia belong to the order of monostromatic green algae; however, their taxonomic delimitation remains controversial at the genus level. This study attempts to address this issue through the combined analysis of the morphology and nuclear-encoded Internal Transcribed Spacer region sequences of monostromatic green algal samples collected in the South China Sea. Our phylogenetic data revealed that the monostromatic specimens were separated into the M. nitidum clade, G. brasiliensis clade, and a single Monostroma sp. clade, and that the inter-genera genetic distance between the Monostroma and Gayralia genera was lower than that observed within the Monostroma genus. All the specimens presented similar morphology in their single cell-layered thallus, with irregularly arranged cells, rounded cell corners, a parietal chloroplast, and predominantly one (>90%) pyrenoid. Their most obvious morphological difference was in thallus thickness and size. Moreover, the monostromatic specimens of the M. nitidum clade corresponded to the morphological description of the M. nitidum-type specimens. The genus Monostroma was erected earlier than the genus Gayralia. Therefore, we propose to assign the genus Gayralia to Monostroma based on the morphological and phylogenetic analysis and genetic distance data presented here.
1. Introduction
Monostromatic green algae with fronds consisting only of horizontally arranged single-cell layers are widely distributed from temperate to tropical seas worldwide [1]. Monostroma Thuret 1854, Gayralia Vinogradova 1969, and Protomonostroma Vinogradova 1969 are among the main monostromatic green algal genera [2,3,4,5]. Monostroma is cosmopolitan and comprises 55 species, of which only 32 are currently taxonomically confirmed [6], while for the Gayralia and Protomonostroma genera, only two species per genus have been confirmed, namely G. brasiliensis and G. oxysperma for the former, and P. undulatum and P. rosulatum for the latter [6]. A number of monostromatic green algal species are attracting global attention due to their economic importance, mainly in the food and cosmetic industries [7,8,9,10]. In addition, chemicals with antiviral and anticoagulant properties were recently isolated from some species of the Monostroma and Gayralia genera [11,12].
Monostroma and Gayralia are the focus of multidisciplinary research involving different fields, such as taxonomy, biology, phylogeny, and biogeography [4,5,10,13,14]. As such, correct species identification and classification are essential to enable further research. Currently, the taxonomy of the Monostroma and Gayralia genera is complex, as there are several inconsistencies among the different classification tools, and experts disagree on the biological features considered relevant for the separation of taxa. Phenotypic variation in monostromatic green algae is well documented. Monostroma, which is characterized by a blade-shaped thallus consisting of one layer of cells, was erected by Thuret in 1854 [15], and it was later lectotypified with M. oxyspermum [16]. Species belonging to this genus are classically defined based on morphological characteristics, such as the size and shape of the cells and the thickness of thalli [17]. Culture studies have been conducted in at least some of the taxa, resulting in taxonomical revisions. For instance, Kornmann (1964) and Bliding (1968) proposed to remove the asexual M. undulatum and M. oxyspermum from the Monostroma genus [18,19]. Subsequently, the Gayraliaceae family—comprising two monotypic genera, Protomonostroma and Gayralia—was erected by Vinogradova (1969) to accommodate the asexual members, respectively [3]. Ulvaria oxysperma and M. oxyspermum have been synonymized with G. oxysperma based on thallus ontogeny and flagellar ultrastructural features [20]. A preliminary molecular analysis of Internal Transcribed Spacer (ITS) sequences revealed that M. nitidum and M. latissimum, the typical species of Monostroma, should be transferred to Gayralia [5]. In contrast, Bast (2011) concluded that Gayralia should be abolished and its species should be transferred back to Monostroma, because no obvious taxonomic distinction was detected between the two genera [4]. Furthermore, the Gayraliaceae family is posterior to the Monostromataceae, Kunieda ex Suneson (1947) and, based on the priority principle, the retention of the latter with the original Monostroma genus would be appropriate [3,4,16]. Thus, considering these contradictions and ambiguities, the current taxonomic status of these two genera warrants clarification.
Morphological differences across specimen types were confirmed as being reliable in taxonomically classifying macroalgae [10,21,22]. To address the taxonomic issues mentioned above, a morphological analysis of the specimen type and targeted monostromatic samples collected from the South China Sea, was conducted. The identification of monostromatic algae based solely upon morphological features is extremely challenging, and studies based on life cycle completion—although they often aid in the identification—are time consuming and difficult [3,18,19]. Molecular phylogenetic analyses compensate for the shortage of morphological studies, and they provide an accurate identification of ITS sequences, which are now available for a large group of marine green algae and have a high degree of variance, even between very closely related algal taxa [23]. Moreover, morphological characteristics, combined with sequence analysis, have also been used to resolve identification issues at the species level [10,21,22,24,25,26,27,28,29,30]. Therefore, in the present study, nuclear-encoded rDNA ITS sequences were obtained from monostromatic green algae collected in the South China Sea and from previously identified Monostroma and Gayralia species to characterize sequence divergence between these two genera.
2. Materials and Methods
2.1. Monostromatic Green Algal Collection
Attached monostromatic algal samples were collected from the following areas of the South China Sea coast in the Guangdong province: Zhanjiang, May 2020; Maoming, February 2020; Yangjiang, November 2019 and February 2020; Zhuhai, March 2021; and Shantou, March 2021 (Figure 1). The samples (Figure 1B–F) were placed in a cooler on ice and were brought to the laboratory. Before identification, sediments and contaminants were removed using filtered seawater and a soft brush. Detailed sample information is listed in Table 1.
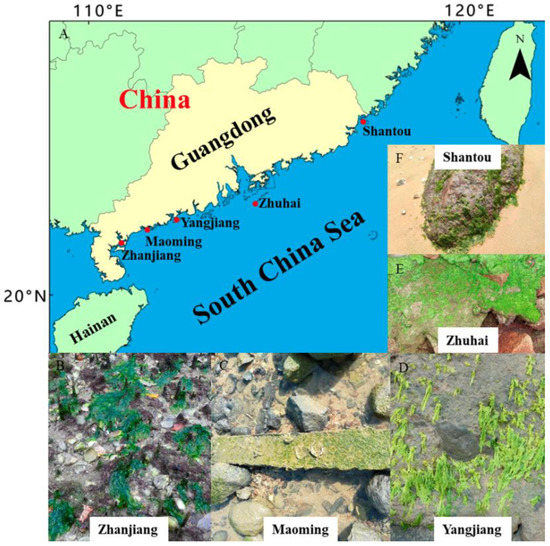
Figure 1.
Sampling sites where monostromatic green algae were collected from the South China Sea. (A) Map of the sampling stations; (B–F) images depicting the monostromatic samples sites.

Table 1.
Sample information concerning the monostromatic species used in this study.
2.2. Morphological Examination
Several intact monostromatic green algae from each sampling station were selected for morphological assessment, and their macroscopic features—including thallus type, size (length × perpendicular width), and color—were recorded. In addition, surface view and transverse section microphotographs of the cells were obtained under a microscope (Olympus CX33, Tokyo, Japan), and they were used to determine the microscopic cellular features, including cell size, shape and arrangement, chloroplast shape, position, and pyrenoid number.
2.3. DNA Extraction
Five monostromatic samples from Yangjiang (strain codes: YJ01, YJ02, YJ03, YJ04, and YJ05), four from Zhanjiang (strain codes: ZJ01, ZJ02, ZJ03, and ZJ04), three from Maoming (strain codes: MM01, MM02, and MM03) and Shantou (strain codes: ST01, ST02 and ST03), and one from Zhuhai (strain code: ZH01), which were randomly selected during the morphological examination, were prepared for DNA extraction. All samples were crushed into a fine powder in liquid nitrogen after being dried under vacuum conditions. Total DNA was extracted from each dried sample using a DNEasy Plant Mini kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. The quality of the extracted DNA was confirmed by electrophoresis on 1% agarose gel in TAE buffer containing ethidium bromide (EtBr) under ultraviolet (UV) light. The extracted DNA was stored at 4 °C until further use.
2.4. PCR Amplification
Polymerase chain reaction amplifications were performed in a total volume of 20 μL containing 2 μL of template DNA, 2 μL of each primer, and 10 μL 2 × San Taq PCR mix (Sangon, Shanghai, China). The primers used to amplify all the ITS sequences were ITS 1 and ITS 4 [23], and the reaction cycles were: initial denaturation at 94 °C for 5 min, followed by 28 cycles at 94 °C for 30 s, 55 °C for 30 s, 72 °C for 60 s, and 10 min at 72 °C as the final extension step. PCR products were checked on 1% TAE agarose gels stained with ethidium bromide, and were sequenced by Shanghai Sangon Corp. (Sangon, Shanghai, China).
2.5. Phylogenetic Analysis
Sequences were aligned with published data (Table 1) using ClustalX [34] and edited in BioEdit [35]. For comparative analyses, sequences (AB298316 and AB298457) downloaded from Genbank were utilized as an outgroup. Finally, 35 nrITS sequences were used for phylogenetic analyses which, for concatenated nrITS, were performed using the maximum likelihood (ML) and neighbor-joining (NJ) methods in Molecular Evolutionary Genetics Analysis (MEGA) v.7.0.21 [36]. The best-fitting model of nrITS for the ML and NJ analyses was the Tamura-Nei + Gamma distribution (G) + Invariant sites (I) and their robustness was tested by bootstrapping with 1000 replicates. Bayesian inference (BI) analysis was performed using MrBayes v3.1.2 [37]. The best partition strategy and model of sequence evolution were selected based on the Bayesian Information Criterion (BIC). Four chains of Markov chain Monte-Carlo iterations were performed for 1,000,000 generations, keeping one tree every 100 generations. Convergence of the runs was checked visually with Tracer v1.6 [38]. A burn-in of 25% was used to avoid suboptimal trees in the final consensus tree. The pairwise distances of nrITS were calculated based on the Maximum Composite Likelihood model using MEGA v.7.0.21 [36].
3. Results
3.1. Phylogenetic Analyses
The ITS 1 and ITS 2 regions that included the 5.8S gene were successfully amplified and sequenced from 16 target samples. Sequences of 546 bp from each of the ZJ01-04, ST01-03, and YJ04-05 samples, and 554 bp from each of the YJ01-03, MM01-03, and ZH01 samples, were used for the phylogenetic analyses. The phylogenetic trees obtained from the ML, NJ, and BI analyses of the nrITS and concatenated data revealed that the monostromatic green algae fell into three distinct clades (Figure 2). Specifically, nine samples (ZJ01-04, ST01-03, and YJ04-05), together with the identified M. nitidum from China, were resolved in the M. nitidum clade (99% in ML and NJ, 0.99 in BI); three samples (YJ01-03), together with the identified G. brasiliensis from Brazil, were resolved in the G. brasiliensis clade (100% in ML and NJ, 1 in BI); and four additional samples (MM01-03, ZH01) were included in a unique Monostroma sp. clade (100% in ML and NJ, 1 in BI). The intraspecific genetic distance of monostromatic green algae in the Monostroma and Gayralia genera was less than 0.6% (Figure 3), while their interspecific genetic distance was undulatory. Surprisingly, the inter-genera genetic distance (between the M. nitidum and G. brasiliensis clades = 0.058–0.062; between the M. nitidum and G. oxysperma clades = 0.208–0.211) was lower than that observed within the Monostroma genus (between the M. nitidum clade and the panmictic Monostroma sp. clade = 0.291–0.313) (Figure 3).
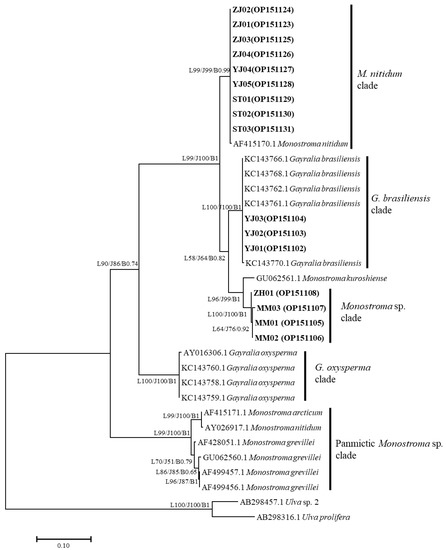
Figure 2.
Phylogenetic tree inferred from the ITS sequences of the monostromatic green algae collected from the South China Sea and those downloaded from GenBank. The specimens from this study are shown in bold. The numbers on the branches indicate bootstrap values from ML (L…, left), NJ (J…, middle), and Bayesian inference posterior probabilities (B…, right). Bootstrap values (>50%) and Bayesian inference posterior probabilities (>0.50) are indicated.
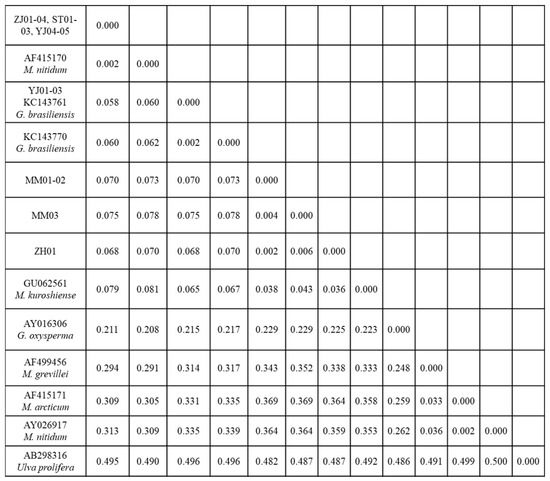
Figure 3.
ITS sequence-based genetic distances between monostromatic green algal strains collected from the South China Sea and those downloaded from GenBank.
3.2. Morphology of Monostromatic Green Algae
Figure 4 shows the morphology of the collected monostromatic green algae. In the target samples, thalli with similar morphology presented a yellowish or light green color, were flat, and had a single-cell layer (Figure 4). In the surface view, cells appeared coupled, irregularly arranged, and triangular or polygonal with three to five rounded corners, partly oval, and paired (Figure 4B,E,H,K,N). A single prominent chloroplast covered most of the outer cell in the surface view and contained predominantly one (≥96%) and occasionally two (≤4%) pyrenoids (Figure 4B,E,H,K,N and Table 2). Cells in transverse sections were circular or quadrangular with rounded corners (Figure 4C,F,I,L,O). However, the thallus shape, cell size, and thallus thickness in the samples presented distinct differences (Figure 4 and Figure 5 and Table 2). Specifically, the thallus of the monostromatic species in the G. brasiliensis clade was approximately 3.64 ± 0.81 cm long and 2.66 ± 1.41 cm wide, which was larger than that of the Monostroma sp. clade (1.12 ± 0.44 cm long and 0.72 ± 0.40 cm wide), and smaller than that of the M. nitidum clade (8.12 ± 2.47 cm long and 4.98 ± 1.69 cm wide). Additionally, as observed for thallus size, the cell size of species within the G. brasiliensis clade (9.06 ± 1.36 μm long and 6.06 ± 0.68 μm wide) was larger than that of the species in the Monostroma sp. clade and smaller than that of the species in the M. nitidum clade. Moreover, for the species within the M. nitidum clade, thallus thickness measured 33.80–34.20 μm, which was thicker than that of the G. brasiliensis and Monostroma sp. clades (Figure 4C,F,I,L,O and Table 2).
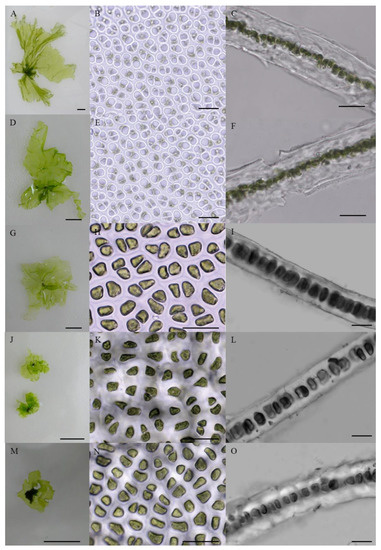
Figure 4.
Morphology of the monostromatic green algae collected from the South China Sea. (A–C) YJ05 wild-living strain; (D–F) ST01 wild-living strain; (G–I) YJ02 wild-living strain; (J–L) MM01 wild-living strain; (M–O) ZH01 wild-living strain; (A,D,G,J,M) wild thallus of each strain; (B,E,H,K,N) cells of each strain in surface view; (C,F,I,L,O) cross-sectional views of each strain. Scale bars in the macro and microphotographs represent 1 cm and 20 μm, respectively.

Table 2.
Thallus morphology of the monostromatic green algae collected from the South China Sea.
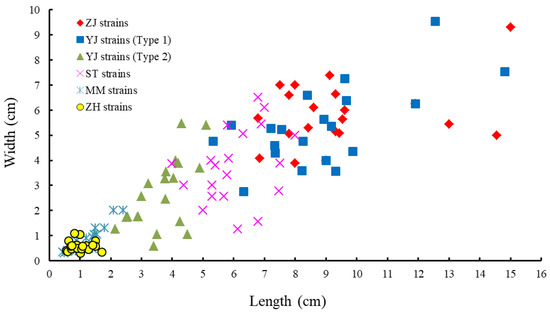
Figure 5.
Thallus size of the monostromatic green algae collected from the South China Sea (n = 20). ZJ strains: Zhanjiang strains, including ZJ01-04 strains; YJ strains (Type 1): Yangjiang strains, including YJ04-05 strains; YJ strains (Type 2): Yangjiang strains, including YJ01-03 strains; ST strains: Shantou strains, including ST01-03 strains; MM strains: Maoming strains, including MM01-03 strains; ZH strains: Zhuhai strains, including the ZH01 strain.
4. Discussion
4.1. Taxonomic History of Monostromatic Green Algae
Monostromatic green algae, characterized by a single cell-layered blade-like thallus, were traditionally grouped under the eponymous genus Monostroma, which comprised M. bullosum (Roth) Wittrock and M. oxyspermum (Kützing) Doty [4,15]. Many species were subsequently added to this genus based on Thuret’s description (Figure 6). For instance, M. nitidum, M. arcticum, M. undulatum, and others, were added to this genus by Wittrock (1866) [17]; M. angicava and M. leptodermum were added by Kjellman (1877) [39]; M. groenlandicum and M. obscurum were added by Agardh (1883) [40]; and M. zostericola was added by Tilden (1900) [41]. Kunieda (1934) erected the family Monostromaceae to accommodate all the related monostromatic green algal species [2]. However, as culture studies were conducted on their life cycle and thallus ontogeny, a number of taxonomic revisions on the genus Monostroma were proposed (Figure 6). Specifically, Gayral (1964) erected a new genus, Ulvopsis, with U. grevillei (synonymous to M. grevillea (Thuret) Wittrock) as a species type based on the sexual life cycle [42]. Bliding (1968) and Tatewaki (1969) recommended that M. bullosum Roth and M. angicava Kjellman should also be transferred to the genus Ulvopsis based on Gayral’s identification [19,43]. Considering the shared life cycle patterns and anatomical features, Bliding also argued that taxa M. leptoderma and M. zostericola should be separated from the genus Monostroma and grouped under the newly erected Kornmannia genus [19]. Vinogradova (1969) proposed to include M. groenlandicum into the genus Capsosiphon based on shared morphology [3], M. oxyspermum into the newly erected genus Gayralia (family Gayraliaceae) based on the typical asexual life cycle and the presence of a “tube” stage in the ontogeny, and M. undulatum into the newly erected genus Protomonostroma (Gayraliaceae) based on the absence of the above-mentioned “tube” stage in thallus ontogeny. After discovering the asexual life cycle of M. latissimum, Bast et al. (2009) questioned the taxonomic credibility of those groups defined based on life cycle type (i.e., sexual vs. asexual) and proposed to abolish the monotypic genus Gayralia and regroup G. oxysperma back to Monostroma as its original lectotype [44]. In contrast, Pellizzari et al. (2013) further increased the members of the genus Gayralia by naming a new species (G. brasiliensis) and also suggested adding M. nitidum from China and M. kuroshiense to this genus, based on morphology and molecular analysis [5]. In summary, there has been confusion and debate throughout the systematic revision history of monostromatic green algae, and it is necessary and important to clarify their current taxonomic status using effective and accepted taxonomic tools. For these reasons, the present study was designed, and it proposes to replace the genus Gayralia with the genus Monostroma.
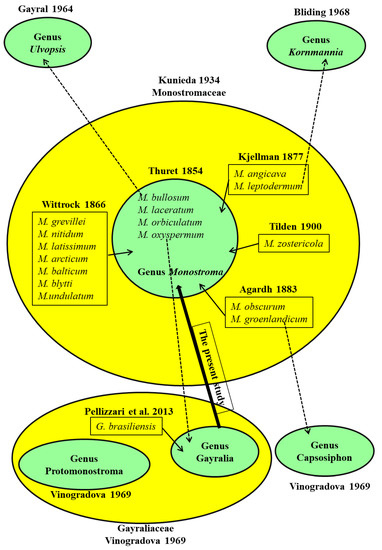
Figure 6.
Taxonomic history of monostromatic green algae. Each taxon enclosed in the light green circles was regarded as a single genus, whereas each taxon enclosed in the yellow circles was regarded as a single family. The solid line arrows show the addition of species to the Monostroma genus; the dotted line arrows show the removal of species from the Monostroma genus; the bold line arrow represents the opinion of the present study [2,3,5,15,17,19,39,40,41,42].
4.2. Taxonomic Assessment of Monostromatic Green Algae
As with many other taxonomic groups of organisms, defining algal species is extremely difficult. Various concepts of the term species exist in systematics based on aspects that can be morphological, biological, genetic, ecological, paleontological, evolutionary, etc. [45]. It is well known that the concept of “morphological species” dominated algal systematics for many years, being based on “taximetrics”, i.e., the overall similarity in morphology. Advancements in microscopy have enabled algal systematists to define new synapomorphies for the algal groups in question. Moreover, the morphology of the type specimens as a classification criterion has been widely used in algal systematics [10,21,22]. Based on the macro and microphotographs obtained in the present study, all the monostromatic green algae collected from the South China Sea showed a yellowish or light green color, were flat, and had a single-cell layer (Figure 4), which corresponds to their original morphological description by Thuret (1854) [15]. Monostroma nitidum was originally described by Wittrock (1866) as being monostromatic and thick, with a yellowish-green and shiny appearance and a frilly and mangled edge [17]. In the surface view, the cells are angular with slightly rounded corners and have an irregular arrangement, whereas in transverse sections, they are almost circular and 30–40 μm thick [17]. These morphological characteristics also match those of monostromatic species in the M. nitidum clade (Figure 4 and Table 2). In addition, the specimen type of G. brasiliensis is characterized by a single, expanded, laminar, monostromatic thallus. The fronds are ca. 10 cm broad, with a thickness of 25.0 ± 1.8 μm, and cell lumen of 9.0 ± 1.0 μm. In the surface view, cells are grouped into pairs, becoming more elongated toward the base. The cells are uninucleate with a large central vacuole, parietal chloroplast, and one or two pyrenoids [5]. These morphological traits are similar to those of monostromatic species in the G. brasiliensis clade, except for thallus size (Figure 4 and Table 2). Thus, the monostromatic green algae in the present study were classified into three different groups (M. nitidum, G. brasiliensis, and Monostroma spp) based on morphological analysis. Moreover, in our target samples, a similar morphology was monostromatic: thallus appearance, cell shape and arrangement, chloroplast position, and pyrenoid number. While the thallus shape, cell size, and thallus thickness of the samples presented distinct differences. These differences should be regarded as the typical morphological features of various Monostroma species. Our results were well supported by the studies of Bliding (1968) [19], Bast (2011) [4], Pellizzari et al. (2013) [5], and Cui et al. (2021) [10].
With the advent of DNA-based molecular barcoding technology, the concept of genetic species—which is based upon the genetic homogeneity of populations—has been increasingly taken into consideration, in addition to other concepts in algal systematics. DNA regions have been differentially used to construct phylogeny at different hierarchical levels, meaning that loci that evolve more slowly are used to analyze higher taxonomic levels, while those that evolve more rapidly are used to analyze relationships between closely related species [4,10,21,22]. ITS sequences were initially proposed for phylogeny reconstruction at or below the species level [46] due to the extensive sequence variation existing between members of closely related taxa. In this study, in line with the results of the morphological analysis, the data from ITS sequences also demonstrated that the monostromatic green algae were divided into three different clades (M. nitidum clade, G. brasiliensis clade, and Monostroma spp. clade) (Figure 2), which was further supported by the intraspecific genetic distance value (less than 0.6%, Figure 3). Monostroma nitidum (AF415170) from China, a reliable and accepted gene sequence, has been widely used as a reference to verify the identity of suspected M. nitidum specimens [4,10,47,48,49]. Thus, the monostromatic green algae (strain codes: ZJ01-04, YJ04-05, and ST01-03) were identified as M. nitidum. Monostroma oxyspermum was separated from the Monostroma genus due to its asexual life cycle and thallus ontogeny, and was subsequently included into the new Gayralia genus, currently comprising G. brasiliensis and G. oxysperma [3]. However, the taxonomic credibility of this species, defined based on sexuality, has been widely discredited [4,21,24]. In addition, asexual G. oxysperma have the same filament-sac-blade ontogeny as that of M. nitidum [43], while asexual G. brasiliensis have the same filament-blade ontogeny as that of M. latissimum [5]. Moreover, this study surprisingly found that the genetic distance between Monostroma and Gayralia was lower than the interspecies distance within the Monostroma genus (Figure 3). Based on these observations, the present study agrees with the opinion of Bast (2011) and suggests that the genus Gayralia should be assigned to its original genus, Monostroma, and G. brasiliensis and G. oxysperma should therefore be renamed as M. brasiliensis and M. oxyspermum, respectively. Accordingly, the monostromatic green algae with the strain code YJ01-03 were identified as M. brasiliensis, and those with strain codes ZH01 and MM01-03 were identified as Monostroma spp.
5. Conclusions
In summary, the results of the present study demonstrated that the collected Monostroma and Gayralia specimens presented similar morphology in their single cell-layered thallus, with irregularly arranged cells, rounded cell corners, a parietal chloroplast, and predominantly one (>90%) pyrenoid. Furthermore, the inter-genera genetic distance between the Monostroma and Gayralia genera was lower than that observed within the Monostroma genus. Considering that the genus Monostroma was erected earlier than the genus Gayralia, it is here proposed to assign the genus Gayralia to the genus Monostroma based on the morphological and phylogenetic analysis and genetic distance data presented here.
Author Contributions
Conceptualization, J.C. and E.X.; methodology, C.C.; software, Y.H.; formal analysis, J.C., X.C. and J.L.; investigation, R.X. and B.H.; data curation, H.T.; writing—original draft preparation, J.C.; writing—review and editing, J.C., C.C. and E.X.; supervision, H.T. and E.X.; funding acquisition, J.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the PhD Start-up Foundation of Guangdong Ocean University, grant number R19049.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Acknowledgments
We would like to thank Dahai Gao for his helpful comments on the phylogenetic analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; De Clerck, O. Taylor & francis online: Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 2012, 31, 1–46. [Google Scholar]
- Kunieda, H. On the life history of Monostroma. Proc. Imp. Acad. 1934, 10, 103–106. [Google Scholar] [CrossRef]
- Vinogradova, K.L. Sistematike poryadka Ulvales (Chlorophyta). Botanicheskii Zhurnal. 1969, 54, 1347–1355. [Google Scholar]
- Bast, F. Monostroma: The Jeweled Seaweed for Future; Lap Lambert Academic Publishing: Chisinau, Moldova, 2011; pp. 1–192. [Google Scholar]
- Pellizzari, F.; Oliveira, M.C.; Amanda, D.S.M.; Yokoya, N.S.; Oliveira, E.C. Morphology, ontogeny, and phylogenetic position of Gayralia brasiliensis sp. nov. (ulotrichales, chlorophyta) from the southern coast of brazil. Bot. Mar. 2013, 56, 197–205. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M.; AlgaeBase. World-Wide Electronic Publication, National University of Ireland, Galway. Available online: http://www.algaebase.org (accessed on 22 September 2021).
- Ohno, M. Cultivation of Monostroma nitidum (Chlorophyta) in a river estuary, southern Japan. J. Appl. Phycol. 1995, 7, 207–213. [Google Scholar] [CrossRef]
- Pellizzari, F.M.; Absher, T.; Yokoya, N.; Oliveira, E.C. Cultivation of the edible green seaweed Gayralia (Chlorophyta) in Southern Brazil. J. Appl. Phycol. 2007, 19, 63–69. [Google Scholar] [CrossRef]
- Pise, N.M.; Verlecar, X.N.; Gaikwad, D.K.; Jagtap, T.G. Nutraceutical properties of the marine macroalga Gayralia oxysperma. Bot. Mar. 2012, 55, 581–589. [Google Scholar]
- Cui, J.J.; Wang, H.; Chen, C.L.; Chen, X.Y.; Chen, Y.H.; Weng, M.X.; Huang, B.W.; Huang, Y.J.; Zhu, D.W.J.; Zhang, N.N.; et al. Formation and early development of the Monostroma nitidum Wittrock. Aquacult. Rep. 2021, 20, 100759. [Google Scholar] [CrossRef]
- Cassolato, J.E.; Noseda, M.D.; Pujol, C.A.; Pellizzari, F.M.; Damonte, E.B.; Duarte, M.E. Chemical structure and antiviral activity of the sulfated heterorhamnan isolated from the green seaweed Gayralia oxysperma. Carb. Res. 2008, 343, 3085–3095. [Google Scholar]
- Zhang, H.J.; Mao, W.J.; Fang, F.; Li, H.Y.; Sun, H.H.; Yin, C.; Qi, X.H. Chemical characteristics and anticoagulant activities of a sulfated polysaccharide and its fragments from Monostroma latissimum. Carbohydr. Polym. 2008, 71, 428–434. [Google Scholar] [CrossRef]
- Chen, Y.C. A simple method for isolating filaments as “algal seed stock” from monostroma latissimum (chlorophyta) germlings, and application for mass cultivation. J. Phycol. 2012, 48, 246–247. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, Y.; Togashi, T. Identification of genomic differences between the sexes and sex-specific molecular markers in Monostroma angicava (Ulvophyceae). J. Phycol. 2021, 57, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Thuret, G. Note sur la synonymie des Ulva lactuca et latissima L. suivie de quelques remarques sur la tribu des Ulvacees. Mem. Soc. Sci. Nat. Cherbourg. 1854, 2, 17–32. (In French) [Google Scholar]
- Papenfuss, G.F. On the genera of Ulvales and the status of the order. Bot. J. Linn. Soc. 1960, 56, 303–318. [Google Scholar] [CrossRef]
- Wittrock, V.B. Försök till en Monographi öfver AlgsläGtet Monostroma. Ph.D. Thesis, Uppsala University, Uppsala, Sweden, 1866; p. 66. (In Swedish). [Google Scholar]
- Kornmann, P. Über Monostroma bullosum (Roth) Thuret und M. oxyspermum (Kützing) Doty. Helgol. Wiss. Meeresuntersuch. 1964, 11, 13–21. (In German) [Google Scholar] [CrossRef]
- Bliding, C.A. Critical survey of European taxa in Ulvales, Part II: Ulva, Ulvaria, Monostroma, Kornmannia. Bot. Not. 1968, 121, 535–629. [Google Scholar]
- South, G.R.; Skelton, P.A. Catalogue of the marine benthic macroalgae of the Fiji, Islands, South Pacific. Aust. Syst. Bot. 2003, 16, 699–758. [Google Scholar] [CrossRef]
- Cui, J.J.; Monotilla, A.P.; Zhu, W.R.; Takano, Y.; Shimada, S.; Ichihara, K.; Matsui, T.; He, P.M.; Hiraoka, M. Taxonomic reassessment of Ulva prolifera (Ulvophyceae, Chlorophyta) based on specimens from the type locality and Yellow Sea green tides. Phycologia 2018, 57, 692–704. [Google Scholar] [CrossRef]
- Calderon, M.S.; Bustamante, D.E.; Gabrielson, P.W.; Martone, P.T.; Hind, K.R.; Huber, S.; Mansilla, A. Type specimen sequencing, multilocus analyses, and species delimitation methods recognize the cosmopolitan Corallina berteroi and establish the northern Japanese C. yendoi sp. nov. (Corallinaceae, Rhodophyta). J. Phycol. 2021, 57, 1659–1672. [Google Scholar] [CrossRef]
- Woolcott, G.W.; King, R.J. Ulvaria (Ulvales, Chlorophyta) in eastern Australia: Morphology, anatomy and ontogeny compared with molecular data. Bot. Mar. 1998, 41, 63–76. [Google Scholar] [CrossRef]
- Hiraoka, M.; Ichihara, K.; Zhu, W.R.; Shimada, S.; Oka, N.; Cui, J.J.; Tsubaki, S.; He, P.M. Examination of species delimitation of ambiguous DNA-based Ulva (Ulvophyceae, Chlorophyta) clades by culturing and hybridisation. Phycologia 2017, 56, 517–532. [Google Scholar] [CrossRef]
- Wang, S.Y.; Huo, Y.Z.; Zhang, J.H.; Cui, J.J.; Wang, Y.; Yang, L.L.; Zhou, Q.Y.; Lu, Y.W.; Yu, K.F.; He, P.M. Variations of dominant free-floating Ulva species in the source area for the world’s largest macroalgal blooms, China: Differences of ecological tolerance. Harmful Algae 2018, 74, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Gaysina, L.A.; Johansen, J.R.; Saraf, A.; Allaguvatova, R.Z.; Pal, S.; Singh, P. Roholtiella volcanica sp. nov., a New Species of Cyanobacteria from Kamchatkan Volcanic Soils. Diversity 2022, 14, 620. [Google Scholar] [CrossRef]
- Jiang, X.; Chen, X.; Pang, W.; Wang, Q. Phylogeny of Trachelomonas and Strombomonas (Euglenaceae) Based on Morphological and Molecular Data. Diversity 2022, 14, 623. [Google Scholar] [CrossRef]
- Lorenzo-Carballa, M.O.; Sanmartín-Villar, I.; Cordero-Rivera, A. Molecular and Morphological Analyses Support Different Taxonomic Units for Asian and Australo-Pacific Forms of Ischnura aurora (Odonata, Coenagrionidae). Diversity 2022, 14, 606. [Google Scholar] [CrossRef]
- Yan, Q.; Dai, Q.; Liu, B.; Liu, G.; Zhu, H. A New Deep-Water Epilithic Green Alga, Ulvella lacustris, from an Alpine Brackish Lake in Qinghai–Tibet Plateau. Diversity 2022, 14, 594. [Google Scholar] [CrossRef]
- Li, F.; Dong, M.; Zhang, N.; Zhang, Y.; Li, Q.; Qian, Z.; Lian, Q.; Luo, J.; Huang, X.; Li, C. Euchlorocystis marina sp. nov. (Oocystaceae, Trebouxiophyceae), a New Species of Green Algae from a Seawater Shrimp Culture Pond. Diversity 2022, 14, 119. [Google Scholar] [CrossRef]
- Su, Q. Molecular Systematic Studies of Several Seaweeds in China. Ph.D. Thesis, Northeast Forestry University, Harbin, China, 2002; p. 81. [Google Scholar]
- Blomster, J.; Back, S.; Fewer, D.P.; Kiirikki, M.; Lehvo, A.; Maggs, C.A.; Stanhope, M.J. Novel morphology of Enteromorpha forming green tides. Am. J. Bot 2002, 89, 1756–1763. [Google Scholar] [CrossRef]
- Shimada, S.; Yokoyama, N.; Arai, S.; Hiraoka, M. Phylogeography of the genus Ulva (Ulvophyceae, Chlorophyta), with special reference to the Japanese freshwater and brackish taxa. J. Appl. Phycol 2008, 20, 979–989. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huellsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Suchard, M.; Drummond, A.J. Tracer, Version 1.6. 2013. Available online: https://tree.bio.ed.ac.uk/software/tracer (accessed on 28 July 2016).
- Kjellman, F.R. Ueber die Algenvegetationen des Murmanschen Meeres av der Westküste von Nowaja Semlja und Wajgatsch. Nova Acta Regiae Soc. Sci. Ups. 1877, 3, 1–86. (In German) [Google Scholar]
- Agardh, J.G. Till algernes systematik. Nya bidrag. (Tredje afdelningen). Lunds Univ. Års-Skr. Afdelningen Math. Och Nat. 1883, 19, 1–177. (In Swedish) [Google Scholar]
- Tilden, J.E. American Algae; University of Michigan: Minneapolis, MN, USA, 1900; Volume 4, pp. 301–400. [Google Scholar]
- Gayral, P. Sur le démembrement de l’actual genre Monostroma Thuret (Chlorophycées, Ulotrichales s.l.). C. R. Acad. Sci. Paris 1964, 258, 2149–2152. [Google Scholar]
- Tatewaki, M. Culture Studies on the Life History of Some Species of the Genus Monostroma; Scientific Papers of the Institute of Algological Research; Hokkaido University: Sapporo, Japan, 1969; Volume 6, pp. 1–56. [Google Scholar]
- Bast, F.; Shimada, S.; Hiraoka, M.; Okuda, K. Asexual life history by biflagellate zoids in Monostroma latissimum (Ulotrichales). Aqua. Bot. 2009, 91, 213–218. [Google Scholar] [CrossRef]
- Mayden, R.L. A hierarchy of species concepts: The denouement in the saga of the species problem. In Species: The Units of Biodiversity; Claridge, M.F., Dawah, H.A., Wilson, M.R., Eds.; Chapman and Hall: London, UK, 1997; pp. 381–424. [Google Scholar]
- Baldwin, B.G. Phylogenetic utility of the Internal Transcribed Spacers of nuclear ribosomal DNA in plants: An example from the Compositae. Mol. Phyl. Evol. 1992, 1, 3–16. [Google Scholar] [CrossRef]
- Karnjanapratum, S.; You, S.G. Molecular characteristics of sulfated polysaccharides from Monostroma nitidum and their in vitro anticancer and immunomodulatory activities. Int. J. Biol. Macromol. 2011, 48, 311–318. [Google Scholar] [CrossRef]
- Kawashima, Y.; Akasaki, T.; Matsumoto, Y.; Yamazaki, Y.; Shimada, S. Species identification of imported and Japanese commercial green algal products based on phylogenetic analyses using the nrITS2 and 5s rDNA spacer regions. Fisheries. Sci. 2013, 79, 521–529. [Google Scholar] [CrossRef]
- Wang, D.; Ma, J.; Du, J.; Xie, E.; Shi, J. Morphological and molecular study of Monostroma nitidum. Acta Agric. Zhejiangensis. 2015, 27, 1593–1600, (In Chinese with English Abstract). [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).